Abstract
A major unmet need in LC-MS/MS-based proteomics analyses is a set of tools for quantitative assessment of system performance and evaluation of technical variability. Here we describe 46 system performance metrics for monitoring chromatographic performance, electrospray source stability, MS1 and MS2 signals, dynamic sampling of ions for MS/MS, and peptide identification. Applied to data sets from replicate LC-MS/MS analyses, these metrics displayed consistent, reasonable responses to controlled perturbations. The metrics typically displayed variations less than 10% and thus can reveal even subtle differences in performance of system components. Analyses of data from interlaboratory studies conducted under a common standard operating procedure identified outlier data and provided clues to specific causes. Moreover, interlaboratory variation reflected by the metrics indicates which system components vary the most between laboratories. Application of these metrics enables rational, quantitative quality assessment for proteomics and other LC-MS/MS analytical applications.
LC-MS/MS provides the most widely used technology platform for proteomics analyses of purified proteins, simple mixtures, and complex proteomes. In a typical analysis, protein mixtures are proteolytically digested, the peptide digest is fractionated, and the resulting peptide fractions then are analyzed by LC-MS/MS (1, 2). Database searches of the MS/MS spectra yield peptide identifications and, by inference and assembly, protein identifications. Depending on protein sample load and the extent of peptide fractionation used, LC-MS/MS analytical systems can generate from hundreds to thousands of peptide and protein identifications (3). Many variations of LC-MS/MS analytical platforms have been described, and the performance of these systems is influenced by a number of experimental design factors (4).
Comparison of data sets obtained by LC-MS/MS analyses provides a means to evaluate the proteomic basis for biologically significant states or phenotypes. For example, data-dependent LC-MS/MS analyses of tumor and normal tissues enabled unbiased discovery of proteins whose expression is enhanced in cancer (5–7). Comparison of data-dependent LC-MS/MS data sets from phosphotyrosine peptides in drug-responsive and -resistant cell lines identified differentially regulated phosphoprotein signaling networks (8, 9). Similarly, activity-based probes and data-dependent LC-MS/MS analysis were used to identify differentially regulated enzymes in normal and tumor tissues (10). All of these approaches assume that the observed differences reflect differences in the proteomic composition of the samples analyzed rather than analytical system variability. The validity of this assumption is difficult to assess because of a lack of objective criteria to assess analytical system performance.
The problem of variability poses three practical questions for analysts using LC-MS/MS proteomics platforms. First, is the analytical system performing optimally for the reproducible analysis of complex proteomes? Second, can the sources of suboptimal performance and variability be identified, and can the impact of changes or improvements be evaluated? Third, can system performance metrics provide documentation to support the assessment of proteomic differences between biologically interesting samples?
Currently, the most commonly used measure of variability in LC-MS/MS proteomics analyses is the number of confident peptide identifications (11–13). Although consistency in numbers of identifications may indicate repeatability, the numbers do not indicate whether system performance is optimal or which components require optimization. One well characterized source of variability in peptide identifications is the automated sampling of peptide ion signals for acquisition of MS/MS spectra by instrument control software, which results in stochastic sampling of lower abundance peptides (14). Variability certainly also arises from sample preparation methods (e.g. protein extraction and digestion). A largely unexplored source of variability is the performance of the core LC-MS/MS analytical system, which includes the LC system, the MS instrument, and system software. The configuration, tuning, and operation of these system components govern sample injection, chromatography, electrospray ionization, MS signal detection, and sampling for MS/MS analysis. These characteristics all are subject to manipulation by the operator and thus provide means to optimize system performance.
Here we describe the development of 46 metrics for evaluating the performance of LC-MS/MS system components. We have implemented a freely available software pipeline that generates these metrics directly from LC-MS/MS data files. We demonstrate their use in characterizing sources of variability in proteomics platforms, both for replicate analyses on a single instrument and in the context of large interlaboratory studies conducted by the National Cancer Institute-supported Clinical Proteomic Technology Assessment for Cancer (CPTAC)1 Network.
EXPERIMENTAL PROCEDURES
Metrics
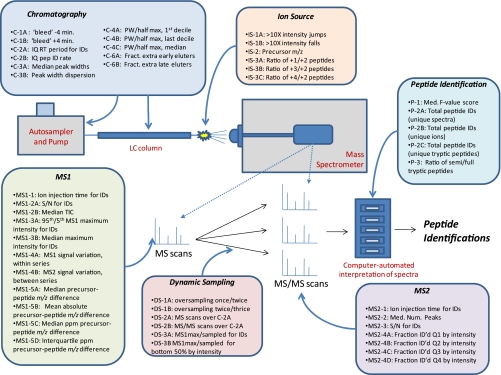
A list of the 46 metrics described in this report is shown schematically in Fig. 1. Short descriptions of the metrics, their assigned reference codes, and the direction indicating improved performance for each can be found in Table I. Although the following section lists the currently used metrics, updates will be described with each release. Additionally, all of the data described in this work can be downloaded from the ProteomeCommons Tranche network at http://cptac.tranche.proteomecommons.org/rudnicketal.html.
Fig. 1.
Schematic representation of performance metrics mapped to LC-MS/MS system elements. PW, peak width; IQ, interquartile; pep, peptide; ID, identification; Med., median; ID'd, identified; Fract., fraction; Num., number.
Table I. Descriptions of metrics.
Th, thomsons; ID, identification; IDed, identified; max, maximum; Betw/in, between/within; Med, median; pctile, percentile; tryp, tryptic; Pep, peptide; interQ, interquartile; fract, fraction.
| Code | Category | Metric group | Metric | Units | Optimal | Purpose/use | Description |
|---|---|---|---|---|---|---|---|
| C-1A | Chromatography | Fraction of repeat peptide IDs with divergent RT | −4 min | Fraction | ↓ | Estimates very early peak broadening | Fraction of all peptides identified at least 4 min earlier than max MS1 for ID |
| C-1B | Chromatography | Fraction of repeat peptide IDs with divergent RT | +4 min | Fraction | ↓ | Estimates very late peak broadening | Fraction of all peptides identified at least 4 min later than max MS1 for ID |
| C-2A | Chromatography | Interquartile retention time period | Period (min) | min | ↑ | Longer times indicate better chromatographic separation | Time period over which 50% of peptides were identified |
| C-2B | Chromatography | Interquartile retention time period | Pep ID rate | Peps/min | ↑ | Higher rates indicate efficient sampling and identification | Rate of peptide identification during C-2A |
| C-3A | Chromatography | Peak width at half-height for IDs | Median value | s | ↓ | Sharper peak widths indicate better chromatographic resolution | Median peak widths for all identified unique peptides (s) |
| C-3B | Chromatography | Peak width at half-height for IDs | Interquartile distance | s | ↓ | Tighter distributions indicate more peak width uniformity | Measure of the distribution of the peak widths; small values indicate consistency |
| C-4A | Chromatography | Peak widths at half-max over RT deciles for IDs | First decile | s | ↓ | Estimates peak widths at the beginning of the gradient | Median peak width for identified peptides in last RT decile (late) |
| C-4B | Chromatography | Peak widths at half-max over RT deciles for IDs | Last decile | s | ↓ | Estimates peak widths at the end of the gradient | Median peak width for identified peptides in first RT decile (early) |
| C-4C | Chromatography | Peak widths at half-max over RT deciles for IDs | Median value | s | ↓ | Estimates peak widths in the middle of the gradient | Median peak width for identified peptides in median RT decile (middle) |
| C-5A | Chromatography | Average elution order differencesa | Between | Percent | ↓ | Estimates peptide elution similarity run to run | Average elution rank order difference for identified peptides between series |
| C-5B | Chromatography | Average elution order differencesa | Betw/in | Ratio | ↓ | Estimates peptide elution similarity between series | Ratio of average rank order difference between series to average rank order difference within a series (low values indicate similarity between series) |
| C-6A | Chromatography | Fraction of extra early eluting peptides in row series (− = fewer)a | Between | Fraction | ↓ | Used to detect differences in the numbers of early peptides | Estimates relative frequency of early eluting peptides |
| C-6B | Chromatography | Fraction of extra late eluting peptides in row series (− = fewer)a | Between | Fraction | ↓ | Used to detect differences in the numbers of late peptides | Estimates relative frequency of late eluting peptides |
| DS-1A | Dynamic sampling | Ratios of peptide ions IDed by different numbers of spectra | Once/twice | Ratio | ↑ | Estimates oversampling | Ratio of peptides identified by 1 spectrum divided by number identified by 2 spectra |
| DS1-B | Dynamic sampling | Ratios of peptide ions IDed by different numbers of spectra | Twice/thrice | Ratio | ↑ | Estimates oversampling | Ratio of peptides identified by 2 spectra divided by number identified by 3 spectra |
| DS-2A | Dynamic sampling | Spectrum counts | MS1 scans/full | Count | ↓ | Fewer MS1 scans indicates more sampling | Number of MS1 scans taken over C-2A |
| DS-2B | Dynamic sampling | Spectrum counts | MS2 scans | Count | ↑ | More MS2 scans indicates more sampling | Number of MS2 scans taken over C-2A |
| DS-3A | Dynamic sampling | MS1 max/MS1 sampled abundance ratio IDs | Median all IDs | Ratio | ↓ | Estimates position on peak where sampled for peptides of all abundances | Ratio of MS1 maximum to MS1 value at sampling for median decile of peptides by MS1 maximum intensity (1 = sampled at peak maxima) |
| DS-3B | Dynamic sampling | MS1 max/MS1 sampled abundance ratio IDs | Med bottom 1/2 | Ratio | ↓ | Estimates position on peak where sampled for least abundant 50% of peptides | Ratio of MS1 maximum to MS1 value at sampling for bottom 50% of peptides by MS1 maximum intensity (1 = sampled at peak maxima) |
| IS-1A | Ion source | MS1 during middle (and early) peptide retention period | MS1 jumps >10× | Count | ↓ | Flags ESI instability | Number of times where MS1 signal greatly decreased between adjacent scans more than 10-fold (electrospray instability) |
| IS-1B | Ion source | MS1 during middle (and early) peptide retention period | MS1 falls >10× | Count | ↓ | Flags ESI instability | Number of times where MS1 signal greatly increased between adjacent scans more than 10-fold (electrospray instability) |
| IS-2 | Ion source | Precursor m/z for IDs | Median | Th | ↓ | Higher median m/z can correlate with inefficient or partial ionization | Median m/z value for all identified peptides (unique ions) |
| IS-3A | Ion source | IDs by charge state (relative to 2+) | Charge 1+ | Ratio | ↓ | High ratios of 1+/2+ peptides may indicate inefficient ionization | Number of 1+ peptides over 2+ peptides |
| IS-3B | Ion source | IDs by charge state (relative to 2+) | Charge 3+ | Ratio | ↓ | Higher ratios of 3+/2+ peptides may preferentially favor longer peptides | Number of 3+ peptides over 2+ peptides |
| IS-3C | Ion source | IDs by charge state (relative to 2+) | Charge 4+ | Ratio | ↓ | Higher ratios of 4+/2+ peptides may preferentially favor longer peptides | Number of 4+ peptides over 2+ peptides |
| MS1-1 | MS1 signal | Ion injection times for IDs | MS1 median | ms | ↓ | Lower times indicate an abundance of ions | MS1 ion injection time |
| MS1-2A | MS1 signal | MS1 during middle (and early) peptide retention period | S/N median | None | ↑ | Higher MS1 S/N may correlate with higher signal discrimination | Median signal-to-noise value (ratio of maximum to median peak height) for MS1 spectra up to and including C-2A |
| MS1-2B | MS1 signal | MS1 during middle (and early) peptide retention period | TIC median | Counts/1,000 | ↑ | Estimates the total absolute signal for peptides (may vary significantly between instruments) | Median TIC value for identified peptides over same time period as used for MS1-2A |
| MS1-3A | MS1 signal | MS1 ID max | 95/5 pctile | Ratio | ↑ | Estimates the dynamic range of the peptide signals | Ratio of 95th over 5th percentile MS1 maximum intensity values for identified peptides (approximates dynamic range of signal) |
| MS1-3B | MS1 signal | MS1 ID max | Median | Counts | ↑ | Estimates the median MS1 signal for peptides | Median maximum MS1 value for identified peptides |
| MS1-4A | MS1 signal | MS1 intensity variation for peptidesa | Within series | Percent | ↓ | Used to monitor relative intensity differences with a series | Average of between series intensity variations for identified peptides |
| MS1-4B | MS1 signal | MS1 intensity variation for peptidesa | Betw/in | Ratio | ↓ | Used to monitor relative intensity differences with a series compared with between series | Ratio of average intensity variation between series to average intensity variation within a series (low values indicate similarity between series) |
| MS1-5A | MS1 signal | Precursor m/z − Peptide ion m/z | Median | Th | ↓ | Measures the accuracy of the identifications | Median real value of precursor errors |
| MS1-5B | MS1 signal | Precursor m/z − Peptide ion m/z | Mean absolute | Th | ↓ | Measures the accuracy of the identifications | Mean of the absolute precursor errors |
| MS1-5C | MS1 signal | Precursor m/z − Peptide ion m/z | ppm median | ppm | ↓ | Measures the accuracy of the identifications | Median real value of precursor errors in ppm |
| MS1-5D | MS1 signal | Precursor m/z − Peptide ion m/z | ppm interQ | ppm | ↓ | Measures the distribution of the real accuracy measurements | Interquartile distance in ppm of the precursor errors |
| MS2-1 | MS2 signal | Ion injection times for IDs | MS2 median | ms | ↓ | MS2 ion injection time | |
| MS2-2 | MS2 signal | MS2 ID S/N | Median | Ratio | ↑ | Higher S/N correlates with increased frequency of peptide identification | Median S/N (ratio of maximum to median peak height) for identified MS2 spectra |
| MS2-3 | MS2 signal | MS2 ID peaks | Median | Count | ↑ | Higher peak counts can correlate with more signal | Median number of peaks in an MS2 scan |
| MS2-4A | MS2 signal | Fraction of MS2 identified at different MS1 max quartiles | ID fract Q1 | Fraction | ↑ | Higher fractions of identified MS2 spectra indicate efficiency of detection and sampling | Fraction of total MS2 scans identified in the first quartile of peptides sorted by MS1 maximum intensity |
| MS2-4B | MS2 signal | Fraction of MS2 identified at different MS1 max quartiles | ID fract Q2 | Fraction | ↑ | Higher fractions of identified MS2 spectra indicate efficiency of detection and sampling | Fraction of total MS2 scans identified in the second quartile of peptides sorted by MS1 maximum intensity |
| MS2-4C | MS2 signal | Fraction of MS2 identified at different MS1 max quartiles | ID fract Q3 | Fraction | ↑ | Higher fractions of identified MS2 spectra indicate efficiency of detection and sampling | Fraction of total MS2 scans identified in the third quartile of peptides sorted by MS1 maximum intensity |
| MS2-4D | MS2 signal | Fraction of MS2 identified at different MS1 max quartiles | ID fract Q4 | Fraction | ↑ | Higher fractions of identified MS2 spectra indicate efficiency of detection and sampling | Fraction of total MS2 scans identified in the last quartile of peptides sorted by MS1 maximum intensity |
| P-1 | Peptide identification | MS2 ID score | Median | fval | ↑ | Higher scores correlate with higher S/N and frequency of identification | Median peptide identification score for all peptides; higher scores generally correlate with increased MS2 S/N |
| P-2A | Peptide identification | Tryptic peptide counts | Identifications | Count | ↑ | Total identifications correlate with high levels of peptide signals, performance | Number of MS2 spectra identifying tryptic peptide ions (total “spectral counts”) |
| P-2B | Peptide identification | Tryptic peptide counts | Ions | Count | ↑ | A good overall performance measure | Number of tryptic peptide ions identified; ions differing by charge state and/or modification state are counted separately |
| P-2C | Peptide identification | Tryptic peptide counts | Peptides | Count | ↑ | A good overall performance measure | Number of unique tryptic peptide sequences identified |
| P-3 | Peptide identification | Peptide counts | Semi/tryp peptides | Ratio | Indicates prevalence of semitryptic peptides in sample; increasing ratios may indicate changes in sample or in source | Ratio of semi/fully tryptic peptide IDs |
a Composite metrics that use values calculated over more than one series.
Chromatography
Metrics C-1A and C-1B report the fraction of peptides with repeat identifications either >4 min earlier (C-1A) or later (C-1B) than the identification nearest the chromatographic peak maximum. Early identifications indicate bleed (typically non-retained, hydrophilic peptides), whereas later identifications arise from peak tailing of either overloaded peptides or peptides with poor chromatographic behavior. These are reported as fractions of all peptide identifications.
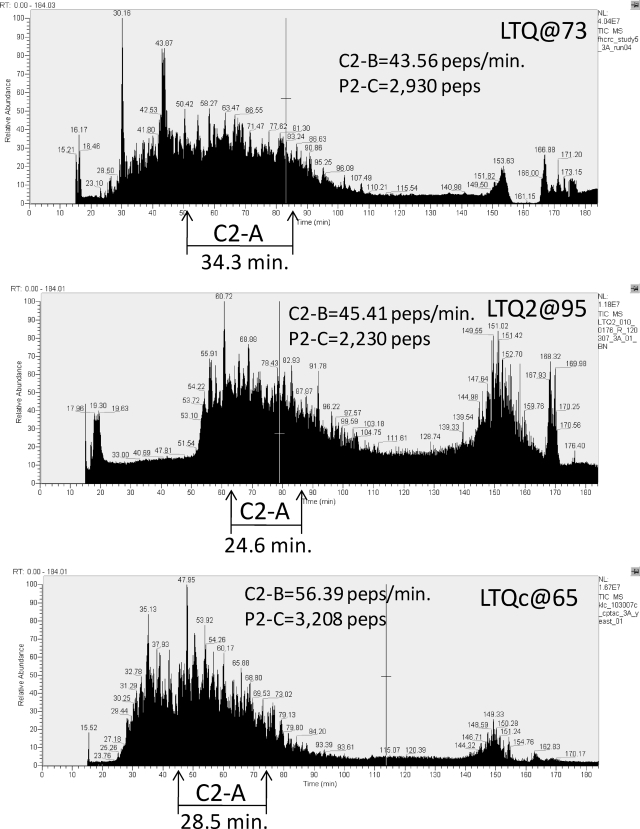
Metric C-2A reports the retention time period over which the middle 50% of the identified peptides eluted, and C-2B is the rate of peptide identification during that period. Sample LC-MS chromatograms from an analysis of a yeast proteome tryptic digest on three LTQ instruments are depicted in Fig. 2. The longer the time period over which peptides elute, the more time is available to acquire MS2 spectra and the greater the number of peptides likely to be sampled and identified is. In this work, this period is defined as the time over which the middle 50% of the identified peptides elute, which corresponds to the difference between the end of the first and beginning of the last retention quartiles (also called “interquartile range”) (C-2A). Various measures described later are computed only during this central period (e.g. C-2B, the identification rate in unique peptides/min) to reject early and later uninformative periods of chromatography and correct for differences in absolute start and end times when comparing chromatograms.
Fig. 2.
Illustration of chromatography metric C-2A applied to LC-MS/MS data from three Thermo LTQ systems in analyses of yeast proteome samples in CPTAC Study 5. Time intervals for elution of the middle quartiles of peptide identifications (C-2A) are indicated as are values for C-2B (peptide identification rate during this interval) and total peptide identifications during the analysis (P-2C). See text for discussion. peps, peptides.
Metrics C-3A, C-4A, and C-4B report chromatographic peak widths for identified peptide peaks. Sharper chromatographic peaks generate higher signal intensities and can reduce oversampling, thereby increasing the diversity of peptides identified. Peak widths (full width at half-maximum) were calculated as RT2 − RT1. Peak width medians (C-3A) and interquartile distances for these values (C-3B) are reported. C-3B is a measure of the distribution of peak widths for all peptides. Smaller values indicate higher degrees of peak uniformity. Peak widths were also calculated on smaller sets of early (decile 1, C-4A) or late (decile 10, C-4B) eluting peptides to measure for peak broadening at the extremes of the gradient.
Peptide elution order can be used to measure elution differences early (hydrophilic) and late (hydrophobic) in the chromatographic gradient. C-6A and C-6B are calculated by first identifying the N(a, b) peptides in common for a pair of runs, a and b, and then sorting all peptides in each run by elution time. If we define R1(a, b) as the rank of the earliest eluting peptide (rank 1) in run a that is also present in run b and R1(b, a) as the equivalent rank of the earliest co-occurring peptide in run b, then R1(a, b) − R1(b, a) is a measure of the number of extra early eluting peptides in run a (C-6A). To make this more robust, we find the maximum of the difference Rn(a, b) − Rn(b, a) from n = 1 to n = N(a, b)/10. C-6B is calculated similarly but for the high ranking, co-occurring peptides. This maximum is divided by the total number of peptides identified in the run, giving the fractional excess (or if negative, deficit) of hydrophobic peptides. For both measures, average differences between all intraseries runs and all interseries runs are reported.
Dynamic Sampling
Metrics DS-1A and DS-1B are measures of peptide ion oversampling, which is controlled by the dynamic exclusion settings in the acquisition software. Ideally, these settings minimize wasteful multiple sampling of a peptide ion by permitting just one MS2 spectrum for a given peptide ion over a chromatographic peak. Ratios of singly to doubly (DS-1A) and doubly to triply (DS-1B) identified peptide ions are reported as measures of oversampling. The ratio of spectrum identifications to peptide ion identifications provides a more general measure of oversampling but mixes the effects of distorted chromatographic peaks and bleed with effects more directly originating from dynamic sampling.
The numbers of MS (DS-2A) and MS2 (DS-2B) spectra acquired during the middle 50% peptide retention period (C2-A) indicate the effective speed of sampling over the most information-rich section of the chromatogram. If, for a given MS spectrum, there is insufficient signal to reach the target threshold or the dynamic exclusion settings are not working as expected, the numbers of MS or MS2 spectra may vary substantially between technical replicates.
Metrics DS-3A and DS-3B describe peak sampling. Ideally, chromatographic peaks will be sampled at their maximum intensity. However, current methods make sampling decisions prior to peak maximization, and larger chromatographic peaks are often sampled well before reaching the maximum. Because an m/z value chosen for MS2 sampling is typically placed on an exclusion list for the theoretical remainder of the peak, it is important that the amount of signal be sufficient to trigger acquisition of an MS2 spectrum with adequate signal to noise (S/N) for successful peptide identification. The trade-off is that the threshold value should not be so high as to prevent lower intensity peaks from being sampled. By monitoring the median ratio of the maximum MS1 intensity value over the MS1 intensity at the sampling time for all identified peptides (DS-3A), it is possible to look for variability in chromatographic peak sampling. To approximate the sampling ratio for less abundant analytes, ratios for peptides in the bottom 50% by MS1 abundance are calculated separately (DS-3B). The reciprocal values for these metrics or the percentage of the total peak height at sampling is also useful and perhaps more intuitive. MS1 maximum peak heights were found by linear interpolation from the extracted ion chromatograms for each precursor m/z using a newly developed method (15–18).
Ion Source
Metrics IS-1A and IS-1B are measures of electrospray stability. Short term electrospray stability is monitored as the median of ratios of MS1 total ion current for each set of adjacent scans, the larger ion current divided by the smaller, over the interquartile time interval C-2A. Electrospray drop-off was monitored by counting the number of events where the total ion current changed more (IS-1A) or less (IS-1B) than 10-fold in adjacent full MS scans. Values greater than zero for either of these metrics indicate spray tip “sputter.”
Instrument setup and tuning, including electrospray optimization, affect distributions of the peptide ions m/z values. This is monitored as the median precursor m/z (IS-2) of the identified peptide ions. This measure is sensitive to sample loading with higher concentrations of peptides generating higher m/z values, presumably at least in part due to the increasing preference for lower charge states with fewer charges available per peptide.
The availability of protons during electrospray ionization, the relative sequence length of peptides, and pH can all affect the charge state ratios of the identified peptides. Because doubly charged peptide ions (2+) represent the majority of the identified species from a typical tryptic digest, the ratios of 1+/2+ (IS-3A), 3+/2+ (IS-3B), and 4+/2+ (IS-3C) allow monitoring of the stability of the distributions. Perturbations in these ratios (or in the median precursor m/z) either run to run or between series may also correlate with decreased overlap of the observed peptide identifications.
MS1 and MS2 Signals and Analysis of Spectra
Reported intensities are the most direct measure of signal strength, although we have found that they can vary substantially between runs and labs for no clear reason. Metrics reporting these values over the C-2A interval are MS1-2B, the median TIC for full MS1 spectra, and MS1-3B, the median of maximum MS1 intensities for peptides. MS1-3A is a measure of “dynamic range,” which is taken as the ratio of the 95th/5th percentile of MS1 maximum intensities for identified peptides over C-2A.
The maximum-to-median signal (a measure of S/N) in both MS1 (MS1-2A) and MS2 (MS2-2) spectra has proven to be a more stable measure of signal strength than absolute intensities. However, because these metrics can depend on thresholding and centroiding, they could vary between classes of instruments and data processing systems. For the data generated in this study with Thermo ion trap instruments, this did not appear as a problem, and this measure served well for monitoring run-to-run variation. The total base peak-normalized abundance provides a related measure but can be sensitive to the m/z range and background ions. Numbers of reported peaks (MS2-3) provide another highly correlated measure and are also reported. We note the general difficulty of determining whether variations in signal intensity arise from amounts injected or changes in instrument sensitivity. In fact, none of the measures described can reliably distinguish between these two factors.
Median ion injection times for MS1 (MS1-1) and MS2 (MS2-1) spectra are also reported. Short times should be associated with high signal levels or low threshold settings. In many cases, the median is also the maximum, so mean ion injection times are also reported. Reduced ion injection times occur when analyzing higher sample loads but should be relatively stable between technical replicates.
Because intensity is used to select ions for fragmentation, intensity variation of the same peptide in different runs is a major source of run-to-run variability. A measure of this variability is the median relative deviation of peptide ion intensities in common between two runs. Absolute differences in peptide ion intensity between the two runs must first be corrected. This is done by sorting an array of the ratios of MS1 maximum intensities for peptide ions found in pairs of runs. The quartile values (Qn is the value for the nth quartile) yielding the median relative deviations are calculated using the following expression.
The average for within series (MS1-4A) and the ratio of within/between series (MS1-4B) are reported.
Another measure of performance is the fraction of identified (scores above threshold) MS2 spectra at different MS1 maximum peptide intensity quartiles (MS2-A–D). These values indicate to what extent intensity variations are sensitive to MS1 signal strength.
An additional class of metrics reports precursor accuracy (i.e. error associated with the identified peptides). All mass error measurements are derived from 2+ peptide ions only. Additionally, mass errors >0.45 m/z were rejected for high resolution Orbitrap and FT MS instruments; they are largely due to incorrect monoisotopic mass assignments at acquisition time. MS1-5A reports the median difference between the theoretical precursor m/z and the measured precursor m/z value as reported in the scan header. Reported monoisotopic values were used if available. MS1-5B is the mean of the absolute differences. MS1-5C is the median value of the real differences in ppm, and MS1-5D is the interquartile distance for the distribution used to determine MS1-5C.
Peptide Identification
Peptide identifications from MS2 spectra, needed for many of the above metrics, were made by matching spectra to those in a reference spectrum library (19). This method, long used in gas chromatography-MS, measures the similarity of reference and search spectra. A “dot product”-based metric measures similarity (20). This function is not only widely used for gas chromatography-MS but has been found to be effective for MS/MS identification and has recently been used in several peptide spectrum-matching search methods (19, 21–23). Spectrum libraries were derived from spectra assigned to peptide ions by sequence search engines (23–25). The yeast library (NIST yeast_consensus_final_ true_lib (June 30, 2008)) contained 79,990 spectra and was derived from a large number of analyses made by many laboratories, including those of the CPTAC Network, and is available on the web. The chicken egg yolk spectral library (NIST chicken_consensus_final_ true_lib (December 5, 2008)) contained 4,437 spectra and was generated from many dozens of runs at NIST. Both libraries are available from the authors (S. E. Stein) on request. Score thresholds were fixed for all analyses to yield an overall false discovery rate of 1% estimated by searching decoy libraries of unrelated organisms and eliminating homologous matches. In cases where a spectrum identified more than one peptide, only the peptide identification with the highest score was used.
ReAdW4Mascot2.exe version 2.1 (ConvVer 20081119b), an extension of ReAdW.exe (Patrick Pedroli, Institute for Systems Biology) was used to extract peak lists to mzXML or Mascot Generic format (MGF). The converter was run using the following arguments: for LTQ data: -sep1 -NoPeaks1 -c -MaxPI; and for Orbitrap data: -sep1 -NoPeaks1 -c -MaxPI -ChargeMgfOrbi -MonoisoMgfOrbi. The search engine used was SpectraST (version 3.0, TPP v4.0 JETSTREAM revision 2, Build 200807011544) (19). For spectral library searches with SpectraST, enzyme specificity (trypsin), numbers of missed cleavages permitted (≤2), fixed modifications (none), and variable modifications (carbamidomethyl-Cys, oxidized Met, N-terminal acetyl, pyro-Glu, and pyro-carbamidomethyl cysteine) were determined by the contents of the spectral library. Mass tolerance for precursor ions was 2 m/z for LTQ data and 1.0 m/z for Orbitrap data. Mass tolerance for fragment ions is not adjustable in SpectraST. SpectraST does not select candidate spectra based on charge. Therefore, charge information in the peak lists is ignored. The cutoff score/expectation value for accepting individual MS/MS spectra was an fval of 0.45. This threshold was based on a global FDR calculation using decoy spectra. FDR was calculated using the formula
where FP is 2 times the number of false positive matches at or above this score and TP is the number of true positive matches at or above this score. Overlapping decoy and target spectra were removed prior to the analysis, and FP was scaled according to the ratio of the target and decoy library size. Decoy spectra were actual peptide spectra from an unrelated organism, and the distribution of random matches between target and decoy libraries was approximately equal.
P-2C, a good overall measure of performance, is defined as the number of distinct identified tryptic peptide sequences, ignoring modifications and charge state. Numbers of unique semitryptic peptides (truncated tryptic peptides) were also counted, and the ratio of semitryptic/tryptic peptides is reported (P-3). Because semitryptic peptides can be formed by sample degradation or in the ionization source, higher total peptide values do not necessarily reflect better performance. This metric should be useful for determining how complete a digest is between preparations or for assessing how variable in-source fragmentation is between runs of the same sample. Also reported are the total numbers of identified spectra (P-2A) and the number of identified precursor ions (P-2B). The median score of identified peptides is also reported (P-1). In the case of the default analysis pipeline, this is the median SpectraST (19) fval for all peptide identifications. A score threshold of 0.45 was applied to all analyses. Relative decreases in the median score can indicate a reduction in MS2 S/N or other problems resulting from a divergence in similarity to consensus library spectra.
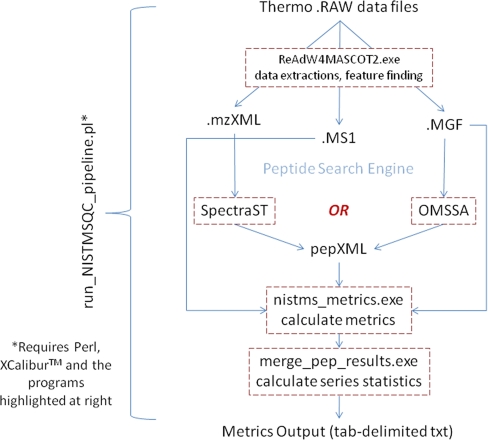
Data Analysis Pipeline for Implementation of Performance Metrics
We have used a newly developed metrics pipeline to calculate all of the values presented in this study (Fig. 3). The software consists of 1) a data extraction/feature-finding algorithm derived from ReAdW.exe (SourceForge) and 2) a peptide identification engine (either SpectraST (19) or open mass spectrometry search algorithm (OMSSA) (24) are currently integrated) followed by 3) a program that calculates all of the metrics from the extracted data and 4) a program that generates statistics for the data series.
Fig. 3.
Schematic representation of software pipeline to generate metrics. See text for discussion. OMSSA, open mass spectrometry search algorithm.
The pipeline is currently used to analyze Thermo Fisher .RAW files, requiring Thermo XcaliburTM to be installed. Updates for handling other vendor-specific files are being developed. The entire work flow is driven by a Perl script that is directed to one or more directories full of raw data files producing an output file in tab-delimited text format. This software is available for download.
Egg Yolk Protein LC-MS/MS Studies
Fresh chicken egg yolk was evaporated in vacuo, and 1-mg samples were stirred in 100 μl of 6 m urea, 0.1 m Tris buffer, pH 8, for 2 h at room temperature. Cysteines were reduced and alkylated with 1 mmol of dithiothreitol (1 h) followed by addition of 4 mmol of iodoacetamide for 1 h at room temperature. Excess iodoacetamide was incubated with excess dithiothreitol (4 mmol) for 1 h. The sample was then diluted with water to a total volume of 1 ml, mixed with 20 μg of Promega sequencing grade modified trypsin, and stirred at 37 °C for 18 h. After digestion, the solution was acidified with 20 μl of 50% formic acid. The digest was divided into three samples. One was refrigerated for 2 weeks and analyzed (sample 1). A second aliquot was frozen for 2 weeks, thawed, and then analyzed (sample 2). The third was evaporated to dryness in vacuo, redissolved in water at the same concentration, frozen for 2 weeks, then thawed, and analyzed (sample 3).
Five technical replicate analyses of each sample were done by LC-MS/MS using an LC-Packings Ultimate 3000 HPLC system (Dionex Corp., Sunnyvale, CA) coupled to a Thermo LTQ linear ion trap mass spectrometer (Thermo Scientific, Waltham, MA). Samples (2 μl) were injected onto a Dionex C18 Acclaim PepMap 300 column (300-μm inner diameter, 15 cm long) and eluted with a gradient of water (A)/acetonitrile (B), each containing 0.1% formic acid as follows: 0–40 min, 0–50% B; 40–45 min, 50–95% B; 45–48 min, 95% B; 48–50 min, 95–0% B; 50–60 min, 0% B. The flow rate was 4 μl/min. The eluent passed through a 15-μm silica tip (New Objective, Woburn, MA) and was sprayed into the LTQ mass spectrometer. Each MS scan was followed by eight MS/MS spectra of the eight most intense peaks, taken in reverse order. A dynamic exclusion time of 20 s was used with a list size of 500. The collision energy was set to 35%.
Yeast Lysate Sample Loading Studies
An aliquot of the tryptic digest of the CPTAC yeast reference material (see supplemental material) was redissolved in 0.1% formic acid in water at a concentration of 1 μg/μl. Solutions at 400 ng/μl, 40 ng/μl, 4 ng/μl, and 400 pg/μl were made by serial dilution and were analyzed by LC-MS/MS using a nanoLC-2D LC pump (Eksigent Technologies, Dublin, CA) coupled to a Thermo LTQ mass spectrometer (Thermo Scientific). For each series of LC-MS/MS runs, samples were analyzed back to back from low to high concentration. Blank gradient runs were done between each series. Samples were loaded onto an Atlantis dC18 trap column at 4.5 μl/min (Waters Corp.) and eluted onto a 100-μm × 10-cm BioBasic C18 IntegraFrit column (New Objective) connected to a 20-μm SilicaTip with a 10-μm tip (New Objective). Peptides were separated at 450 nl/min with the following gradient: 2–30.5% B over 90 min, 30.5–90% B over 10 min, 90% B for 2 min, 90–2% B over 3 min, and 2% B for 10 min (A = 0.1% formic acid in water, and B = 0.1% formic acid in acetonitrile). Except for the LC analysis described above, all other instrument settings and parameters were according to the CPTAC Network Study 6 standard operating procedure (SOP) (see the supplemental material).
Additional Methods
Complete descriptions of the CPTAC yeast protein materials, their preparation, and digestion and of the SOPs for CPTAC Network Studies 5 and 6 described here are presented in the supplemental material.
RESULTS
Overview of Performance Metrics
We developed 46 performance metrics, which map to LC-MS/MS system components as shown in Fig. 1. A list annotated with brief descriptions of each metric is provided in Table I; in the text, we refer to them by category and code. The metrics map to functional system components including the liquid chromatography system, the MS instrument (electrospray source and MS1 and MS2 signal intensities), the MS instrument control system (dynamic sampling), and the data analysis system (peptide identification outputs after database searching).
The metrics were first derived empirically through examination of run-to-run or lab-to-lab differences in data sets. In some cases, characteristics of the data sets suggested relevant metrics. For example, it was noted that on one LC-MS/MS system early eluting peptides were absent. Based on this observation, an algorithm was written to calculate the median peptide elution rank order differences in the early part of the chromatogram. Further logical examination of variability in the chromatography led to a focus on quantification of the observed variation and thus to the related chromatography metrics. In another example, average precursor m/z values for identified peptides were calculated for each run because the data were easily accessible. Differences between instruments for this metric were later attributed to differences in tuning protocols. Variations of this iterative “observe, quantify, evaluate, and refine” approach have yielded over 100 metrics of which 46 are reported here. Further development of performance metrics is a continuing focus of our work (at NIST).
Evaluation of Variability in Replicate Analyses of Similar Samples
We initially evaluated the behavior of the metrics in replicate analyses of three similar samples on a single LC-MS/MS system. An egg yolk protein extract was digested with trypsin and stored refrigerated for 2 weeks (preparation A, six LC-MS/MS replicates); frozen for 2 weeks, thawed, and analyzed (preparation B, six replicates); or evaporated in vacuo, redissolved at the same concentration, stored frozen for 2 weeks, thawed, and analyzed (preparation C, five runs). The replicates were each analyzed on a Thermo LTQ linear ion trap mass spectrometer. The median intraseries (replicates within a preparation) and interseries (between preparations) deviations are given in Table II. The %dev values represent the median absolute difference between all pairs of measurements expressed as a percentage of the median value. The average %dev within replicates for each preparation was less than 2% under these conditions and less than 3% across all runs for the three preparations. As expected, variation was greater on average between preparations A, B, and C than between replicate analyses. For example, the variability between preparations in the number of tryptic peptide counts (P-2A) was nearly 3 times greater than the average intraseries variability. This reflects the expected behavior of this metric in which modest differences between the three preparations should give rise to varying peptide identifications.
Table II. Variation of metrics in replicate analyses of egg yolk protein digest.
Avg., average; intra/inter, intraseries/interseries; med., median; pep(s)., peptide(s); IQ, interquartile; abund., abundance; IDs, identifications; ID'd, identified; fract., fraction.
| Metric | Avg. value | Avg. intraseries %dev | %Dev of intraseries %dev | Interseries %dev | Ratio avg. intra/inter |
|---|---|---|---|---|---|
| C-1A (bleed −4 min) | 0.0082 | 34.88 | 3.9 | 42.87 | 1.23 |
| C-1B (bleed +4 min) | 0.1499 | 4.84 | 3.98 | 0.63 | 0.13 |
| C-2A (IQ pep. RT period) | 11.56 | 2.43 | 1.4 | 0.3 | 0.12 |
| C-2B (peptides/min) | 41.9 | 2.59 | 1.21 | 0.79 | 0.31 |
| C-3A (med. peak width) | 11.34 | 0.35 | 1.92 | 0.14 | 0.41 |
| C-3B (IQ for peak widths) | 2.19 | 3.04 | 1.59 | 3.2 | 1.05 |
| DS-1A (oversampling; once/twice) | 3.31 | 4.35 | 2.52 | 3.11 | 0.71 |
| DS-1B (oversampling; twice/thrice) | 2.51 | 7.24 | 1.88 | 0.25 | 0.03 |
| DS-2A (MS1 scans over C-2A) | 345.3 | 1.06 | 7.63 | 2.53 | 2.39 |
| DS-2B (MS2 scans over C-2A) | 1,899 | 2.39 | 1.45 | 0.66 | 0.27 |
| DS-3A (med. MS1 max/MS1 sampled all IDs) | 2.48 | 2.44 | 2.49 | 2.01 | 0.82 |
| DS-3B (med. MS1 max/MS1 sampled for bottom 50% by abund.) | 1.44 | 1.45 | 1.41 | 1.08 | 0.75 |
| IS-1A (MS1 >10× jumps) | 0 | 0 | 0 | 0 | 0 |
| IS-1B (MS1 >10× falls) | 0 | 0 | 0 | 0 | 0 |
| IS-2 (med. precursor m/z) | 689.61 | 0.32 | 2.03 | 0.52 | 1.66 |
| IS-3A (ratio IDs 1+/2+) | 0.263 | 1.65 | 1.98 | 4.98 | 3.02 |
| IS-3B (ratio IDs 3+/2+) | 0.393 | 1.29 | 1.91 | 4.18 | 3.24 |
| IS-3C (ratio IDs 4+/2+) | 0.105 | 1.94 | 1.73 | 6.46 | 3.33 |
| MS1-1 (ion injection (ms) for IDs) | 5.6 | 3.17 | 0 | 13.86 | 4.36 |
| MS1-2A (S/N) | 317.8 | 1.77 | 1.42 | 6.6 | 3.73 |
| MS1-2B (med. TIC/1e3 over C-2A) | 5,470.6 | 1.76 | 11.98 | 15.31 | 8.68 |
| MS1-3A (dynamic range 95th/5th for IDs) | 87.2 | 5.21 | 1.28 | 6.16 | 1.18 |
| MS1-3B (med. MS1 signal for IDs) | 66,265 | 2 | 2.37 | 13.33 | 6.66 |
| MS2-1 (ion injection (ms) for IDs) | 100 | 0 | 0 | 0 | 0 |
| MS2-2 (S/N for IDs) | 220.5 | 1.44 | 5.15 | 2.04 | 1.41 |
| MS2-3 (med. no. peaks for IDs) | 314.6 | 1.84 | 1.61 | 2.46 | 1.34 |
| MS2-4A (fract. ID'd Q1) | 0.748 | 1.62 | 1.26 | 1.85 | 1.15 |
| MS2-4B (fract. ID'd Q2) | 0.585 | 1.7 | 1.15 | 2.2 | 1.29 |
| MS2-4C (fract. ID'd Q3) | 0.453 | 1.48 | 1.28 | 3.52 | 2.37 |
| MS2-4D (fract. ID'd Q4) | 0.368 | 3.79 | 1.54 | 6.17 | 1.63 |
| P-1 (med. f-value score for IDs) | 0.891 | 0.24 | 3.91 | 0.38 | 1.54 |
| P-2A (total IDs) | 2,767.7 | 1.11 | 2.6 | 3.01 | 2.71 |
| P-2B (unique ion IDs) | 1,260.5 | 0.59 | 1.7 | 1.6 | 2.71 |
| P-2C (unique peptide IDs) | 859.5 | 0.55 | 1.85 | 0.87 | 1.59 |
| P-3 (semi/fully tryptic peps.) | 0.13 | 3.33 | 1.42 | 3.28 | 0.99 |
| Median values | 1.76 | 1.7 | 2.2 | 1.29 |
The average ratio of interseries to intraseries %dev in Table II was 1.29, which indicates that the metrics vary slightly more due to modest sample differences than to variation between replicate analyses. Thus, the results in Table II indicate that the metrics are stable across analytical replicates and provide estimates of normal ranges for the metrics for typical laboratory handling.
Monitoring System Changes in Response to Controlled Operational Variation
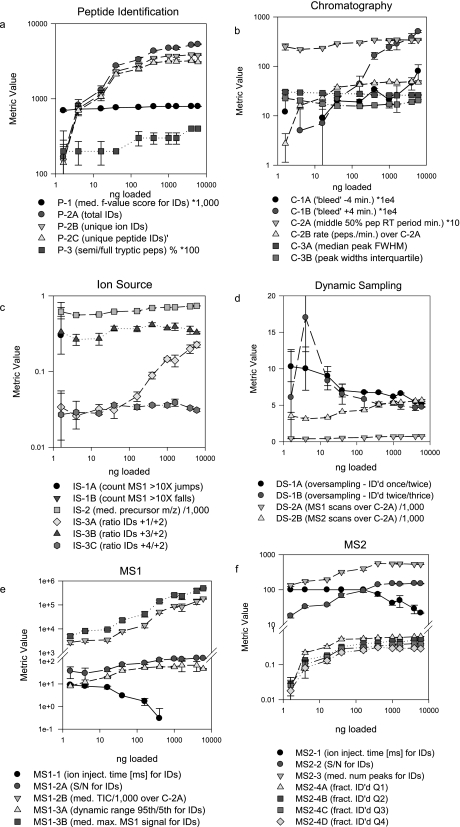
To evaluate the response of the metrics to controlled experimental variation, we analyzed a tryptic digest of the NCI-CPTAC yeast proteome reference material in triplicate at 10 sample loads ranging from 1.6 to 6,000 ng. Patterns of variation of the metrics with sample load illustrate their response characteristics (Fig. 4). Tryptic peptide identification metrics P-2A, P-2B, and P-2C all increased with load, but increases were marginal at loads above 100 ng (Fig. 4a). This was paralleled by increases in MS1 signal intensity parameters MS1-2B and MS1-3B together with a concomitant drop-off in ion injection time (MS1-1) (Fig. 4e) and an increase in median S/N for identified MS1 (MS1-2A) and MS2 spectra (MS2-2) for identified peptides (Fig. 4f). These metrics quantitatively illustrate the well known relationship between higher sample load, greater MS1 and MS2 signal intensities, higher spectral quality, and greater numbers of successful database matches. The increased variation in the metric values at lower sample loads is also presumably due to the diminishing concentration of the analytes. The fraction of identified MS2 spectra in each of the four MS1 quartiles also increased with sample loading up to 100 ng (MS2-4A through MS2-4D) (Fig. 4f) as expected. Metrics of ion source performance indicated that, although the median precursor m/z for identified peptides (IS-2) increased only slightly with sample load, the number of 1+ species identified relative to 2+ species (IS-3A) increased sharply at loads above 160 ng (Fig. 4c). This may reflect limited availability of protons at the source at higher analyte concentrations.
Fig. 4.
Stability and variation of metrics over a range of sample injection amounts. Serial dilutions of a tryptic digest of the CPTAC yeast reference proteome were analyzed in triplicate by LC-MS/MS on an LTQ instrument. Median values for each series were plotted according to the categories in Fig. 1. Error bars represent ±median error. Some values have been scaled as indicated in the panel legends. pep, peptide; IDs, identifications; med., median; ID'd, identified; fract., fraction; num, number; FWHM, full width at half-maximum; inject., injection.
Some of the chromatography metrics exhibited striking dependence on sample load (Fig. 4b). Most notably, C-2B (peptide identification rate) increased only at lower loading concentrations, whereas the “bleed” metric C-1A increased across the entire range. This latter metric was not accompanied by corresponding changes in peak width metrics (C-3A and C-3B), which suggests that C-1A simply reflects load rather than loss of column performance. This is consistent with the observation that peptide identifications increased across the load range, albeit more slowly at the highest concentrations.
These data demonstrate that the metrics correctly represent well understood relationships among system components and demonstrate sensitivity to “real world” variables (e.g. sample load). In this example, the practical inference drawn from the combined metrics is that system performance improves dramatically with incremental sample loading increases below 160 ng as reflected collectively by identifications, chromatography, and MS signal intensities, whereas improvement at higher loads is not as significant. This illustrates the value of the metrics for rational, quantitative optimization of system performance.
System Variability in Interlaboratory Studies
The CPTAC interlaboratory studies (CPTAC Studies 5 and 6) involved analyses of a common yeast proteome reference sample on multiple Thermo LTQ and LTQ-Orbitrap instruments in several laboratories. The yeast extract was digested with trypsin, and aliquots were distributed for analysis. Although the participating laboratories used different LC systems and autosamplers with these MS instruments, they also used an SOP, which standardized sample loading, chromatography, MS instrument tuning, and dynamic sampling (see supplemental material).
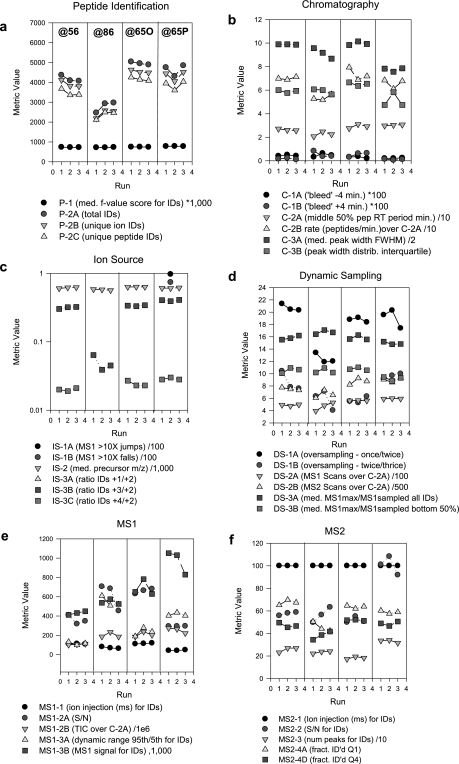
As part of Study 6, three replicate analyses of the yeast digest were analyzed on four different LTQ-Orbitrap systems in three different laboratories (Fig. 5). Inspection of the peptide identification summary (Fig. 5a) indicates an approximately 40% reduction in the number of peptide identifications by LTQ-Orbitrap@86 compared with the other three Orbitraps. Inspection of the other metrics indicates that dynamic oversampling parameters DS-1A and DS-1B were lower for LTQ-Orbitrap@86 than for the others (Fig. 5d), indicating excessive repeat sampling of peptide ion signals for MS/MS. This would have the observed effect of lowering peptide identifications. Another major characteristic of underperformance of instrument “@86” is the reduced value for ratios of 3+/2+ charge states (IS-3B) and 4+/2+ charge states (IS-3C) for identified peptides (Fig. 5c). (No 4+ peptide ions were identified for this instrument.) This may indicate a shift in the distribution of all peptide ion charge states and may have led to an increased fraction of 1+ peptide ions, which were excluded from MS/MS in the Orbitraps. Compliance with SOP dynamic exclusion settings was verified by inspection of the data file headers. However, further investigation identified inadequate formic acid content in the LC mobile phase as the likely cause, which is consistent with both a decreased proportion of higher charge state ions (IS-3B and IS-3C) and increased oversampling (fall in the value of DS-1A).
Fig. 5.
Performance metrics for triplicate analyses of a tryptic digest of the CPTAC yeast reference proteome on four LTQ-Orbitraps at three different sites in CPTAC Study 6. Instruments labeled @56, @86, and @65O are LTQ-Orbitraps; the instrument labeled @65P is an LTQ-XL-Orbitrap. a–f display metrics according to category; values for each of the three runs are represented by a symbol. Low values for peptide identifications for instrument LTQ- Orbitrap@86 (a) coincide with low metric values for peptide ion charge states (metric IS-3B; c) and dynamic sampling (metric DS-1A; d) (see text for discussion). pep, peptide; IDs, identifications; med., median; ID'd, identified; fract., fraction; num, number; FWHM, full width at half-maximum; inject., injection.
The Study 6 data also identified performance metrics whose variation had little or no impact on peptide identifications, including the MS1 and MS2 signal intensity metrics (Fig. 5, c and e) for the case of “Orbitrap@86.” Also noteworthy is the modest dip in peptide identifications in the second run for instrument LTQ-Orbitrap@65P (Fig. 5a). This occurred together with evidence of electrospray instability as indicated by 10-fold jumps or drops in MS1 signal intensity (IS-1A and IS-1B; Fig. 5c) in adjacent full scans. Manual inspection of the corresponding data file revealed a periodic “sawtooth” profile for the base peak chromatogram, indicating electrospray instability.
Another noteworthy aspect of the Study 6 data is the high degree of reproducibility of the chromatography metrics across instruments. Although the four Orbitraps used three different combinations of LC and autosampler systems, the SOP specified stationary phase, column measurements, flow rates, and injection parameters. Peptide elution periods, peak widths, and chromatographic bleed were generally consistent across instruments and replicate analyses, illustrating the feasibility of SOP-driven studies of LC-MS platforms, even when different hardware components may be used.
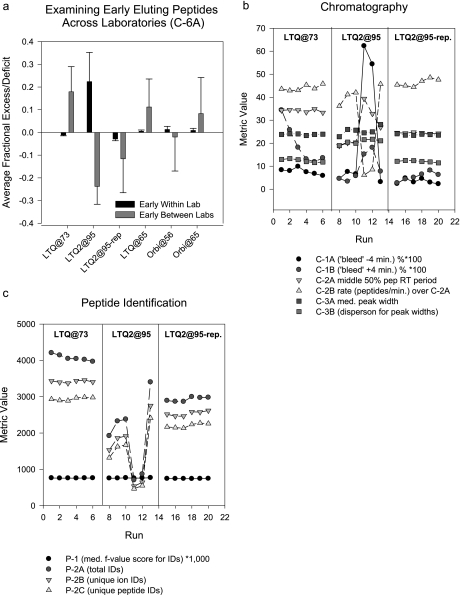
In CPTAC Study 5, the yeast digest was analyzed in six replicates on three LTQ and three Orbitrap instruments in five laboratories. The metric C-6A, which detects differences in the relative numbers of early and late eluting peptides within and between labs, identified large differences within replicates for “LTQ2@95” (Fig. 6a). These differences were reflected in increased values for chromatographic bleed metrics C-1A and C-1B and decreased peptide identification rate (C-2B) (Fig. 6b) and lowered numbers of peptide identifications (Fig. 6c) in the fourth and fifth runs in the series. This enabled identification of wash solvent cross-contamination of the sample injection loop as a probable cause. Correction of the problem partially restored the metrics and peptide identifications to values comparable to the other systems (see Fig. 6, “LTQ2@95-rep”).
Fig. 6.
Performance metrics for six replicate analyses of a tryptic digest of the yeast reference proteome on LTQ and Orbitrap instruments in CPTAC Study 5. Instruments labeled LTQ@73, LTQ@65, and LTQ@95 are LTQ instruments; the instrument labeled Orbi@56 is an LTQ-Orbitrap; the instrument labeled Orbi@65 is an LTQ-XL- Orbitrap. Marked variations in the relative number of early and late eluting peptides (a), chromatography metrics (b, middle section), and identifications (c, middle section) for instrument LTQ2@95 led to diagnosis and resolution of the problem (see text for discussion) as indicated by a second set of analyses on this instrument (LTQ2@95-rep). Instrument LTQ@73 is included in both panels as representative of the performance of the other instruments. The Orbitraps were included in the analysis and in a as a source of diversity from different laboratories and to demonstrate the usefulness of the chromatographic metrics across instrument platforms. Error bars represent median error. med., median; IDs, identifications.
Performance of Metrics across Systems and Laboratories
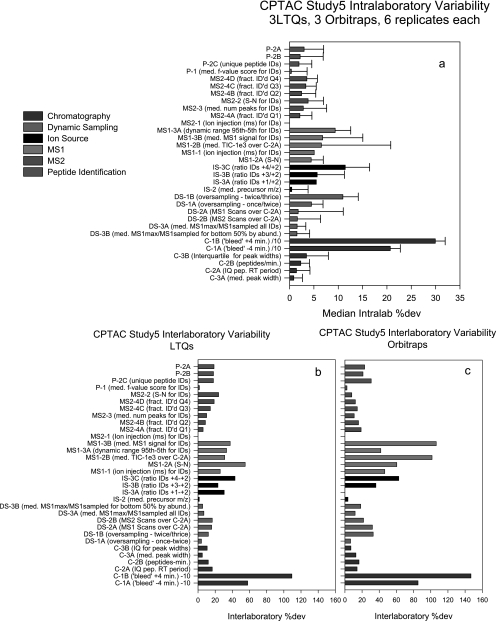
Whereas Table II describes variation in the metrics for a single instrument in replicate analyses, the CPTAC interlaboratory studies provided a means to evaluate variability across multiple laboratories, instruments, and analyses. Fig. 7a displays average intralab %dev values for the metrics for all six instruments (three LTQs and three Orbitraps) in CPTAC Study 5 (six replicate yeast digest analyses). The metrics have been sorted within categories from lowest to highest and the error bars represent the %dev of the intralab %dev (i.e. they estimate the range of %dev values across the labs). The plotted size of the colored bar represents variation within a laboratory; the relative size of the accompanying error bar indicates variation (%dev) of the intralab %dev across laboratories. These values as displayed are not intended to represent interlab variability, which would require comparing measurements between instrument classes directly, but to approximate the variability of the intralab %dev values. For most of the metrics, these values are comparable and average less than 10%. The most highly variable metrics describe MS1 signal intensity, dynamic sampling, and chromatographic bleed. Indeed, this latter metric was the most variable both within and between labs, also consistent with the egg yolk studies presented in Table I.
Fig. 7.
Summary of intralaboratory and interlaboratory variation for metrics for three LTQ and three LTQ-Orbitrap instruments in six replicate analyses of a tryptic digest of CPTAC yeast reference proteome in CPTAC Study 5. The instruments labeled LTQ are all LTQ model instruments; the instruments labeled Orbis include two LTQ-Orbitraps and one LTQ-XL-Orbitrap. Metrics are grouped by system category and ranked by code for comparison between panels. Intralaboratory variation in %dev for each metric and variation in %dev are shown in a. b and c show interlaboratory variation in metrics for LTQ and Orbitrap instruments, respectively. IQ, interquartile; pep, peptide; IDs, identifications; med., median; ID'd, identified; fract., fraction; num, number.
This comparison can be extended to interlab values for the LTQs and Orbitraps (Fig. 7, b and c). Whereas Fig. 6a depicts the variation within labs, Fig. 7, b and c, reveal values that are most irregular between labs. The main finding is that interlaboratory variation in peptide identifications and most other performance metrics are comparable between the LTQ and Orbitrap systems, even though there are large differences in the average values for some metrics between them (not shown). Differences in these mass analyzers result in different MS1 signal intensities, but this also results in greater variation in both MS1 and dynamic sampling metrics for the Orbitraps (Fig. 7, b versus c). This could also be reflected by the fact that one of the Orbitraps outperformed the others in the study by more than 20% in the number of unique tryptic peptide identifications for this study (data not shown). Nevertheless, variations in MS2 metrics appear comparable for LTQs and Orbitraps.
Fig. 7, a–c, also provide a broad perspective on which analytical techniques pose the greatest challenges to intralaboratory and interlaboratory standardization. For example, the metrics with the highest variation describe peptide chromatography (e.g. C-1A and C-1B), which is subject to more variable influences than any other component of the LC-MS/MS system. Even modest variations in mobile phase composition, gradient delivery, flow control stability, sample contaminants, and the composition of previous samples can influence peptide elution. We note that the relative variability of C-1A and C-1B corresponds to the typical experience in chromatography where peak tailing (C-1B) is a more common problem than peak bleed or “fronting” (C-1A).
DISCUSSION
Troubleshooting poor system performance in LC-MS/MS-based proteomics typically involves a combination of experience, intuition, and a highly subjective evaluation of limited data (e.g.“what does the chromatogram look like?”). Although causes of commonly encountered system problems are well known and can be rationalized in retrospect, the combinatorial possibilities of malfunction among multiple components may preclude a systematic approach to diagnosis. The 46 performance metrics described here enable the implementation of a quantitative, integrated approach to system troubleshooting. These metrics enable diagnosis of poor system performance, such as the ∼40% decline in peptide identifications by one Orbitrap compared with others in an interlaboratory study (Fig. 5).
However, performance metrics can help diagnose much more subtle, yet important, problems. These metrics provide the first effective means to deal with modest decrements in system performance, which are frequently encountered yet difficult to diagnose. The metrics are quite sensitive to small changes in performance: most have %dev values of less than 10%. The sensitivity of these metrics enabled detection of electrospray instability as a contributing cause of modestly diminished peptide identification performance in one replicate analysis of a yeast proteome digest in CPTAC Study 6 (Fig. 5, LTQ-Orbitrap@65P).
A relatively large number of metrics (46 presented here) could be considered excessive for purposes of troubleshooting system performance. However, the availability of over 40 metrics does not imply that all metrics are always used together for diagnostic purposes. Indeed, this would probably always be unnecessary. In the examples we present here, specific system malfunctions are indicated by changes in smaller subsets of metrics. On the other hand, the advantage of the relatively large number of metrics is that they reflect diverse components of the system.
A complete record of system performance should become a critical element of quality control documentation for LC-MS data sets. This requirement for documentation of system performance is important in many applications in proteomics, particularly where analysis and comparison of LC-MS data sets provide the basis for identifying distinct characteristics of biological systems. Apparent differences between phenotypes are detected by comparing data sets from multiple technical replicate analyses of the corresponding samples. A key assumption underlying this approach is that observed differences represent true proteomic differences rather than variability in system performance. This is particularly important when biological differences between samples are relatively modest as analyses must be able to discern differences comprising a small subset of proteome components. The metrics we describe here could provide an unambiguous basis for quantitatively defining platform stability and could enable identification of outlier data that would otherwise confound biological comparisons.
We also have shown here in the context of the CPTAC interlaboratory studies that these metrics display stability in behavior across multiple laboratories and systems. These observations not only further define the utility and normal ranges of the metrics but provide insights into the aspects of multilaboratory studies that provide the greatest barriers to platform standardization. However, the implementation of an SOP in CPTAC Study 6 effectively normalized key features of the chromatography (Fig. 5c), a remarkable achievement in view of the use of different LC systems by the participating laboratories. We note that SOPs were used in the CPTAC studies to enable comparisons under conditions where key system variables were held constant, thus enabling identification of sources of variability in peptide and protein detection. The SOPs represented a balance between performance optimization for peptide detection and practical considerations for interlaboratory studies. They do not represent fully optimized methods and are not intended as prescriptive for the proteomics research community.
This work on performance metrics was done with Thermo LTQ and LTQ-Orbitrap instruments, which are commonly used for LC-MS/MS proteomics and were the principal instruments available in our laboratories (at NIST) and in the participating CPTAC laboratories. Although a few of the metrics we describe (e.g. ion injection times) are not applicable to other instruments, such as quadrupole-time of flight instruments, most of these metrics can be applied to any electrospray LC-MS/MS instrument platform used for proteomics. Extension of the metrics to these other systems will require software to extract data from instrument data files and reference data sets to define the behavior of individual metrics in different instrument platforms.
The metrics described here represent a subset of a larger body of measurements explored in our studies of LC-MS systems. Although these metrics were applied to data-dependent LC-MS/MS analyses, subsets of these metrics and variations thereof could be applied similarly to high resolution LC-MS “MS1 profiling” systems or to LC-multiple reaction monitoring-MS on triple quadrupole and quadrupole-ion trap instruments. Indeed, many of the metrics are applicable to the analysis of LC-MS systems for non-peptide analytes, including metabolites, lipids, carbohydrates, and other molecule classes. Implementation of these performance metrics will be facilitated by the distribution of software for extracting the metrics directly from raw data files and by development of graphical user interfaces and integration with standard proteome analysis work flows. We are continuing development and evaluation of new performance metrics and will make available software to facilitate their implementation in the analytical community.
Supplementary Material
Footnotes
* This work was supported, in whole or in part, by an interagency agreement between the NCI, National Institutes of Health and the National Institute of Standards and Technology and by National Institutes of Health Grants U24CA126476, U24CA126485, U24CA126480, U24CA126477, and U24CA126479 as part of the NCI Clinical Proteomic Technologies for Cancer (http://proteomics.cancer.gov) initiative. A component of this initiative is the Clinical Proteomic Technology Assessment for Cancer (CPTAC) teams, which include the Broad Institute of MIT and Harvard (with the Fred Hutchinson Cancer Research Center, Massachusetts General Hospital, the University of North Carolina at Chapel Hill, the University of Victoria, and the Plasma Proteome Institute), Memorial Sloan-Kettering Cancer Center (with the Skirball Institute at New York University), Purdue University (with Monarch Life Sciences, Indiana University, Indiana University-Purdue University Indianapolis, and the Hoosier Oncology Group), University of California, San Francisco (with the Buck Institute for Age Research, Lawrence Berkeley National Laboratory, the University of British Columbia, and the University of Texas M. D. Anderson Cancer Center), and Vanderbilt University School of Medicine (with the University of Texas M. D. Anderson Cancer Center, the University of Washington, and the University of Arizona). Contracts from NCI, National Institutes of Health through SAIC to Texas A&M University funded statistical support for this work. A complete listing of the CPTAC Network can be found at http://proteomics.cancer.gov.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material, including detailed descriptions of the preparation and characterization of the yeast protein extracts used in these studies and standard operating procedures for LC-MS/MS analyses in CPTAC Studies 5 and 6.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material, including detailed descriptions of the preparation and characterization of the yeast protein extracts used in these studies and standard operating procedures for LC-MS/MS analyses in CPTAC Studies 5 and 6.
1 The abbreviations used are:
- CPTAC
- Clinical Proteomic Technology Assessment for Cancer
- MS1
- full MS scan
- MS2
- tandem MS scan
- SOP
- standard operating procedure
- LTQ
- linear trap quadrupole
- RT
- retention time
- S/N
- signal to noise
- TIC
- total ion current
- NIST
- National Institute of Standards and Technology
- FDR
- false discovery rate
- %dev
- percent deviation.
REFERENCES
- 1.Yates J. R., 3rd ( 2004) Mass spectral analysis in proteomics. Annu. Rev. Biophys. Biomol. Struct 33, 297– 316 [DOI] [PubMed] [Google Scholar]
- 2.Aebersold R., Mann M. ( 2003) Mass spectrometry-based proteomics. Nature 422, 198– 207 [DOI] [PubMed] [Google Scholar]
- 3.Liu H., Lin D., Yates J. R., 3rd ( 2002) Multidimensional separations for protein/peptide analysis in the post-genomic era. BioTechniques 32, 898, 900,, 902 [DOI] [PubMed] [Google Scholar]
- 4.Eriksson J., Fenyö D. ( 2007) Improving the success rate of proteome analysis by modeling protein-abundance distributions and experimental designs. Nat. Biotechnol 25, 651– 655 [DOI] [PubMed] [Google Scholar]
- 5.Whiteaker J. R., Zhang H., Zhao L., Wang P., Kelly-Spratt K. S., Ivey R. G., Piening B. D., Feng L. C., Kasarda E., Gurley K. E., Eng J. K., Chodosh L. A., Kemp C. J., McIntosh M. W., Paulovich A. G. ( 2007) Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J. Proteome Res 6, 3962– 3975 [DOI] [PubMed] [Google Scholar]
- 6.Alldridge L., Metodieva G., Greenwood C., Al-Janabi K., Thwaites L., Sauven P., Metodiev M. ( 2008) Proteome profiling of breast tumors by gel electrophoresis and nanoscale electrospray ionization mass spectrometry. J. Proteome Res 7, 1458– 1469 [DOI] [PubMed] [Google Scholar]
- 7.Sandhu C., Hewel J. A., Badis G., Talukder S., Liu J., Hughes T. R., Emili A. ( 2008) Evaluation of data-dependent versus targeted shotgun proteomic approaches for monitoring transcription factor expression in breast cancer. J. Proteome Res 7, 1529– 1541 [DOI] [PubMed] [Google Scholar]
- 8.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J. ( 2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190– 1203 [DOI] [PubMed] [Google Scholar]
- 9.Guo A., Villén J., Kornhauser J., Lee K. A., Stokes M. P., Rikova K., Possemato A., Nardone J., Innocenti G., Wetzel R., Wang Y., MacNeill J., Mitchell J., Gygi S. P., Rush J., Polakiewicz R. D., Comb M. J. ( 2008) Signaling networks assembled by oncogenic EGFR and c-Met. Proc. Natl. Acad. Sci. U.S.A 105, 692– 697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jessani N., Niessen S., Wei B. Q., Nicolau M., Humphrey M., Ji Y., Han W., Noh D. Y., Yates J. R., 3rd, Jeffrey S. S., Cravatt B. F. ( 2005) A streamlined platform for high-content functional proteomics of primary human specimens. Nat. Methods 2, 691– 697 [DOI] [PubMed] [Google Scholar]
- 11.Elias J. E., Haas W., Faherty B. K., Gygi S. P. ( 2005) Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods 2, 667– 675 [DOI] [PubMed] [Google Scholar]
- 12.Resing K. A., Meyer-Arendt K., Mendoza A. M., Aveline-Wolf L. D., Jonscher K. R., Pierce K. G., Old W. M., Cheung H. T., Russell S., Wattawa J. L., Goehle G. R., Knight R. D., Ahn N. G. ( 2004) Improving reproducibility and sensitivity in identifying human proteins by shotgun proteomics. Anal. Chem 76, 3556– 3568 [DOI] [PubMed] [Google Scholar]
- 13.Omenn G. S., States D. J., Adamski M., Blackwell T. W., Menon R., Hermjakob H., Apweiler R., Haab B. B., Simpson R. J., Eddes J. S., Kapp E. A., Moritz R. L., Chan D. W., Rai A. J., Admon A., Aebersold R., Eng J., Hancock W. S., Hefta S. A., Meyer H., Paik Y. K., Yoo J. S., Ping P., Pounds J., Adkins J., Qian X., Wang R., Wasinger V., Wu C. Y., Zhao X., Zeng R., Archakov A., Tsugita A., Beer I., Pandey A., Pisano M., Andrews P., Tammen H., Speicher D. W., Hanash S. M. ( 2005) Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics 5, 3226– 3245 [DOI] [PubMed] [Google Scholar]
- 14.Liu H., Sadygov R. G., Yates J. R., 3rd ( 2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem 76, 4193– 4201 [DOI] [PubMed] [Google Scholar]
- 15.Hamming R. W. ( 1998) Digital Filters, 3rd Ed., Dover Publications, Mineola, NY [Google Scholar]
- 16.Rice S. O. ( 1945) Mathematical analysis of random noise, part III. Bell Syst. Tech. J 24, 46– 156 [Google Scholar]
- 17.Lawson C. L., Hanson R.J. ( 1987) Solving Least Squares Problem, Society for Industrial and Applied Mathematics, Philadelphia [Google Scholar]
- 18.Ghoudi K., Kulperger R. J., Remillard B. ( 2001) A nonparametric test of serial independence for time series and residuals. J. Multivariate Anal 79, 191– 218 [Google Scholar]
- 19.Lam H., Deutsch E. W., Eddes J. S., Eng J. K., King N., Stein S. E., Aebersold R. ( 2007) Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 7, 655– 667 [DOI] [PubMed] [Google Scholar]
- 20.Stein S. E., Scott D. R. ( 1994) Optimization and testing of mass-spectral library search algorithms for compound identification. J. Am. Soc. Mass Spectrom 5, 859– 866 [DOI] [PubMed] [Google Scholar]
- 21.Liu J., Bell A. W., Bergeron J. J., Yanofsky C. M., Carrillo B., Beaudrie C. E., Kearney R. E. ( 2007) Methods for peptide identification by spectral comparison. Proteome Sci 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eng J. K., McCormack A. L., Yates J. R. ( 1994) An approach to correlate tandem mass-spectral data of peptides with amino-acid sequences in a protein database. J. Am. Soc. Mass. Spectrom 5, 976– 989 [DOI] [PubMed] [Google Scholar]
- 23.Craig R., Beavis R. C. ( 2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466– 1467 [DOI] [PubMed] [Google Scholar]
- 24.Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. ( 2004) Open mass spectrometry search algorithm. J. Proteome Res 3, 958– 964 [DOI] [PubMed] [Google Scholar]
- 25.Clauser K. R., Baker P., Burlingame A. L. ( 1999) Role of accurate mass measurement (+/−10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem 71, 2871– 2882 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.