Abstract
Mammalian hibernation involves complex mechanisms of metabolic reprogramming and tissue protection. Previous gene expression studies of hibernation have mainly focused on changes at the mRNA level. Large scale proteomics studies on hibernation have lagged behind largely because of the lack of an adequate protein database specific for hibernating species. We constructed a ground squirrel protein database for protein identification and used a label-free shotgun proteomics approach to analyze protein expression throughout the torpor-arousal cycle during hibernation in arctic ground squirrels (Urocitellus parryii). We identified more than 3,000 unique proteins from livers of arctic ground squirrels. Among them, 517 proteins showed significant differential expression comparing animals sampled after at least 8 days of continuous torpor (late torpid), within 5 h of a spontaneous arousal episode (early aroused), and 1–2 months after hibernation had ended (non-hibernating). Consistent with changes at the mRNA level shown in a previous study on the same tissue samples, proteins involved in glycolysis and fatty acid synthesis were significantly underexpressed at the protein level in both late torpid and early aroused animals compared with non-hibernating animals, whereas proteins involved in fatty acid catabolism were significantly overexpressed. On the other hand, when we compared late torpid and early aroused animals, there were discrepancies between mRNA and protein levels for a large number of genes. Proteins involved in protein translation and degradation, mRNA processing, and oxidative phosphorylation were significantly overexpressed in early aroused animals compared with late torpid animals, whereas no significant changes at the mRNA levels between these stages had been observed. Our results suggest that there is substantial post-transcriptional regulation of proteins during torpor-arousal cycles of hibernation.
Hibernation is a survival strategy of regulated metabolic suppression adopted by a wide range of mammalian species to survive environmental conditions of low or unpredictable food availability (1, 2). Through complex cellular and molecular reorganization, hibernators have evolved the ability to sustain and spontaneously reverse profoundly low levels of body temperature and rates of oxygen consumption. For example, during torpor the arctic ground squirrel, Urocitellus parryii, adopts a core body temperature of −2.9 °C and a minimum metabolic demand that is 2% of its normal, basal metabolic rate (3, 4). However, during the hibernation season, arctic ground squirrels, like all other small mammalian hibernators, alternate between prolonged torpor (to 24 days) and periodic arousal episodes when they spontaneously rewarm to euthermic body temperatures (36–37 °C) for 10–15 h before returning to torpor. The genetic and molecular mechanisms of regulation and tolerance of hibernation and the functional significance of arousal episodes remain central questions in hibernation research. In addition, understanding the molecular mechanisms of hibernation has the potential of leading to the development of novel preventive and therapeutic approaches to the treatment of human maladies such as trauma, cardiovascular disease, stroke, and ischemia/reperfusion injury.
Nearly two decades of gene expression studies on hibernating mammals have suggested that the hibernation phenotype results from the differential expression of existing genes rather than the evolution of novel genes (1, 5, 6). Recently, several laboratories have utilized large scale genomics approaches to investigate differential gene expression during hibernation in different ground squirrel species (7, 8). In initial studies of the arctic ground squirrel (AGS),1 we compared mRNA levels in brown adipose tissue using mouse microarrays and identified 625 genes that were differentially expressed between torpid and summer active animals (9). We subsequently designed Illumina BeadArrays that measured expression of about 1,400 genes in five AGS tissues using probes based on pooled mRNA sequences of three ground squirrel species, comparing four hibernation stages during spontaneous torpor-arousal cycles with non-hibernating AGS. Using the BeadArrays as well as real time PCR assays, we identified significant differences in hibernating animals compared with non-hibernating animals in mRNA levels of genes involved in metabolism and tissue protection and genes related to circadian rhythm and cell growth during the torpor-arousal cycle (10).
Transcripts of mRNA are protected while translation is inhibited during torpor in ground squirrels (11, 12). Therefore, protein variety and abundance can be very different from corresponding gene expression at the mRNA level, and differential protein expression may more directly reflect regulatory changes related to hibernation. Martin et al. (13) observed a slow loss of protein integrity during prolonged torpor in golden-mantled ground squirrels (Callospermophilus lateralis) and proposed that replenishment of proteins may be a cause for arousal episodes. These results call for investigating hibernation at the proteomic in addition to transcriptomic levels. However, because of the difficulty of developing high throughput proteomics technologies for non-model organisms, large scale hibernation proteomics has lagged behind hibernation transcriptomics (14). Initial hibernation proteomics studies were based on two-dimensional gel electrophoresis. Using two-dimensional gels followed by MS/MS, Epperson et al. (15) compared protein expression in the livers of golden-mantled ground squirrels entering torpor and summer active animals and identified 68 differentially expressed proteins. Using a similar approach, Martin et al. (16) then identified 27 different proteins showing significant differential expression in the intestines of thirteen-lined ground squirrels (Ictidomys tridecemlineatus) sampled during interbout arousal compared with summer active animals. However, two-dimensional gel-based proteomics approaches have intrinsic problems such as limited coverage, low sensitivities, and unidentifiable spots (17, 18). Another obstacle in hibernation proteomics is the lack of a protein database for any hibernating species. Toward overcoming this problem, Russeth et al. (14) combined multiple software packages to search for proteins in metazoan protein databases to analyze the electrospray ionization and matrix-assisted laser desorption ionization MS results of seven spots that showed differential expression between active and hibernating animals on two-dimensional gels of skeletal muscle and heart of thirteen-lined ground squirrels.
More quantitative MS-based methods have been developed, including stable isotope labeling such as isotope-coded affinity tag, stable isotope labeling by amino acids in cell culture, and isobaric tags for relative and absolute quantitation. Unfortunately, these methods are expensive and time-consuming. Recently, an alternative label-free shotgun proteomics strategy based on spectral counting has become available (19). In this method, relative protein abundance is estimated from the number of spectral matches for a given protein species across samples. The low abundance proteins that may be randomly picked up in the experiment can be filtered out. Appropriate statistical methods have been developed to analyze such label-free spectral data (20, 21). Although considered less accurate than the isotope labeling methods, this approach has an advantage of higher proteome coverage, higher dynamic range, and a simpler experimental protocol and is therefore more convenient for global protein expression studies (22).
Here we apply a label-free shotgun proteomics approach for the first time on a hibernating species. We collected MS spectra using LC-MS/MS, and results were searched against a ground squirrel protein database that we constructed by combining Ensembl annotation of the newly available thirteen-lined ground squirrel genome along with pooled expressed sequence tag (EST) sequences from three closely related ground squirrel species. We then compared protein expression results with our previously published mRNA results using the same tissue samples (10). We also designed additional real time PCR assays for mRNA of newly identified proteins in the high throughput proteomics study. Selected differentially expressed proteins identified in our approach were further validated by Western blot analyses. Our analysis results indicate the potentially significant role of post-transcriptional regulation in torpor-arousal cycles during hibernation.
EXPERIMENTAL PROCEDURES
Construction of Ground Squirrel Protein Database
The thirteen-lined ground squirrel genome and annotations for 17,920 protein-coding and non-coding genes containing splice site information as well as 14,830 protein sequences of protein-coding genes were downloaded from Ensembl release 49 (speTri1, June 2006). EST sequences of golden-mantled ground squirrel (8,803 sequences) and thirteen-lined ground squirrel (5,256 sequences) were obtained from NCBI. Arctic ground squirrel EST sequences (13,505 sequences) were obtained from the EST sequencing project at University of Alaska Fairbanks. These ground squirrel EST sequences were aligned to the thirteen-lined ground squirrel genome using the blastn program (23) to identify the genomic contigs to which the ESTs belong, using a minimum alignment score of 160 as the criterion. To identify the precise splice sites, the EST sequences were realigned to the corresponding genomic contigs using the sim4 program (24). To identify the human and mouse homologous genes in the thirteen-lined ground squirrel genome that could have been missed in Ensembl gene annotations, we further aligned the human and mouse RefSeq mRNA sequences (25) to the thirteen-line ground squirrel genome by the same procedure. In the sim4 alignments, we required that the mapped portion of the EST or RefSeq alignment is at least 50% of the full sequence and that match identities were higher than 95% for ground squirrel EST sequences and 85% for human and mouse RefSeq sequences. We clustered the EST alignments, RefSeqs alignments, and Ensembl gene annotations on the same contig into “gene clusters” based on the splice site information and the mutual overlaps; i.e. they were clustered if they shared at least one common splice site or they overlapped for at least 50% of the length of the shorter sequence. The ESTs that could not be reliably aligned to the genome were clustered into “unaligned clusters” by the blastclust program. To remove the redundancy of gene clusters due to genome duplication or incomplete assembly, gene clusters were further clustered to “unique gene clusters” if a gene cluster shared at least 80% of its members with another gene cluster.
If the unique gene cluster contained Ensembl gene annotations, the protein sequences from Ensembl annotations were used directly. There were gaps in some Ensembl protein sequences because of incomplete genome sequences. In this case, we aligned the ground squirrel ESTs onto the protein sequences with the blastx program and used the protein translation of EST sequences to fill in the gaps in Ensembl protein sequences. In this way, a total of 1,246 Ensembl protein sequences were “patched.” If the unique gene cluster did not have any Ensembl annotation but had human RefSeq alignment, the genomic regions aligned to the human RefSeq were translated into protein sequence according to the open reading frames in human. If the unique gene cluster had neither Ensembl annotation nor human RefSeq alignment but had mouse RefSeq alignment, the genomic regions aligned to the mouse RefSeq were translated into protein sequence. If the unique gene cluster had none of the above, the genomic regions aligned to the ESTs in that cluster to the maximum spanning length were obtained and aligned to the nr database by the blastx program. The protein sequences were obtained by translating the aligned genomic regions using the open reading frame of the aligned nr proteins. For the unaligned clusters, we also aligned the EST sequences to the nr database and selected the longest translated protein sequence in that cluster. All protein sequences were merged to form the ground squirrel protein database. They were further aligned to human RefSeq protein sequences by the blastp program to obtain their corresponding human gene symbols where a minimum alignment score of 100 was used as criteria. The scheme of the ground squirrel protein database construction is shown in supplemental Fig. S1. The protein amino acid sequences of the ground squirrel protein database and their annotations are provided as supplemental Tables S1 and S2.
Animals
Animals and tissues used in this proteomics study were the same as those analyzed in our previous microarray study (10). We included two more postreproductive animals in the Western blot validation. Briefly, U. parryii were trapped in July near the Toolik Field Station in northern Alaska (68.6° north 149.4° west; elevation, 809 m) and transported to the University of Alaska Fairbanks. Animals were housed at 20 ± 2 °C with a 16:8-h light:dark photoperiod and provided with Mazuri rodent chow and water ad libitum. Hibernating animals were implanted with temperature-sensitive radiotransmitters in their abdominal cavities and maintained at a 4:20-h photoperiod. Body temperature (Tb) was monitored for the precise stage of torpor and arousal by an automated telemetry system that measures and records Tb every 10 min (4). Animals late in a torpor bout (late torpor (LT); n = 4) were collected after 80–90% of the duration of the previous torpor bout (8–12 days). Animals sampled early after spontaneously arousing from torpor (early arousal (EA); n = 4) were collected 1–2 h after core Tb had increased above 30 °C during rewarming. Postreproductive summer euthermic animals sampled in May and June 1–2 months after ending hibernation (postreproduction (PR); n = 4) were used as non-hibernating controls. These animals had completed reproductive regression as assessed by external inspection of gonads and genitalia and had entered molt. Torpid animals were euthanized by decapitation without anesthesia. Aroused and postreproductive animals were deeply anesthetized with isoflurane and decapitated. Liver tissue was rapidly dissected, frozen in liquid N2, and stored at −80 °C. Frozen tissues were transported to Shanghai, China on dry ice where proteomics and real time PCR analyses were conducted.
Protein Digestion
Liver samples (1 mg) were homogenized in a Dounce glass grinder using 1 ml of lysis buffer consisting of 9.5 m urea, 4% CHAPS (w/v), 65 mm Tris, 10 mm DTT, and a protease inhibitor mixture (100:1; Merck). The crude tissue extracts were centrifuged for 45 min at 15,000 × g to remove the undissolved pellets. The tissue lysates (1 mg) were reduced for 2 h at room temperature by addition of 1 m DTT to a final concentration of 10 mm DTT and then alkylated for 45 min by addition of 1 m iodoacetamide to a final concentration of 50 mm. After reduction and alkylation, the lysates were precipitated using 5 volumes of ice-cold TCA/acetone. The pellets were then washed three times with ice-cold pure acetone and resuspended in 200 μl of 100 mm NH4HCO3. For in-solution digestion, 20 μg of modified trypsin (sequencing grade, Promega) were added, and the mixtures were digested at 37 °C for 1,000 rpm overnight in a Thermomixer compactTM (Eppendorf). Trypsin activity was quenched by acidification using TFA to a final concentration of 1%. The mixed digests were further ultrafiltered by MicroconTM (10 kDa; Millipore) to remove the large molecules.
Strong Cation Exchange Chromatography
The strong cation exchange fractionation protocol mainly followed a previous report (26) with slight modification. Briefly, the mixed digests were loaded onto a 2.1-mm × 10-cm strong cation exchange column (Column Technology, Inc.) equilibrated with 0.1% formic acid, 20% acetonitrile, and 5 mm ammonium formate (NH4HCO2) using an HPLC 1090 system. The peptides were separated by a linear gradient of NH4HCO2 from 5 to 100 mm with a flow rate of 0.5 ml/min. A total of six fractions including flow-through were collected, and each fraction was dried with ConcentratorTM (Eppendorf). The pellets were resuspended by adding 100 μl of 0.1% formic acid before the LC-MS run.
Mass Spectrometric Analysis
The fractionated digests were analyzed by C18 reversed-phase microscale liquid chromatography-tandem mass spectrometry. In this study, EttanTM MDLC instrument controlled by UNICORNTM software (GE Healthcare), a system for automated multidimensional chromatography, was used for desalting and separation of peptides prior to LTQ analyses. The mass spectrometer was operated in the data-dependent mode to automatically switch between MS and MS/MS acquisition. A Finnigan LTQ linear ion trap mass spectrometer equipped with a microspray interface was used as the detector for the peptides separated on the reversed-phase chromatography column. Each scan cycle consisted of one full-scan mass spectrum (m/z 400–1800) followed by 10 MS/MS events of the most intense ions with the following dynamic exclusion settings: repeat count, 2; repeat duration, 30 s; and exclusion duration, 90 s (27). The samples were loaded onto the trap column first with 10 μl/min flow rate, and then the desalted samples were eluted at a flow rate of 1300 nl/min in the MDLC instrument by applying a linear gradient of 0–50% buffer (0.1% formic acid, 84% ACN) for 180 min. The Finnigan LTQ linear ion trap mass spectrometer was used for the MS/MS experiment with an ion transfer capillary of 160 °C and iSpray voltage of 3 kV. Normalized collision energy was 35.0%.
Assigning Peptide Sequences Using SEQUEST
Peak lists were generated using Bioworks 3.1 (Thermo) with default parameters. All acquired MS/MS spectra from 12 AGS samples were searched against 19,297 proteins in the ground squirrel protein database using SEQUEST (version 3.1). Searches were performed using the “trypsin enzyme” parameter of the software. Methionine oxidation was only specified as a variable modification, and cysteine carboxamidomethylation was specified as the fixed modification. The peptide database search was carried out with the following parameters: peptide mass tolerance, 3.0 Da; mass tolerance for fragment ions, 0 Da; maximum number of internal cleavage sites, 2; and number of allowed errors in matching autodetected peaks, match peak tolerance, 1. SEQUEST results were filtered using the following parameter: Xcorr > 1.55 and ΔCn > 0.15 for the peptides in 1+ charged state, Xcorr > 1.85 and ΔCn > 0.16 for the peptides in 2+ charged state, and Xcorr > 2.6 and ΔCn > 0.22 for the peptides in 3+ charged state (28). These thresholds were determined to maximize the number of peptides identified while satisfying a false discovery rate (FDR) of ≤0.05. The FDR was calculated based on the following formula: FDR = nrev/ntar (29). nrev is the number of peptide hits matched to the “reverse” protein, and ntar is the number of peptide hits matched to the “target” protein. Peptides exactly matched to more than one protein in our database were included in all proteins onto which they mapped. All output results were combined using an in-house software named buildsummary.
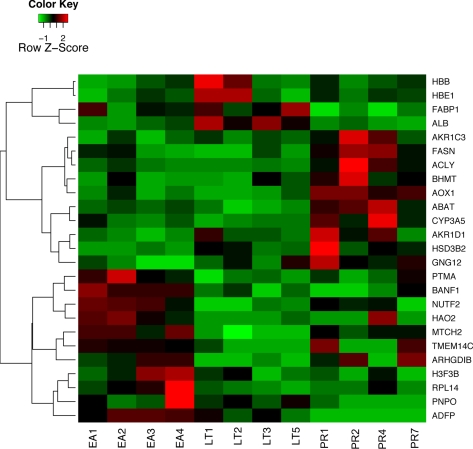
Protein Data Analysis
The SEQUEST program identified 3,594 AGS proteins that had at least two unique peptide hits to ground squirrel proteins in our database (supplemental Table S3). 3,537 of these AGS proteins have homologous human gene symbols. The remaining 57 AGS proteins were manually annotated by aligning with nr database by blastp program. When we removed redundant proteins corresponding to the same gene symbols, we kept the one with the highest number of spectral counts. After removing this redundancy, we obtained 3,104 unique proteins with at least two unique peptide counts. For reliable protein quantification, we selected 1,209 unique proteins satisfying peptide counts ≥2 for every animal sample in at least one physiological state (EA, LT, and PR). The total numbers of spectral hits of peptides within each protein in the ground squirrel protein database were used to quantify the protein expression level in each sample. Spectral counts in each sample were divided by protein length to obtain the spectral abundance factor (SAF). Normalized spectral abundance factors (NSAFs) were obtained by further dividing SAF values by the sum of SAF values in each sample according to Pavelka et al. (30). The dynamic range of our NSAF values was computed by the base 10 logarithm of the ratio between the 99.95th percentile and the 0.05th percentile after removing the zero values according to Pavelka et al. (30). NSAF values were analyzed by the power law global error model (PLGEM)-signal to noise (StN) method (Bioconductor package) (31) to compute the statistical significance of protein expression differences in pairwise comparisons among EA, LT, and PR stages. An FDR-adjusted p value <0.05 was used as the criterion for statistical significance. A heat map of the 25 most significant proteins (FDR-adjusted p value <0.001) was generated by heatmap2 in the gplots package (Bioconductor package) (31). The red-green colors represented scaled NSAF values. The hierarchical clustering of genes was based on the distances defined as one subtracts Pearson correlation coefficients (Fig. 2). Log2-transformed protein changes were calculated as log2-based ratios of mean NSAF values in EA, LT, and PR stages. Spectral count data were also analyzed using the Qspec program obtained from Choi et al. (20). A Bayes factor >10 and a |fold change| >1.5 were used as the criteria for statistical significance in pairwise comparisons. The combined PLGEM and Qspec results are listed in supplemental Table S4. Functional analyses using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (32) program were conducted using the default background gene set.
Fig. 2.
Heat map of NSAF values of 25 most significant differentially expressed proteins in Table I. Red color corresponds to high levels of expression, and green color corresponds to low levels of expression.
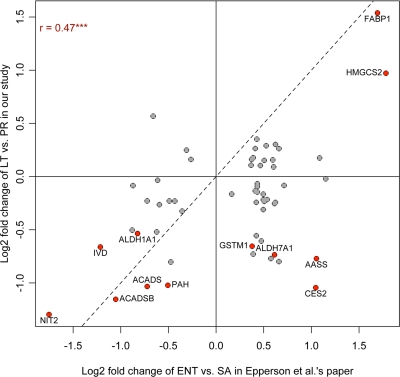
To compare our results with those of Epperson et al. (15), we extracted fold changes of significant proteins shown comparing animals during entrance into torpor (Ent) versus non-hibernating animals sampled in summer (SA) from Tables I and II in their study. Homologous human gene symbols were obtained from the accession number of identified proteins listed in the tables. Fold changes of proteins with the same gene symbols were averaged. MDH1 protein was removed because it appeared in both over- and underexpressed protein lists. Fold changes of both Epperson et al. (15) Ent versus SA and our LT versus PR comparisons were log2-transformed and plotted in Fig. 4.
Table I. 25 most significant differentially expressed proteins in PLGEM-StN analysis.
The 25 most significant differentially expressed proteins in PLGEM-StN analysis (FDR-adjusted p value <0.001) are shown. Original spectral counts in 12 samples and FDR-adjusted p values in EA versus LT, EA versus PR, and LT versus PR comparisons are also included. Bold values indicate they are also significant in the Qspec result. Pro ID, protein identity.
| AGS Pro ID | ENSEMBL Pro ID | Gene symbol | UPCa | PCb | EA1 | EA2 | EA3 | EA4 | LT1 | LT2 | LT3 | LT5 | PR1 | PR2 | PR4 | PR7 | p valuec EA vs. LT | p value EA vs. PR | p value LT vs. PR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | |||||||||||||||||||
| AGS_1431 | ENSSTOP00000004452 | ABAT | 12 | 60 | 16 | 22 | 19 | 18 | 11 | 2 | 7 | 14 | 29 | 41 | 48 | 23 | 1.13e−02 | 3.17e−03 | 7.01e−04 |
| AGS_11191 | ENSSTOP00000005420 | ACLY | 21 | 26 | 5 | 11 | 1 | 0 | 0 | 0 | 2 | 0 | 10 | 47 | 23 | 12 | 4.51e−03 | 4.63e−04 | 5.79e−04 |
| AGS_5977 | ENSSTOP00000007189 | ADFP | 8 | 27 | 5 | 8 | 8 | 6 | 4 | 2 | 5 | 2 | 0 | 0 | 0 | 0 | 4.86e−02 | 7.28e−05 | 4.16e−03 |
| AGS_590 | ENSSTOP00000003215 | AKR1C3 | 17 | 71 | 33 | 95 | 28 | 59 | 63 | 39 | 79 | 77 | 77 | 201 | 132 | 61 | 5.89e−02 | 2.38e−04 | 3.43e−03 |
| AGS_5442 | ENSSTOP00000000753 | AKR1D1 | 5 | 37 | 3 | 2 | 0 | 2 | 7 | 3 | 4 | 3 | 11 | 6 | 9 | 2 | 5.88e−03 | 5.26e−04 | 7.07e−02 |
| AGS_15305 | None | ALB | 26 | 79 | 111 | 109 | 143 | 142 | 238 | 179 | 314 | 258 | 81 | 124 | 86 | 77 | 2.98e−04 | 1.85e−01 | 3.21e−04 |
| AGS_13709 | ENSSTOP00000006409 | AOX1 | 27 | 31 | 5 | 24 | 1 | 8 | 1 | 0 | 3 | 17 | 37 | 49 | 32 | 36 | 5.82e−02 | 7.94e−04 | 1.21e−03 |
| AGS_15506 | None | ARHGDIB | 5 | 45 | 2 | 3 | 5 | 4 | 0 | 0 | 1 | 0 | 2 | 5 | 0 | 5 | 4.57e−04 | 9.50e−01 | 3.19e−03 |
| AGS_1099 | ENSSTOP00000007259 | BANF1 | 3 | 40 | 6 | 5 | 5 | 4 | 1 | 2 | 0 | 2 | 0 | 0 | 1 | 3 | 1.42e−03 | 8.21e−04 | 4.56e−01 |
| AGS_14254 | ENSSTOP00000005260 | BHMT | 16 | 41 | 42 | 116 | 39 | 36 | 28 | 29 | 110 | 79 | 89 | 183 | 68 | 88 | 3.21e−01 | 9.20e−04 | 3.69e−03 |
| AGS_15328 | None | CYP3A5 | 10 | 50 | 20 | 9 | 7 | 11 | 2 | 5 | 8 | 9 | 24 | 17 | 41 | 15 | 1.79e−02 | 2.12e−03 | 7.78e−04 |
| AGS_2065 | ENSSTOP00000004232 | FABP1 | 11 | 84 | 252 | 103 | 206 | 162 | 185 | 114 | 205 | 351 | 33 | 100 | 39 | 103 | 3.11e−02 | 1.09e−04 | 4.96e−05 |
| AGS_931 | ENSSTOP00000009235 | FASN | 48 | 25 | 87 | 104 | 29 | 17 | 9 | 10 | 74 | 35 | 92 | 191 | 163 | 91 | 2.82e−02 | 1.65e−03 | 8.90e−04 |
| AGS_8158 | ENSSTOP00000004030 | GNG12 | 3 | 57 | 2 | 1 | 0 | 0 | 1 | 2 | 1 | 4 | 4 | 3 | 2 | 3 | 5.72e−03 | 5.03e−04 | 6.88e−02 |
| AGS_2985 | ENSSTOP00000013949 | H3F3B | 4 | 56 | 4 | 7 | 16 | 14 | 4 | 7 | 0 | 3 | 3 | 1 | 6 | 0 | 3.38e−03 | 8.07e−04 | 1.78e−01 |
| AGS_7005 | ENSSTOP00000003811 | HAO2 | 5 | 16 | 5 | 6 | 4 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 5 | 0 | 4.47e−04 | 1.22e−02 | 2.67e−02 |
| AGS_16132 | None | HBB | 12 | 73 | 112 | 143 | 244 | 229 | 444 | 359 | 172 | 180 | 205 | 133 | 182 | 229 | 1.75e−04 | 4.40e−02 | 2.65e−03 |
| AGS_18963 | None | HBE1 | 2 | 16 | 2 | 13 | 25 | 17 | 43 | 46 | 16 | 4 | 19 | 11 | 20 | 19 | 7.54e−04 | 3.05e−02 | 1.94e−02 |
| AGS_17287 | None | HSD3B2 | 3 | 17 | 1 | 1 | 2 | 1 | 4 | 4 | 2 | 3 | 10 | 3 | 5 | 4 | 4.47e−03 | 3.97e−04 | 5.92e−02 |
| AGS_17578 | None | MTCH2 | 7 | 48 | 8 | 9 | 6 | 8 | 2 | 0 | 2 | 2 | 5 | 4 | 5 | 5 | 7.61e−04 | 1.27e−01 | 9.86e−03 |
| AGS_7129 | ENSSTOP00000002574 | NUTF2 | 4 | 63 | 4 | 4 | 4 | 2 | 0 | 0 | 1 | 1 | 2 | 2 | 2 | 0 | 5.33e−04 | 2.95e−02 | 1.94e−02 |
| AGS_7834 | ENSSTOP00000005555 | PNPO | 4 | 21 | 4 | 1 | 2 | 9 | 2 | 3 | 2 | 5 | 1 | 0 | 0 | 0 | 2.41e−01 | 4.76e−04 | 7.32e−03 |
| AGS_1897 | ENSSTOP00000009373 | PTMA | 5 | 37 | 12 | 19 | 11 | 7 | 0 | 3 | 5 | 3 | 5 | 2 | 4 | 9 | 3.14e−04 | 7.76e−03 | 2.72e−02 |
| AGS_15424 | None | RPL14 | 5 | 27 | 5 | 9 | 11 | 17 | 3 | 2 | 2 | 1 | 2 | 3 | 7 | 2 | 8.44e−04 | 5.04e−03 | 1.24e−01 |
a Unique peptide counts.
b Peptide coverage.
c p value, FDR-adjusted p value.
Table II. Enriched GO categories in five differentially expressed protein groups.
Enriched GO categories identified by the DAVID program (FDR < 0.05) in five differentially expressed protein groups are shown: EA > LT, EA < LT, EA > PR, EA < PR, and LT < PR. No significantly enriched GO category was identified in the LT > PR group.
| GO term | Protein count | p value | FDR |
|---|---|---|---|
| EA > LT | |||
| Oxidoreductase | 33 | 1.48e−11 | 2.36e−08 |
| Protein biosynthesis | 19 | 2.11e−10 | 3.37e−07 |
| Proteasome | 12 | 9.48e−10 | 1.51e−06 |
| Phosphoprotein | 113 | 7.96e−11 | 1.27e−07 |
| Ribosome | 10 | 3.95e−07 | 6.29e−04 |
| Ribonucleoprotein | 17 | 1.78e−06 | 2.8e−03 |
| EA < LT | |||
| Steroid metabolic process | 7 | 1.10e−05 | 0.02 |
| Lipid metabolic process | 11 | 3.01e−05 | 0.04 |
| EA > PR | |||
| Translation | 28 | 1.61e−10 | 3.07e−07 |
| Phosphoprotein | 78 | 3.95e−08 | 6.29e−05 |
| EA < PR | |||
| Oxidoreductase | 36 | 1.10e−29 | 1.75e−26 |
| Electron transport | 24 | 1.85e−15 | 3.61e−12 |
| Lipid metabolic process | 25 | 4.22e−12 | 8.07e−09 |
| Response to chemical stimulus | 17 | 2.92e−07 | 5.59e−04 |
| Amino acid metabolic process | 12 | 1.34e−06 | 2.57e−03 |
| LT < PR | |||
| Electron transport | 26 | 5.06e−14 | 9.68e−11 |
| Lipid metabolic process | 29 | 6.88e−12 | 1.32e−08 |
| Amine metabolic process | 19 | 2.21e−08 | 4.24e−05 |
| Response to chemical stimulus | 20 | 2.94e−07 | 5.62e−04 |
Fig. 4.
Comparison of log2-transformed protein expression fold changes between our LT versus PR result and Ent versus SA result of Epperson et al. (15). 58 significant proteins in the Epperson et al. (15) results are shown. Red color represents the proteins also significant in our LT versus PR result, and their gene symbols are listed. The dashed line represents y = x; *** represents p < 0.001.
mRNA Data Analysis
We used normalized gene expression values from our previously published Illumina BeadArray experiment (10). For real time PCR analysis, total RNA was prepared from frozen tissues by homogenizing in liquid N2 and TRIzol reagent (Invitrogen) with mortar and pestle. Total RNA quantities were measured by a Nanodrop spectrometer. RNA quality was assessed by an Agilent Bioanalyzer. 500 ng of total RNA from each sample was reverse transcribed using random hexamer and reverse transcriptase (Invitrogen) in a 20-μl reaction. 5 μl of RT product (1:10 diluted) and SYBR Green I Master Mix (Roche Applied Science) in a 20-μl reaction were used in real time PCR on a LightCycler 480 (Roche Applied Science). The 18S rRNA was used as reference in real time PCR. The primer sequences of genes tested in real time PCR are listed in supplemental Table S5. The specificity of PCR was checked by melting curve analysis. The critical threshold (CT) value is the PCR cycle number where the PCR growth curve crosses a defined threshold in the linear range of the reaction. Gene expression values can be calculated from CT by the formula: 235 − CT. In every real time PCR assay, 18S rRNA was used as the control for significant bias of starting materials across samples. The CT values are listed in supplemental Table S6. We again used the PLGEM-StN method to analyze the Illumina BeadArray and real time PCR gene expression data. An FDR-adjusted p value <0.05 was used as the criterion for statistical significance. Log2-transformed mRNA changes were calculated as log2-based ratios of mean expression values in EA, LT, and PR stages for Illumina BeadArray and real time PCR data.
Western Blot Validation
Liver samples of arctic ground squirrels were washed with ice-cold PBS three times to remove blood and then homogenized in lysis buffer (8 m urea, 4% CHAPS, 65 mm DTT, 40 mm Tris with protease inhibitor mixture, 200 mg of wet tissue/ml) with a motor-driven homogenizer (Glas-Col) until no sample pieces were visible. The homogenate was sonicated for a total of 3 min followed by centrifugation at 25,000 × g for 1 h at 4 °C. Proteins in the supernatant were separated by SDS-PAGE and transferred to nitrocellulose (NC) membranes. Each NC membrane was incubated with blotting buffer (5% milk, 150 mm NaCl, 20 mm Tris·HCl (pH 8.0), 0.05% (v/v) Tween 20) with gentle shaking at room temperature for 1 h. The NC membrane was then incubated with primary antibody in fresh blotting buffer at the concentration specified by the manufacturer with gentle shaking at 4 °C for 12 h. Primary antibodies were: goat anti-COPG (C-19) polyclonal antibody (sc-14167, Santa Cruz Biotechnology), mouse anti-PRP8 monoclonal antibody (clone E-5) (sc-55533, Santa Cruz Biotechnology), goat anti-antithrombin III (C-18) polyclonal antibody (sc-32453, Santa Cruz Biotechnology), goat anti-albumin (ALB) (P-20) polyclonal antibody (sc-46293, Santa Cruz Biotechnology), rabbit anti-human ornithine carbamoyltransferase (OTC) precursor polyclonal antibody (LS-C31865, Lifespan Biosciences), goat anti-mitochondrial hydroxymethylglutaryl-CoA synthase (HMGCS) (C-20) polyclonal antibody (sc-32426, Santa Cruz Biotechnology), goat anti-human liver fructose-1,6-bisphosphatase (C-17) polyclonal antibody (sc-32432, Santa Cruz Biotechnology), mouse anti-NDUFS3 (17D95) monoclonal antibody (sc-58393, Santa Cruz Biotechnology), and mouse anti-human factor H (L20/3) monoclonal antibody (sc-47686, Santa Cruz Biotechnology). After incubation with the horseradish peroxidase-conjugated secondary antibody in fresh blotting buffer at 1:10,000 for 1 h, the NC membranes were detected using the Pierce chemiluminescence kit. To ensure the signals were in the linear range of the x-ray film, immunoblots were exposed for variable lengths of time. Images were captured with the luminescent image analyzer LAS-4000 (FUJIFILM). All bands were quantified using Multi Gauge Software (FUJIFILM). Protein expression was measured in arbitrary densitometric units. Significant differences in protein expression among different stages of hibernation were determined by ANOVA and compared with LC-MS/MS data.
RESULTS
Ground Squirrel Protein Database
Three ground squirrel species well studied in hibernation research, the thirteen-lined ground squirrel, golden-mantled ground squirrel, and arctic ground squirrel, are closely related and share high mRNA sequence identities at the nucleotide level (9). We obtained 14,830 unique ground squirrel proteins from genome annotations of the thirteen-lined ground squirrel genome in the Ensembl database. To extract any proteins missed in the Ensembl genome annotation, we mapped all human and mouse RefSeq mRNA sequences onto the thirteen-lined ground squirrel genome, and after removing overlaps with Ensembl-annotated ground squirrel proteins, we obtained 624 additional ground squirrel proteins. We then mapped all available EST sequences of the three ground squirrel species onto the thirteen-lined ground squirrel genome and obtained 473 additional ground squirrel proteins. Taking into account the incompleteness of the thirteen-lined ground squirrel genome, we further clustered the ESTs that had failed to be mapped onto this genome and obtained 3,370 more ground squirrel proteins. We thus created a ground squirrel protein database of in total 19,297 ground squirrel proteins with their sequences and annotations available in supplemental Tables S1 and S2. The detailed scheme of the ground squirrel protein database construction is described under “Experimental Procedures” and shown in supplemental Fig. S1.
Arctic Ground Squirrel Liver Proteome
To survey hibernation-related protein expression profiles in AGS, we conducted a large scale hibernation proteomics study using label-free LC-MS/MS. We studied liver tissues from 12 AGS sampled in three physiological stages: LT after 8–12 days of continuous torpor during winter, EA in euthermic animals early after spontaneous arousal from torpor during winter, and PR in euthermic, posthibernation, and postreproductive animals sampled in May and June, which are used as non-hibernating controls. These three stages were chosen because they showed the most significant differences in across group comparisons in our previous gene expression study (10). Four animals were sampled from each stage as Pavelka et al. (30) suggested that four or five replicates represent a reasonable compromise between cost and accuracy of label-free experiments. To be able to make direct comparisons, we sampled from the same tissues analyzed in the previous experiment. Six fractionations prior to MS/MS analysis were collected to increase the coverage and dynamic range of our data. We chose liver for this initial study because of its important role in energy metabolism and the large amplitude of change in metabolic rate that hibernators display. The spectra generated from LC-MS/MS were searched against our ground squirrel protein database using the SEQUEST program (28) with an FDR <0.05. The complete results are listed in supplemental Table S3.
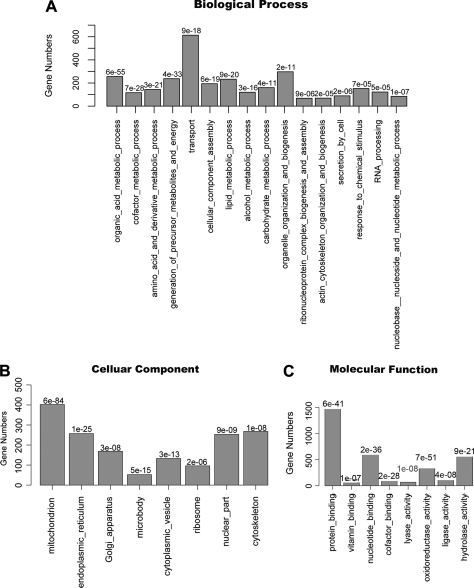
We detected 3,104 unique AGS proteins with at least two unique peptide counts in the ground squirrel protein database. We annotated these proteins by the gene symbols of their homologous human proteins. Enriched gene ontology (GO) categories of ground squirrel liver proteins as indicated by the DAVID program are shown in Fig. 1, A–C (FDR < 10−4) (32, 33). We compared our result with the mouse liver protein expression data obtained by a similar label-free LC-MS/MS method (19). In the mouse liver data set, we were able to annotate 1,591 mouse proteins with homologous human gene symbols, and 1,010 of these overlapped with AGS liver proteins in our study. There is a general consistency of biological functions of the liver proteomes of mouse and ground squirrel.
Fig. 1.
Number of proteins in enriched GO categories in AGS liver proteome related to biological processes (A), cellular components (B), and molecular functions (C) identified by DAVID (FDR < 10−4) are shown. Numbers above the bars are FDRs for enriched GO categories.
Differential Protein Expression during Hibernation
To reliably quantify levels of protein expression across 12 AGS liver samples, we selected 1,209 proteins with spectral counts larger than or equal to 2 in every animal sample in at least one physiological group (EA, LT, or PR). Under these conditions, zero values only consisted of 4.94% of the total data. To correct the bias due to large proteins leading to more spectral matches, spectral counts in each sample were divided by protein length to obtain an SAF. NSAFs were obtained after further dividing SAF values by the sum of SAF values in each sample. Our NSAF values spanned a 3.61 order of magnitude, according to the dynamic range defined by Pavelka et al. (30). NSAF values have been shown to carry statistical properties similar to those of microarray data and can be modeled by PLGEM. Instead of the traditional StN method, we applied the PLGEM-StN method on NSAF values to assess the statistical significance of protein expression differences among EA, LT, and PR stages. PLGEM-StN analysis identified 517 proteins that showed significant differences (FDR-adjusted p value <0.05) in at least one pairwise comparison: 188 proteins between LT and PR, 279 proteins between EA and PR, and 325 proteins between EA and LT. Statistics for the 25 most significant proteins (FDR-adjusted p value <0.001) in at least one pairwise comparison are shown in Table I, and their NSAF values are displayed as a heat map in Fig. 2.
We also analyzed our results with an alternative statistical method, Qspec, which was recently developed to analyze spectral count data in label-free proteomics (20). It is based on a Poisson model with generalized linear mixed effects where the parameters are estimated by a hierarchical Bayesian approach. We used the original spectral counts and protein lengths as direct inputs for Qspec and identified 216 proteins showing significant differences in at least one pairwise comparison: 117 proteins between LT and PR, 98 proteins between EA and PR, and 82 proteins between EA and LT showing significant differences by default parameters. The PLGEM-StN and Qspec results are shown side by side in supplemental Table S4. 107 (91%, LT versus PR), 98 (100%, EA versus PR), and 81 (99%, EA versus LT) Qspec-identified significant proteins were also identified as significant by PLGEM-StN. We chose the PLGEM-StN result for our subsequent analyses.
We then analyzed GO enrichment among six groups of differentially expressed proteins in pairwise comparisons of differences in abundance (EA > LT, EA < LT, EA > PR, EA < PR, LT > PR, and LT < PR) by the DAVID program using an FDR <0.05 as the criteria. No significantly enriched GO categories were identified in the LT > PR group, presumably due to the small number of proteins overexpressed in LT compared with PR. The significantly enriched GO categories in the other five groups are listed in Table II.
Comparison between Protein and mRNA Expression
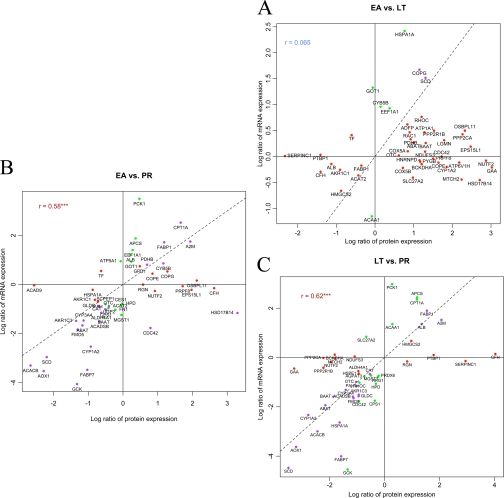
We have previously reported mRNA levels measured in liver tissues from the same animals used in this study, measured using Illumina BeadArray and real time PCR (10). In the present study, we conducted new real time PCR assays on 53 additional genes that showed differential protein expression in our LC-MS/MS results (supplemental Table S5). Combining these with previous data, we obtained real time PCR measurements for 188 genes (supplemental Table S6). Illumina BeadArray and real time PCR data were analyzed by the PLGEM-StN method. There were 104 genes shared between LC-MS/MS and real time PCR data. We compared the log2-transformed protein expression fold changes and mRNA expression fold changes for the genes significantly differentially expressed either at the protein or the mRNA level in pairwise comparisons among EA, LT, and PR stages. The Pearson correlation coefficients between protein and mRNA changes for the same genes are 0.065 between EA versus LT, 0.58 between EA versus PR, and 0.62 between LT versus PR (Fig. 3, A–C). We further computed Pearson correlation coefficients and their statistical significances between protein and mRNA levels across the same set of animals for each gene. There were 30 genes (28.8%) that were significantly correlated (p < 0.05) between two data sets, and all were positively correlated except SLC27A2. We also compared the protein levels measured by LC-MS/MS with the mRNA levels measured by the previous Illumina BeadArray study. There were 85 genes shared between LC-MS/MS and the Illumina BeadArray experiment. The comparisons of log2-transformed protein expression fold changes and mRNA expression fold changes also showed high correlations in the LT versus PR (r = 0.57) and EA versus PR (r = 0.58) comparisons but again low correlation in the EA versus LT (r = 0.098) comparison (supplemental Fig. S2, A–C). There were 17 genes (20%) that were significantly correlated between the two data sets (p < 0.05), and all were positively correlated. Thus, there is overall good correlation between mRNA and protein levels among AGS liver genes.
Fig. 3.
Log2-transformed protein expression fold changes and mRNA expression fold changes for genes significantly differentially expressed either at protein or mRNA level in pairwise comparisons among EA, LT, and PR stages. The mRNA levels were measured using real time PCR. A, EA versus LT. B, EA versus PR. C, LT versus PR. Red color represents significance at the protein level but not at the mRNA level. Green color represents significance at the mRNA level but not at the protein level. Purple color represents significance at both levels. The dashed line represents y = x; *** represents p < 0.001.
Comparison with Previous Proteomics Studies
Epperson et al. (15) identified 68 differentially expressed proteins in the livers of golden-mantled ground squirrels during Ent compared with SA animals. 58 of these are quantified in our study. We plotted the log2-fold changes of these proteins in the LT versus PR comparison in our study versus those in Ent versus SA in the Epperson et al. study (15) (Fig. 4). We observed a significant positive Pearson correlation (r = 0.47, p = 0.0002) between the log2-fold changes. In our analysis, HMGCS2 and liver-specific fatty acid-binding protein (FABP1) were significantly overexpressed in LT versus PR, whereas ACADS, ACADSB, ALDH1A1, IVD, NIT2, and PAH were significantly underexpressed, consistent with the Epperson et al. (15) result. However, AASS, ALDH7A1, CES2, and GSTM1 were significantly underexpressed in our result but overexpressed in the Epperson et al. (15) study. The animals we sampled were in different hibernation stages than in the Epperson et al. (15) study. Ent animals used in the Epperson et al. (15) study were sampled during the entrance into torpor when body temperatures were 16–30 °C, whereas our LT animals were sampled after 8–12 days of continuous torpor with body temperatures near 4 °C. SA animals in their study were non-hibernating euthermic animals but not necessarily in the postreproductive stage. The limited overlap of proteins that were found to be differentially expressed in the two studies could be due to these differences in sample times as well as the different ground squirrel species and technologies used.
Western Blot Validation
We conducted Western blot validation on nine selected ground squirrel proteins that showed significant differential expression in our study using antibodies derived from non-ground squirrel species (see “Experimental Procedures”). Multiple bands were present in the blotting of PRPF8 and CFH indicating nonspecific bindings, and HMGCS2 hybridized at a band with a different molecular weight. These results were not pursued. Six ground squirrel proteins, OTC, FBP1, ALB, COPG, NDUFS3, and SERPINC1, bound to their corresponding antibodies at a unique band with the correct molecular weight. The gel images and quantitative densitometric analysis are shown in Fig. 5, A and B, respectively. In the Western blot analyses, ALB showed significant (p < 0.05, ANOVA) overexpression in both EA and LT versus PR. COPG showed significant overexpression in EA versus PR. OTC showed significant underexpression in both EA and LT versus PR. FBP1 showed significant underexpression in EA versus LT. SERPINC1 showed significant overexpression LT versus PR. Although the overexpression of NDUFS3 in EA versus LT is not considered as significant (p = 0.11), its Western result still correlated well with its NSAF values across the same 12 animals with Pearson correlation r = 0.71. Thus, these semiquantitative Western blot results were in general consistent with our LC-MS/MS results.
Fig. 5.
Western blot gel images (A) and quantitative densitometric analysis (B) for ALB, COPG, OTC, NDUFS3, FBP1, and SERPINC1 protein expression in 14 AGS liver samples. * represents p < 0.05, and ** represents p < 0.01 in ANOVA.
DISCUSSION
Metabolic Shift
A consistent result in molecular studies of hibernation has been the observation of a shift of metabolic fuel use from glucose to fatty acids as animals enter hibernation season. In this study, in support of these previous findings, glucokinase (GCK), catalyzing the irreversible reaction in glycolysis, was significantly underexpressed at the protein level in livers of arctic ground squirrels sampled in EA versus PR, consistent with its mRNA changes. Many proteins involved in fatty acid catabolism were significantly overexpressed during compared with after hibernation. Carnitine palmitoyltransferase 1A (CPT1A) was significantly overexpressed in EA versus PR. FABP1, responsible for the intracellular transport of fatty acids, was significantly overexpressed in both EA and LT versus PR at both mRNA and protein levels. FABP1 was also significantly underexpressed in EA versus LT. However, brain-specific fatty acid binding protein (FABP7) was significantly underexpressed in both EA and LT versus PR at both mRNA and protein levels and showed no significant difference between EA and LT. Several types of fatty acid-binding proteins exist in mammals that have different ligand binding affinities (34). FABP1 is the predominant form in liver and can bind two fatty acids simultaneously, whereas FABP7 is expressed at a lower level in liver and binds only one fatty acid. The opposite changes of FABP1 and FABP7 protein expression may increase the efficiency of fatty acid transport during hibernation and facilitate fatty acid β-oxidation in liver. Acyl-CoA dehydrogenases (ACAD) catalyze the dehydrogenation of acyl-CoA derivatives as the first step of fatty acid β-oxidation. ACADS and ACADSB, acting on short chained and short branched chained fatty acids, respectively, were both significantly underexpressed at the protein level in both EA and LT versus PR, consistent with the Epperson et al. (15) results. ACADM and ACADL, acting on medium and long chained fatty acids, respectively, however, were not differentially expressed in our study. El Kebbaj et al. (35) observed that the activity of ACADM decreased in the livers of hibernating jerboa (Jaculus orientalis) compared with euthermic active animals but increased compared with prehibernating animals. Epperson et al. (15) showed that ACADL was significantly overexpressed in the livers of thirteen-lined ground squirrel in Ent versus SA. Therefore, the effect of protein expression changes of ACAD proteins on fatty acid β-oxidation during hibernation is still unclear and deserves further studies.
On the other hand, proteins involved in fatty acid biosynthesis such as acetyl-CoA carboxylase (ACACA and ACACB) were underexpressed in both EA and LT during the hibernation season versus PR animals sampled after hibernation had ended. These results were consistent with the mRNA level changes of ACACA and ACACB in our microarray experiment. A decrease in enzyme activities involved in fatty acid biosynthesis was also observed by Wang et al. (36) in hibernating compared with summer active golden-mantled ground squirrel. Proteins involved in amino acid metabolism such as branched chain ketoacid dehydrogenase (BCKDHA and BCKDHB) were underexpressed in LT versus PR. BCKDHA was also overexpressed in EA versus LT. Those proteins involved in the urea cycle such as carbamoyl-phosphate synthetase I (CPS1) were underexpressed in EA versus PR. Ornithine aminotransferase (OAT) involved in the pathway that converts arginine and ornithine into glutamate and γ-aminobutyric acid and 4-aminobutyrate aminotransferase (ABAT) responsible for catabolism of γ-aminobutyric acid were both underexpressed in EA and LT compared with PR but overexpressed in EA compared with LT.
As an important enzyme in ketone body formation in the mitochondrion, HMGCS2 was significantly overexpressed at both protein and mRNA levels in LT versus PR and underexpressed in EA versus LT, returning in EA to a level similar to that in PR. The overexpression of HMGCS2 during hibernation was also observed in the Epperson et al. (15) study. During starvation and diabetes, gluconeogenesis and ketone body production are up-regulated. Ketone bodies including acetoacetate and d-β-hydroxybutyrate are exported from liver as an energy source to heart, skeletal muscle, kidney, and brain. Andrews et al. (37) recently demonstrated that d-β-hydroxybutyrate reaches the highest level in serum during torpor in thirteen-lined ground squirrel and is selectively metabolized in the brain and heart. Use of ketone as a metabolic fuel may be protective for the brain and other organs during ischemia and hypoxia (38).
Detoxification and Tissue Protection
Aldehyde oxidase (AOX1) produces hydrogen peroxide and catalyzes the formation of superoxide. AOX1 was significantly underexpressed in both EA and LT versus PR. Superoxide dismutase 2 (SOD2), which binds superoxide, was also significantly underexpressed in LT versus PR, overexpressed in EA versus LT, and in EA returned to a level similar to that in PR. Cytochrome P450 family proteins (CYP1A2, CYP2C70, CYP3A4, and CYP3A5), catalyzing drug metabolism and synthesis of cholesterol, steroids, and other lipids, were all significantly underexpressed in LT versus PR. All except CYP2C70 were also underexpressed in EA versus PR. All except CYP3A4 showed overexpression in EA versus LT. It is appealing to speculate that the drastically reduced basal metabolic rate during torpor may have rendered antioxidant protection and detoxification less necessary. Hibernators may mainly require protection against reactive oxidative species generated during arousal (39) when the metabolic rate rapidly returns to normal. This is consistent with our observation that the levels of antioxidant proteins such as SOD2 in EA indeed returned to levels similar to those in PR.
Regucalcin (RGN), or senescence marker protein-30, was significantly overexpressed in both EA and LT versus PR. RGN shows decreased expression in the livers of aging rats (40). It has been proposed that RGN regulates calcium homeostasis in cells (41), suppresses cell proliferation (42), and promotes cell survival (43). RGN may play an important role for tissue protection against damages in liver during hibernation.
Blood Coagulation
α2-Macroglobulin (A2M) was the first gene shown to be differentially expressed during hibernation at both mRNA and protein levels (6). It is suggested that A2M plays an important role in preventing blood clotting in hibernators, although A2M also plays a major role as a proteinase inhibitor (44). In our study, A2M was also overexpressed in both EA and LT versus PR. In addition to A2M, serpin peptidase inhibitor (SERPINC1) and histidine-rich glycoprotein (HRG) have serine-type endopeptidase inhibitor activity and are involved in blood coagulation. Both SERPINC1 and HRG were overexpressed in LT versus PR, underexpressed in EA versus LT, and in EA returned to a level similar to that in PR.
Torpor-Arousal Cycle
Although the metabolic shift and tissue-protective changes during the hibernation season are largely known, the functional significance of spontaneous arousal episodes during hibernation and how they are regulated are still unclear. Our previous gene expression study showed that a subset of metabolic genes decreased in expression during the transition from late torpor to early arousal. In this proteomics study, we observed this pattern not only in metabolic enzymes such as HMGCS2 but also in transport proteins such as ALB. The expression of these proteins increased in LT versus PR but in EA decreased to a level similar to that in PR. This is unlikely to be a simple temperature-dependent effect as the expression of many other metabolic proteins involved in fatty acid catabolism remained high during arousal. Our previous gene expression study also suggested that the circadian rhythm and cell cycle were suspended during torpor and resumed during arousal. Although we did not identify any key circadian rhythm proteins showing differential expression in liver in this study, we observed that cell division cycle 42 (CDC42), involved in cell proliferation, was significantly overexpressed in EA versus LT and EA versus PR, and protein phosphatase (PPP2CA), which prompts cell growth, was underexpressed in LT versus PR but overexpressed in EA versus LT.
In contrast to the results at the mRNA level in our previous study, we observed a large number of proteins showing differential expression between EA and LT, many of which have not shown changes at the mRNA level. Proteins involved in protein translation and degradation, mRNA processing, and oxidative phosphorylation were significantly overexpressed in EA compared with LT during the torpor-arousal cycle, whereas such changes were not significant at the mRNA level. Seven eukaryotic translation initiation factors, 13 ribosomal proteins, and 12 subunits of the proteasome were significantly overexpressed in EA versus LT. Proteins involved in mRNA processing such as two pre-mRNA processing factors, three heterogeneous nuclear ribonucleoproteins, and six splicing factors were all significantly overexpressed in EA versus LT. Proteins involved in oxidative phosphorylation and the electron transport chain including seven subunits of ATP synthases, two subunits of cytochrome c oxidase, and seven subunits of NADH dehydrogenase were also significantly overexpressed in EA versus LT. However, the mRNA levels of these proteins as measured by real time PCR and Illumina BeadArray did not show any significant changes between EA and LT. The disassociation of change in protein from change in mRNA during the LT to EA transition is also reflected in the poor correlation between log ratios of protein and mRNA expression in Fig. 3A. This may indicate significant post-transcriptional modifications leading to rapid protein turnover, mRNA processing, and oxidative phosphorylation during the torpor-arousal cycle.
Conclusion
The availability of the thirteen-lined ground squirrel genome and various ground squirrel EST resources enabled us to construct a ground squirrel protein database with minimal redundancy. Despite its incompleteness compared with the protein databases in model organisms, searching the spectra generated from label-free shotgun LC-MS/MS in the constructed ground squirrel protein database allowed us to identify thousands of ground squirrel proteins in AGS liver with unprecedented coverage and depth. Considering the noisy nature of LC-MS/MS data, we carefully filtered out the low spectral count data and applied established statistical methods in protein quantification. We identified hundreds of differentially expressed proteins comparing early aroused, late torpid, and non-hibernating animals. Furthermore, we were able to compare global protein expression with global gene expression across the same set of AGS samples. The positive correlations with gene expression data on the same set of animals supported that our protein data were reproducible and that our results reflected the real biological differences among animal groups. Consistent with our previous gene expression study, we observed the significant protein changes that suggested down-regulation of glycolysis, fatty acid synthesis, and the urea cycle but up-regulation of gluconeogenesis, fatty acid catabolism, and ketone body metabolism in the hibernating season. Our proteomics results are also in support of our previous hypotheses put forward in our gene expression study that a subset of metabolic genes decreased their expression during the transition from late torpor to early arousal and that the cell cycle was suspended during torpor and resumed during arousal. For these genes, the changes at the mRNA level have clearly penetrated to the protein levels.
Our finding of Pearson correlations of r ∼ 0.6 (r2 ∼ 0.4) between significant mRNA and protein changes in two pairwise comparisons (LT versus PR and EA versus PR) is compatible with the estimate in other studies that differences in mRNA expression can only explain about 40% of protein differences (45). In contrast, the Pearson correlation of r ∼ 0.1 (r2 ∼ 0.01) in the EA versus LT comparison is strikingly low. This may be largely because expression of proteins involved in protein turnover, mRNA processing, and oxidative phosphorylation is increased at the protein level from LT to EA, but no significant changes at the mRNA level are shown. Global post-transcriptional modifications could be crucial for torpor-arousal cycles during hibernation as transcriptional changes at the mRNA level may not be fast enough to respond in such a short time. This is also consistent with the hypothesis that animals arouse to synthesize new proteins during hibernation (12). RNA-binding proteins bind specific mRNA transcripts and influence their stabilities and translation at the post-transcriptional level. One of them, RBM3, has previously been shown to be significantly overexpressed during hibernation (10). Systematic identification of its binding targets will help to explain some of the discrepancies between the mRNA and protein results. miRNAs might represent another important post-transcriptional regulatory mechanism in hibernation. Combining gene expression, protein expression, and miRNA expression with bioinformatics approaches will enable the identification of regulatory targets of miRNAs and further the understanding of molecular mechanism of hibernation at the systems level.
Supplementary Material
Acknowledgments
We thank Loren Buck and Martin A. Carrasco for helpful discussions, Nathan Stewart for help in arctic ground squirrel EST sequencing, and Mehmet Somel and anonymous referees for suggestions on the manuscript. The Qspec program was kindly provided by Alexey I. Nesvizhskii and Damian Fermin.
Footnotes
* This work was supported by National Basic Research Program of China Grant 2006CB910700, Shanghai Science and Technology Committee Grant 08QA1407500 (to J. Y.), and National Science Foundation Grant 0732755 and United States Army Medical Research and Materiel Command Grant 05178001 (to B. M. B.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1 and S2 and Tables S1–S6.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1 and S2 and Tables S1–S6.
1 The abbreviations used are:
- AGS
- arctic ground squirrel
- EA
- early arousal
- LT
- late torpor
- PR
- postreproduction
- SAF
- spectral abundance factor
- NSAF
- normalized spectral abundance factor
- PLGEM
- power law global error model
- StN
- signal to noise
- FDR
- false discovery rate
- GO
- gene ontology
- EST
- expressed sequence tag
- nr
- non-redundant
- LTQ
- linear trap quadrupole
- DAVID
- Database for Annotation, Visualization, and Integrated Discovery
- Ent
- entrance into torpor
- SA
- summer active
- NC
- nitrocellulose
- ALB
- albumin
- OTC
- ornithine carbamoyltransferase
- HMGCS
- hydroxymethylglutaryl-CoA synthase
- ANOVA
- analysis of variance
- COPG
- coatomer protein complex, subunit γ
- ACAD
- acyl-CoA dehydrogenase
- FABP
- fatty acid-binding protein
- ACAC
- acetyl-CoA carboxylase
- BCKDH
- branched chain ketoacid dehydrogenase
- RGN
- regucalcin
- A2M
- α2-macroglobulin
- miRNA
- microRNA
- GCK
- glucokinase
- ABAT
- 4-aminobutyrate aminotransferase
- HRG
- histidine-rich glycoprotein.
REFERENCES
- 1.Carey H. V., Andrews M. T., Martin S. L. ( 2003) Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol. Rev 83, 1153– 1181 [DOI] [PubMed] [Google Scholar]
- 2.Buck C. L., Barnes B. M. ( 1999) Temperatures of hibernacula and changes in body composition of arctic ground squirrel over winter. J. Mammal 80, 1264– 1276 [Google Scholar]
- 3.Barnes B. M. ( 1989) Freeze avoidance in a mammal: body temperatures below 0 degree C in an arctic hibernator. Science 244, 1593– 1595 [DOI] [PubMed] [Google Scholar]
- 4.Buck C. L., Barnes B. M. ( 2000) Effects of ambient temperature on metabolic rate, respiratory quotient, and torpor in an arctic hibernator. Am. J. Physiol. Regul. Integr. Comp. Physiol 279, R255– R262 [DOI] [PubMed] [Google Scholar]
- 5.Boyer B. B., Barnes B. M. ( 1999) Molecular and metabolic aspects of mammalian hibernation. BioScience 49, 713– 724 [Google Scholar]
- 6.Srere H. K., Wang L. C., Martin S. L. ( 1992) Central role for differential gene expression in mammalian hibernation. Proc. Natl. Acad. Sci. U.S.A 89, 7119– 7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams D. R., Epperson L. E., Li W., Hughes M. A., Taylor R., Rogers J., Martin S. L., Cossins A. R., Gracey A. Y. ( 2005) Seasonally hibernating phenotype assessed through transcript screening. Physiol. Genomics 24, 13– 22 [DOI] [PubMed] [Google Scholar]
- 8.Brauch K. M., Dhruv N. D., Hanse E. A., Andrews M. T. ( 2005) Digital transcriptome analysis indicates adaptive mechanisms in the heart of a hibernating mammal. Physiol. Genomics 23, 227– 234 [DOI] [PubMed] [Google Scholar]
- 9.Yan J., Burman A., Nichols C., Alila L., Showe L. C., Showe M. K., Boyer B. B., Barnes B. M., Marr T. G. ( 2006) Detection of differential gene expression in brown adipose tissue of hibernating arctic ground squirrels with mouse microarrays. Physiol. Genomics 25, 346– 353 [DOI] [PubMed] [Google Scholar]
- 10.Yan J., Barnes B. M., Kohl F., Marr T. G. ( 2008) Modulation of gene expression in hibernating arctic ground squirrels. Physiol. Genomics 32, 170– 181 [DOI] [PubMed] [Google Scholar]
- 11.van Breukelen F., Martin S. L. ( 2002) Reversible depression of transcription during hibernation. J. Comp. Physiol. B 172, 355– 361 [DOI] [PubMed] [Google Scholar]
- 12.Knight J. E., Narus E. N., Martin S. L., Jacobson A., Barnes B. M., Boyer B. B. ( 2000) mRNA stability and polysome loss in hibernating Arctic ground squirrels (Spermophilus parryii). Mol. Cell. Biol 20, 6374– 6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin S. L., Dahl T. A., Epperson L. E. ( 2004) in Life in the Cold: Evolution, Mechanisms, Adaptation, and Application: Twelfth International Hibernation Symposium ( Barnes B. M., Carey H. V. eds.) pp. 199– 208, University of Alaska, Fairbanks, Alaska [Google Scholar]
- 14.Russeth K. P., Higgins L., Andrews M. T. ( 2006) Identification of proteins from non-model organisms using mass spectrometry: application to a hibernating mammal. J. Proteome Res 5, 829– 839 [DOI] [PubMed] [Google Scholar]
- 15.Epperson L. E., Dahl T. A., Martin S. L. ( 2004) Quantitative analysis of liver protein expression during hibernation in the golden-mantled ground squirrel. Mol. Cell. Proteomics 3, 920– 933 [DOI] [PubMed] [Google Scholar]
- 16.Martin S. L., Epperson L. E., Rose J. C., Kurtz C. C., Ané C., Carey H. V. ( 2008) Proteomic analysis of the winter-protected phenotype of hibernating ground squirrel intestine. Am. J. Physiol. Regul. Integr. Comp. Physiol 295, R316– R328 [DOI] [PubMed] [Google Scholar]
- 17.Wolff S., Otto A., Albrecht D., Zeng J. S., Büttner K., Glückmann M., Hecker M., Becher D. ( 2006) Gel-free and gel-based proteomics in bacillus subtilis: a comparative study. Mol. Cell. Proteomics 5, 1183– 1192 [DOI] [PubMed] [Google Scholar]
- 18.Roe M. R., Griffin T. J. ( 2006) Gel-free mass spectrometry-based high throughput proteomics: tools for studying biological response of proteins and proteomes. Proteomics 6, 4678– 4687 [DOI] [PubMed] [Google Scholar]
- 19.Kislinger T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M. S., Gramolini A. O., Morris Q., Hallett M. T., Rossant J., Hughes T. R., Frey B., Emili A. ( 2006) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125, 173– 186 [DOI] [PubMed] [Google Scholar]
- 20.Choi H., Fermin D., Nesvizhskii A. I. ( 2008) Significance analysis of spectral count data in label-free shotgun proteomics. Mol. Cell. Proteomics 7, 2373– 2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian W. J., Liu T., Monroe M. E., Strittmatter E. F., Jacobs J. M., Kangas L. J., Petritis K., Camp D. G., 2nd, Smith R. D. ( 2005) Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J. Proteome Res 4, 53– 62 [DOI] [PubMed] [Google Scholar]
- 22.Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. ( 2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem 389, 1017– 1031 [DOI] [PubMed] [Google Scholar]
- 23.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. ( 1990) Basic local alignment search tool. J. Mol. Biol 215, 403– 410 [DOI] [PubMed] [Google Scholar]
- 24.Florea L., Hartzell G., Zhang Z., Rubin G. M., Miller W. ( 1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res 8, 967– 974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruitt K. D., Maglott D. R. ( 2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res 29, 137– 140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. ( 2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635– 648 [DOI] [PubMed] [Google Scholar]
- 27.Li R. X., Chen H. B., Tu K., Zhao S. L., Zhou H., Li S. J., Dai J., Li Q. R., Nie S., Li Y. X., Jia W. P., Zeng R., Wu J. R. ( 2008) Localized statistical quantification of human serum proteome associated with type 2 diabetes. PLoS ONE 3, e3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eng J. K., McCormack A. L., Yates J. R. ( 1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass. Spectrom 5, 976– 989 [DOI] [PubMed] [Google Scholar]
- 29.Elias J. E., Gygi S. P. ( 2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207– 214 [DOI] [PubMed] [Google Scholar]
- 30.Pavelka N., Fournier M. L., Swanson S. K., Pelizzola M., Ricciardi-Castagnoli P., Florens L., Washburn M. P. ( 2008) Statistical similarities between transcriptomics and quantitative shotgun proteomics data. Mol. Cell. Proteomics 7, 631– 644 [DOI] [PubMed] [Google Scholar]
- 31.Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y., Zhang J. ( 2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang D. W., Sherman B. T., Lempicki R. A. ( 2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44– 57 [DOI] [PubMed] [Google Scholar]
- 33.Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. ( 2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4, P3. [PubMed] [Google Scholar]
- 34.Richieri G. V., Ogata R. T., Zimmerman A. W., Veerkamp J. H., Kleinfeld A. M. ( 2000) Fatty acid binding proteins from different tissues show distinct patterns of fatty acid Interactions. Biochemistry 39, 7197– 7204 [DOI] [PubMed] [Google Scholar]
- 35.El Kebbaj Z., Andreoletti P., Mountassif D., Kabine M., Schohn H., Dauça M., Latruffe N., El Kebbaj M. S., Cherkaoui-Malki M. ( 2009) Differential regulation of peroxisome proliferator-activated receptor (PPAR)-{alpha}1 and truncated PPAR{alpha}2 as an adaptive response to fasting in the control of hepatic peroxisomal fatty acid {beta}-oxidation in the hibernating mammal. Endocrinology 150, 1192– 1201 [DOI] [PubMed] [Google Scholar]
- 36.Wang P., Walter R., Bhat B., Florant G., Coleman R. ( 1997) Seasonal changes in enzymes of lipogenesis and triacylglycerol synthesis in the golden-mantled ground squirrel (Spermophilus lateralis). Comp Biochem. Physiol. B Biochem. Mol. Biol 118, 261– 267 [DOI] [PubMed] [Google Scholar]
- 37.Andrews M. T., Russeth K. P., Drewes L. R., Henry P. G. ( 2009) Adaptive mechanisms regulate preferred utilization of ketones in the heart and brain of a hibernating mammal during arousal from torpor. Am. Physiol. Regul. Integr. Comp. Physiol 296, R383– R393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki M., Suzuki M., Kitamura Y., Mori S., Sato K., Dohi S., Sato T., Matsuura A., Hiraide A. ( 2002) Beta-hydroxybutyrate, a cerebral function improving agent, protects rat brain against ischemic damage caused by permanent and transient focal cerebral ischemia. Jpn. J. Pharmacol 89, 36– 43 [DOI] [PubMed] [Google Scholar]
- 39.Tøien Ø., Drew K. L., Chao M. L., Rice M. E. ( 2001) Ascorbate dynamics and oxygen consumption during arousal from hibernation in Arctic ground squirrels. Am. J. Physiol. Regul. Integr. Comp. Physiol 281, R572– R583 [DOI] [PubMed] [Google Scholar]
- 40.Maruyama N., Ishigami A., Kuramoto M., Handa S., Kubo S., Imasawa T., Seyama K., Shimosawa T., Kasahara Y. ( 2004) Senescence marker protein-30 knockout mouse as an aging model. Ann. N.Y. Acad. Sci 1019, 383– 387 [DOI] [PubMed] [Google Scholar]
- 41.Fujita T., Shirasawa T., Maruyama N. ( 1999) Expression and structure of senescence marker protein-30 (SMP30) and its biological significance. Mech. Ageing Dev 107, 271– 280 [DOI] [PubMed] [Google Scholar]
- 42.Yamaguchi M., Daimon Y. ( 2005) Overexpression of regucalcin suppresses cell proliferation in cloned rat hepatoma H4-II-E cells: involvement of intracellular signaling factors and cell cycle-related genes. J. Cell. Biochem 95, 1169– 1177 [DOI] [PubMed] [Google Scholar]
- 43.Matsuyama S., Kitamura T., Enomoto N., Fujita T., Ishigami A., Handa S., Maruyama N., Zheng D., Ikejima K., Takei Y., Sato N. ( 2004) Senescence marker protein-30 regulates Akt activity and contributes to cell survival in Hep G2 cells. Biochem. Biophys. Res. Commun 321, 386– 390 [DOI] [PubMed] [Google Scholar]
- 44.Sottrup-Jensen L. ( 1989) Alpha-macroglobulins: structure, shape, and mechanism of proteinase complex formation. J. Biol. Chem 264, 11539– 11542 [PubMed] [Google Scholar]
- 45.Tian Q., Stepaniants S. B., Mao M., Weng L., Feetham M. C., Doyle M. J., Yi E. C., Dai H., Thorsson V., Eng J., Goodlett D., Berger J. P., Gunter B., Linseley P. S., Stoughton R. B., Aebersold R., Collins S. J., Hanlon W. A., Hood L. E. ( 2004) Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol. Cell. Proteomics 3, 960– 969 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.