Abstract
Detection of proteins released in the bloodstream from tissues damaged by disease can promote early detection of pathological conditions, differential diagnostics, and follow-up of therapy. Despite these prospects and a plethora of candidate biomarkers, efforts in recent years to establish new protein diagnostic assays have met with limited success. One important limiting factor has been the challenge of detecting proteins present at trace levels in complex bodily fluids. To achieve robust, sensitive, and specific detection, we have developed a microparticle-based solid-phase proximity ligation assay, dependent on simultaneous recognition of target proteins by three antibody molecules for added specificity. After capture on a microparticle, solid-phase pairs of proximity probes are added followed by washes, enabling detection and identification of rare protein molecules in blood while consuming small amounts of sample. We demonstrate that single polyclonal antibody preparations raised against target proteins of interest can be readily used to establish assays where detection depends on target recognition by three individual antibody molecules, recognizing separate epitopes. The assay was compared with state-of-the-art sandwich ELISAs for detection of vascular endothelial growth factor, interleukin-8 and interleukin-6, and it was found to be superior both with regard to dynamic range and minimal numbers of molecules detected. Furthermore, the assays exhibited excellent performance in undiluted plasma and serum as well as in whole blood, producing comparable results for nine different antigens. We thus show that solid-phase proximity ligation assay is suitable for validation of a variety of protein biomarkers over broad dynamic ranges in clinical samples.
Analyses of the plasma proteome, its protein content, their modifications, and interactions, hold great promise to improve detection, classification, and prognostication of pathological conditions such as cancer (1). The attraction of serum or plasma biomarkers lies in their potential to reveal disease processes throughout the body and to guide selection of therapy and follow-up using minimally invasive blood sampling.
This optimism is tempered by the molecular complexity of plasma and the fact that the abundance of known plasma proteins varies over at least 12 orders of magnitude (1), posing great challenges for immunoassays used to investigate the plasma proteome. Thus, new assay formats are needed that can offer improved sensitivity and specificity over a broad dynamic range with good precision to assess new protein biomarkers for analysis in plasma, serum, or whole blood.
The proximity ligation assay (PLA),1 first described by Fredriksson et al. (2) in 2002, is an immunoassay for detection of protein molecules via DNA ligation and amplification, offering high specificity and sensitivity. In PLA, pairs of affinity probes directed against the same target molecule are modified by attaching short single-stranded DNA molecules, creating so-called PLA probes. Upon proximal binding of a pair of PLA probes to a target molecule, the DNA strands are brought in close proximity and allowed to hybridize to a connector oligonucleotide. The DNA strands can then be joined by enzymatic ligation, forming a reporter DNA molecule. This new DNA sequence can be quantified by sensitive and specific nucleic acid detection techniques, such as quantitative real time PCR (q-PCR). The first form of PLA was a homogeneous-phase assay where the antigen was recognized by DNA aptamers in solution before ligation and amplification with real time detection. The assay has also been performed on solid supports by immobilizing antibodies directly on the walls of PCR tubes (2) or by immobilizing biotinylated antibodies on the surface of streptavidin-coated tubes (3). The PLA technique has been implemented for a wide variety of applications, including to visualize proteins in situ (4), to reveal infectious agents (3) and protein-DNA interactions (5), and for biomarker detection in both singleplex (6, 7) and multiplex (9, 10).
Microparticles are commonly used as solid supports in immunoreactions (11, 12) to capture and separate target molecules. Here, we report the development of a generally useful solid-phase PLA protocol (SP-PLA) (Fig. 1) based on paramagnetic microparticles for robust and highly sensitive protein detection in complex biologic material. We used this solid-phase PLA to detect nine different proteins in plasma and serum, demonstrating very low limits of detection and broad working dynamic ranges. In addition, we compared the performance of SP-PLA with that of homogenous-phase PLA and state-of-the-art sandwich ELISAs. The sensitivity of detection of SP-PLA was shown to be clearly superior to that of ELISA. When compared with the homogenous-phase PLA described previously (9) where proteins are detected in solution by a pair of oligonucleotide-labeled antibodies without solid support capture, SP-PLA performed equally in terms of minimal numbers of VEGF molecules detected while exhibiting a broader dynamic range. In addition, we evaluated the performance of SP-PLA for the detection of the cardiac marker protein GDF-15 in clinical samples obtained from two groups of patients, each consisting of 20 individuals, and from 20 healthy controls. The assay exhibited very good interassay correlation and agreement with available clinical data as measured in a conventional ELISA. Furthermore, the levels of the protein were shown to differ significantly between patient and control samples, supporting the value of this protein as a marker of coronary artery disease. Accordingly, SP-PLA is excellently suited for analyses of proteins present at low concentrations in complex biologic material such as undiluted plasma or serum or whole blood.
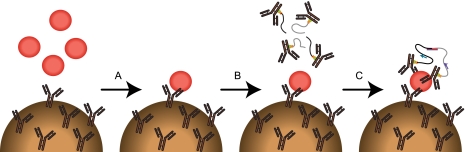
Fig. 1.
Schematic description of SP-PLA method. A, samples are incubated with antibodies preimmobilized on microparticles. B, next, microparticles are washed and incubated with pairs of PLA probes. C, finally, oligonucleotides on PLA probes are ligated upon proximal binding of a common antigen and addition of a connector oligonucleotide. This is followed by amplification and detection of the ligated products by quantitative real time PCR, the primers of which are indicated by arrows.
EXPERIMENTAL PROCEDURES
Proteins and Oligonucleotides
Antigens, affinity-purified polyclonal antibodies, and their biotinylated forms for VEGF, IL-6, IL-8, IL-4, TNFα, PSA, p53, ICAM-1, and GDF-15 were purchased from R&D Systems. The oligonucleotide-streptavidin conjugates SLC1 (5′-CGCATCGCCCTTGGACTACGACTGACGAACCGCTTTGCCTGACTGATCGCTAAATCGTG-3′) and SLC2 (5′-TCGTGTCTAAAGTCCGTTACCTTGATTCCCCTAACCCTCTTGAAAAATTCGGCATCGGTGA-3′) were purchased from Solulink (San Diego, CA).
Forward primer (Biofwd) (5′-CATCGCCCTTGGACTACGA-3′), reverse primer (Biorev) (5′-GGGAATCAAGGTAACGGACTTTAG-3′), and connector oligonucleotide (5′-TACTTAGACACGACACGATTTAGTTT-3′) were obtained from Biomers. A TaqMan probe labeled with the FAM flourophore (5′-FAM-TGACGAACCGCTTTGCCTGA-quencher-3′) was from Applied Biosystems.
Clinical Samples
Plasma samples were collected from 20 patients within 24 h after onset of unstable angina or minor myocardial infarction (group A) or collected upon arrival at the hospital from 20 patients with sudden total thrombotic occlusion of a coronary artery as evidenced by electrocardiographic ST segment elevation, motivating reperfusion therapy (group B). A further 20 samples were collected from age-matched individuals with no signs or symptoms of coronary disease.
Preparation of PLA Probes
Before being combined with the antibodies, a 100 nm concentration of each of the streptavidin-oligonucleotide conjugates was separately mixed with 100 nm streptavidin from Streptomyces avidinii (Sigma-Aldrich) at a 1:4 volume ratio of conjugate to streptavidin followed by incubation at 65 °C for 30 min to dissociate the streptavidin tetramers before these reformed upon return to lower temperatures (supplemental Fig. S1a). This was done to reduce the number of oligonucleotide moieties per streptavidin tetramer. The above treatment allowed us to use 4-fold less streptavidin-oligonucleotide conjugates while maintaining assay performance. Heat-treated oligonucleotide-streptavidin conjugates could be stored at 4 °C for several months without negatively affecting assays.
For preparation of PLA probes and capture antibodies, biotinylated polyclonal antibodies were each divided in three aliquots. One aliquot was immobilized on streptavidin-coated microparticles to serve as capture antibody (see below). The other two were separately mixed with 100 nm heat-treated streptavidin-conjugated oligonucleotides SLC1 or SLC2 at a ratio of 1:1 (supplemental Fig. S1b). The mixtures were incubated for 1 h at room temperature (RT) to allow antibodies to bind streptavidin-oligonucleotide conjugates. Prior to use, the pairs of PLA probes were then combined in the PLA probe mixture at a final concentration of 250 pm for each probe.
Sample Depletion
Prior to analysis of 100% serum or plasma samples, we depleted the samples from potentially interfering substances by preincubation with microparticles coated with nonspecific goat IgG. 10 pmol of biotinylated goat IgG was added per mg of Dynabeads® MyOneTM Streptavidin T1 microparticles (Invitrogen) and incubated for 1 h at RT with constant rotation. The microparticles were then washed twice with washing buffer (1× PBS, 0.05% Tween 20 (Sigma-Aldrich)) and reconstituted with 200 μl of storage buffer (1× PBS, 0.1% purified BSA (New England Biolabs)). For depletion of samples, we used 0.05 μl of the stored microparticles for every μl of sample. Microparticles and samples were incubated for 3–5 h at RT before the microparticles used for depletion were removed.
Antibody Immobilization on Microparticles
To coat microparticles with specific antibodies, 1 mg of Dynabeads, washed twice with 500 μl of washing buffer, was combined with 200 μl of 50 nm (1.5 μg) biotinylated antibody and incubated for 1 h at RT under rotation. Next, the microparticles were washed twice with 500 μl of washing buffer and reconstituted with 200 μl of storage buffer (1× PBS, 0.1% purified BSA (New England Biolabs)). Microparticles on which antibodies have been immobilized can be stored at 4 °C for up to 2 months without noticeable loss of activity.
Solid-phase PLA
For every PLA reaction, we used 1 μl of antibody-coated microparticles, which corresponds to 5 μg of microparticles and 7.5 ng of antibody. The storage buffer was removed, and the microparticles were reconstituted in 5 μl of PLA buffer (1 mm d-biotin (Invitrogen), 0.1% purified BSA (New England Biolabs), 0.05% Tween 20 (Sigma-Aldrich), 100 nm goat IgG (Sigma-Aldrich), 0.1 μg/μl salmon sperm DNA (Invitrogen), 5 mm EDTA, 1× PBS) prior to mixing the microparticles with the sample.
Dilution series of antigens were prepared in PLA buffer, 10 or 100% chicken serum (Invitrogen), 10 or 100% chicken plasma (Rockland), and 10% chicken whole blood (Innovative Research). 10% chicken plasma, serum, and whole blood were prepared by diluting 100% chicken plasma, serum, or whole blood in PLA buffer. Each dilution series contained a negative control where no protein was included to determine background noise.
The assays were initiated by mixing 45 μl of each sample with 5 μl of microparticles in microtiter wells followed by incubation for 1.5 h at RT under rotation. Next, reaction wells with microparticles were allowed to settle on a 96-well plate magnet (PerkinElmer Life Sciences). The particles were washed twice with washing buffer, and 50 μl of PLA probe mixture was added to each well and incubated for 1.5 h at RT while rotating. Thereafter, the microparticles were again collected on a 96-well plate magnet and washed twice with washing buffer. Finally, 50 μl of ligation/PCR mixture (1× PCR buffer (Invitrogen), 2.5 mm MgCl2 (Invitrogen), a 0.1 μm concentration of each primer Biofwd and Biorev, 0.22 μm TaqMan probe, 0.08 mm ATP, 100 nm connector oligonucleotide, 0.2 mm dNTPs (containing dUTP) (Fermentas), 1.5 units of Platinum Taq polymerase (Invitrogen), 0.5 units of T4 DNA ligase (Fermentas), 0.1 unit of uracil-N-glycosylase (Fermentas)) were added to each well followed by detection of PLA DNA ligation products by q-PCR. A dNTP mixture containing dUTP and uracil-N-glycosylase was used to minimize the risk of PCR contamination. The thermocycling program used includes an initial incubation for 2 min at 95 °C followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. All q-PCRs were performed on an Mx-3000 instrument (Stratagene). Solid-phase PLAs using tubes as solid support were performed as previously described by Ericsson et al. (13).
Sandwich ELISA
The R&D Quantikine® ELISA kits for VEGF, IL-8, GDF-15, and IL-6 were used according to the manufacturer's specifications with the exception that for VEGF, IL-8, and IL-6 ELISAs the proteins were spiked in 10% calibrator diluents to ensure a fair comparison between SP-PLA and ELISA. Information regarding limits of detection (LODs) of ELISAs for ICAM-1, GDF-15, TNFα, IL-4, and PSA was from R&D Quantikine ELISA kit; information for p53 was from the Invitrogen immunoassay kit for detection of total p53.
Data Analysis
The q-PCR data were analyzed with MxPro software (Stratagene), and the recorded Ct values were exported and further analyzed with Microsoft Excel software. The statistical significance of the difference in levels of the GDF-15 protein between patient group B and healthy controls determined by SP-PLA was calculated using a two-sample Wilcoxon rank sum test in R.
Definitions of Terms
The LOD was defined as the concentration of protein corresponding to CtLOD = CtN − 2 × SN where CtN is the average Ct obtained for the background noise and SN is the standard deviation of this value. LOD values for the detection of proteins in plasma and serum are calculated in molar concentrations of the proteins in 50 μl of 10% plasma or serum.
Coefficient of variation percent (CV%) was calculated for each data point using the following formula: CV%a = Sa/Ma where Sa is the standard deviation and Ma is the average number of starting PCR amplicons for point a. Ct values were converted to numbers of starting PCR amplicons using the equation Ma = 2(38 − Cta) where Cta is the Ct of point a. This formula assumes that 38 is the number of PCR cycles required to bring a single amplicon to the threshold for fluorescence detection. Recovery was calculated using the following formula: Recovery (%) = Measured concentration (molar)/Spiked concentration (molar).
RESULTS
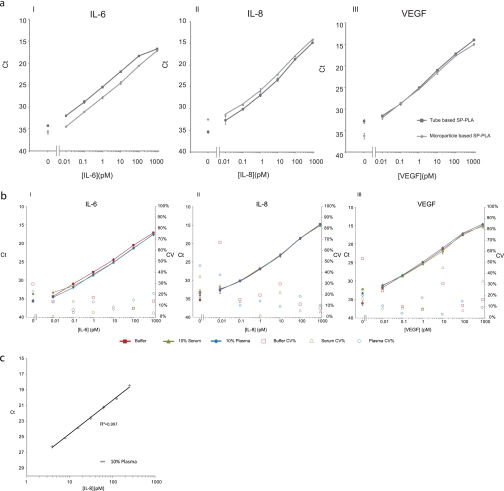
We compared the microparticle-based protocol with the use of polycarbonate tubes as described by Ericsson et al. (13) for detection of VEGF, IL-8, and IL-6. The results demonstrated that the two protocols performed equally for detecting proteins in buffer (Fig. 2a). The LOD of both assays was in the low fm range for all three analytes, and they exhibited a broad dynamic range of up to 6 orders of magnitude. The microparticle-based assay used significantly less antibody, however, and assays could be completed in 3 h compared with 6 h when tubes were used as a solid support, probably as a result of enhanced binding kinetics to microparticles.
Fig. 2.
Performance of microparticle-based SP-PLA and comparison with tube-based assay. a, comparison of microparticle-based (squares) and tube-based assay (diamonds) for detection of IL-6 (I), IL-8 (II), and VEGF (III) in buffer. All measurements were performed at least in triplicates. Error bars indicate SD. b, SP-PLA performance for detection of IL-6 (I), IL-8 (II), and VEGF (III) in buffer (squares), 10% serum (triangles), and 10% plasma (diamonds) with corresponding CV% values for every data point for buffer (open squares), 10% serum (open triangles), and 10% plasma (open diamonds). c, 2-fold dilutions of IL-8 in 10% plasma to estimate precision. The y axes display threshold cycle values of real time PCR assays; the x axes display concentrations of the investigated proteins in pm.
To investigate assay performance in more complex matrices, we used the microparticle-based SP-PLA for detection of the three analytes, VEGF, IL-8, and IL-6, in dilution series prepared either in 10% chicken plasma or in 10% chicken serum (Fig. 2b). Chicken plasma and serum were used to represent the complex composition of human plasma or serum while ensuring the absence of the investigated human target proteins. The assay performance in plasma and serum was comparable to that in buffer, demonstrating that any interfering substances in such biological materials could be avoided by target capture on the particle supports. Assay characteristics in buffer, 10% plasma, and 10% serum such as LOD, linear range, interassay variation, recovery, and R2 are summarized in Table I. For all three analytes, the assays exhibited very low limits of detection and broad dynamic ranges, suitable to study protein concentrations that may vary widely among different samples as is often the case in pathological conditions. Furthermore, we assessed the precision of the assay by performing 2-fold dilutions of IL-8 in 10% plasma, ranging from 3.9 to 250 pm. The results shown in Fig. 2c demonstrate that the assay is readily able to discern concentration differences of the investigated protein of less than 2-fold.
Table I. Summary of SP-PLA performance characteristics for detection of VEGF, IL-8, and IL-6.
The measurements were performed in buffer, 10% chicken serum, or chicken plasma.
| VEGF |
IL-8 |
IL-6 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | Serum | Plasma | Buffer | Serum | Plasma | Buffer | Serum | Plasma | |
| LOD (fm) | 1.9 | 10.7 | 5.2 | 2.8 | 20 | 15.6 | 8.9 | 20 | 11 |
| Linear range | 106 | 105 | 105 | 106 | 106 | 106 | 106 | 106 | 106 |
| Intra-assay variation (CV%) | 21.3 | 7.8 | 10.6 | 24.8 | 16.1 | 21 | 16 | 7.5 | 13.8 |
| R2 | 0.998 | 0.986 | 0.996 | 0.988 | 0.990 | 0.989 | 0.999 | 0.984 | 0.997 |
| Recovery (%) | 105.6 | 100.1 | 94.8 | 99.4 | 107.6 | 107.1 | 100.7 | 113 | 102.4 |
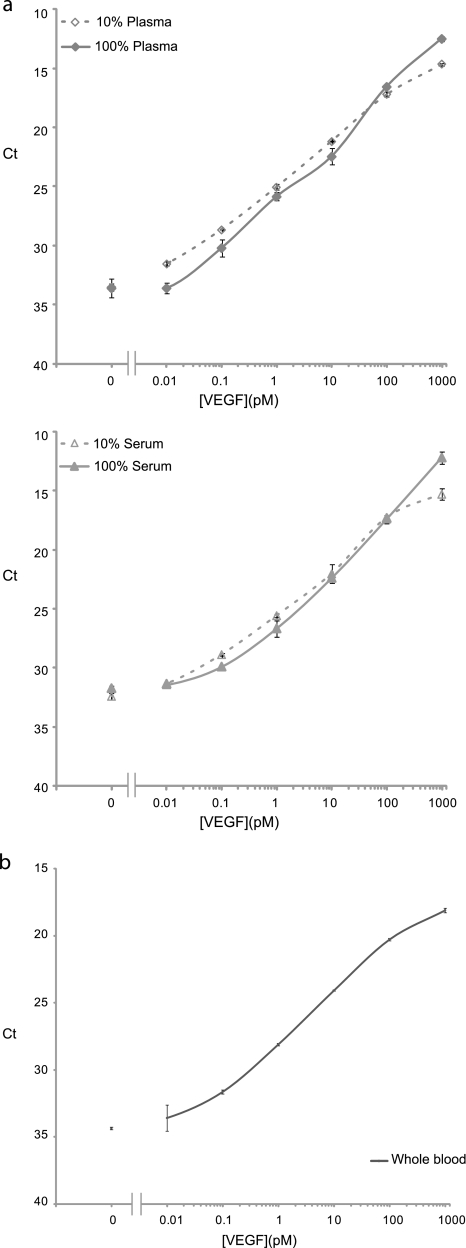
In addition, we validated the assay for detection of VEGF in undiluted (100%) chicken plasma and serum. Here, we used an initial depletion step to remove unknown interfering agents that had been shown to give rise to increased background in bodily fluids but without removing the antigens of interest from the sample (data not shown). Prior to analysis, samples were incubated with microparticles coated with unspecific IgG from goat. The assay performance in 100% plasma and serum was very similar to that in 10% dilutions with LODs of 25 and 28 fm for plasma and serum, respectively (Fig. 3a), and excellent recovery for both plasma and serum. It is also of interest to ascertain whether the assays can perform well in whole blood, such as when there is a need for rapid procedures with minimal sample preparation, for example in point-of-care applications. To investigate the effect of whole blood, we measured VEGF in 10% whole chicken blood. The volume of blood used was 5 μl, ∼– of that obtained from a finger prick. We found that the assay performance was not affected by the presence of whole blood, and VEGF was detected with an LOD of 10 fm (Fig. 3b).
Fig. 3.
Performance of SP-PLA in complex biological samples. a, comparison of performance of SP-PLA in 100% serum (open triangles) and plasma (open diamonds) and 10% serum (triangles) and plasma (diamonds) for the detection of VEGF. b, detection of VEGF by SP-PLA in whole blood. Error bars indicate SD.
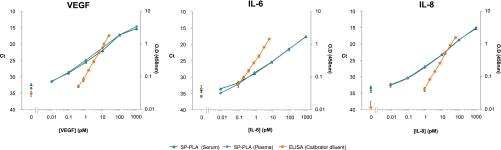
We furthermore validated the technology by comparing it with state-of-the-art ELISAs. The results are summarized in Fig. 4. SP-PLA was capable of detecting ∼100 times lower concentrations of each of the three investigated antigens compared with sandwich ELISAs and with a broader dynamic range by up to 2 further orders of magnitude. Moreover, SP-PLAs only required 5 μl, whereas the corresponding ELISAs consumed up to 100 μl of sample.
Fig. 4.
Comparison of SP-PLA with sandwich ELISA for detection of VEGF, IL-6, and IL-8. For SP-PLA, the proteins were spiked in 10% serum (triangles) and 10% plasma (diamonds), and for ELISA, the proteins were spiked in the 10% calibrator diluent for serum and plasma provided by the manufacturer (circles). The primary y axes display threshold cycle values of real time PCR for SP-PLA; the secondary y axes display OD measured at 450 nm for ELISA. x axes display concentration of the analyzed proteins in the 10-fold dilutions of each protein in the 50 μl SP-PLA reactions and the 200 μl ELISAs. All measurements were performed at least in triplicates. Error bars indicate SD.
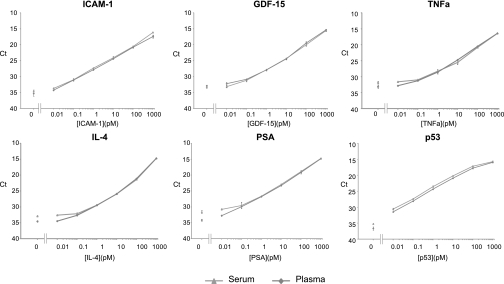
We demonstrated the assay performance for detection of an additional six proteins in serum and plasma: p53, ICAM-1, GDF-15, PSA, TNFα, and IL-4. Results confirmed that the assays perform equally well for a number of protein targets using polyclonal antibodies raised against the whole proteins (Fig. 5 and Table II). SP-PLA exhibited an LOD in the fm range for all nine different proteins, and the dynamic ranges of all assays extended over 5 or 6 orders of magnitude.
Fig. 5.
Detection of ICAM-1, GDF-15, TNFα, IL-4, PSA, and p53 with SP-PLA in 10% serum (triangles) and 10% plasma (diamonds). y axes display threshold cycle values of real time PCR assays; x axes display concentration of each antigen in pm.
Table II. Comparison of LOD between SP-PLA and ELISA for detection of ICAM-1, GDF-15, TNFα, IL-4, PSA, and p53.
| LOD |
|||
|---|---|---|---|
| ELISA | SP-PLA |
||
| Serum | Plasma | ||
| pm | |||
| ICAM-1 | 1.993421 | 0.014 | 0.02 |
| GDF-15 | 0.071786 | 0.05 | 0.06 |
| TNFα | 0.091429 | 0.16 | 0.08 |
| IL-4 | 0.714286 | 0.16 | 0.08 |
| PSA | 1.071429 | 0.07 | 0.009 |
| p53 | 1.162791 | 0.005 | 0.0003 |
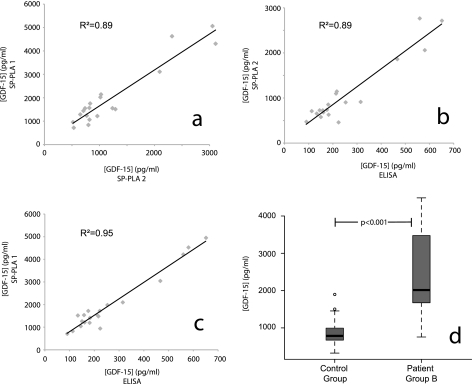
To investigate the SP-PLA test in a practical application, we evaluated the performance of the assay for detection of GDF-15 in human plasma samples obtained from 40 patients belonging to two different patient groups (A and B) and from 20 healthy controls. GDF-15 has emerged as a blood biomarker for recurrence of myocardial infraction and increased mortality in patients with non-ST elevation acute coronary syndrome (14). We first estimated the levels of GDF-15 in the 20 patients belonging to group A on two separate occasions (assay 1 and assay 2). The intra-assay CV% for the two experiments was 15–16%. The linear correlation coefficient (R2) between the two measurements of GDF-15 for the 20 samples of patient group A was 0.89 (Fig. 6a), reflecting a good agreement between the two data sets. The interassay CV% for the two experiments was found to be 35%. To assess the validity of the results generated by SP-PLA, we compared the results with those obtained by ELISA for patient group A. The linear correlation coefficient (R2) between the measured values in the two types of assays was between 0.89 and 0.95 (Fig. 6, b and c), verifying the validity of the values measured by SP-PLA. We then used SP-PLA to assess the concentration of GDF-15 in 20 samples from patient group B and 20 healthy controls. The GDF-15 was found to be significantly elevated (p < 0.001, two-sample Wilcoxon rank sum test) in patient group B compared with the healthy control group as shown in Fig. 6d.
Fig. 6.
Measurements of GDF-15 in patient samples as analyzed in two independent SP-PLA tests and in one sandwich ELISA. a, GDF-15 was measured by SP-PLA in 20 human plasma samples obtained from patient group A on two separate occasions (SP-PLA1 and SP-PLA2). Values of GDF-15 in pg/ml obtained in assay 1 are plotted on the y axis, whereas values obtained from assay 2 are plotted on the x axis. b and c, scatter plots showing the correlation between ELISA and each of the two SP-PLA tests (SP-PLA1 and SP-PLA2) for data from the same 20 human plasma samples. Values of GDF-15 in pg/ml obtained from SP-PLA are plotted on the y axis, whereas ELISA values are plotted on the x axis. d, box plots showing differences in levels of GDF-15 in patients belonging to patient group B and in healthy controls. The p value was calculated using a two-sample Wilcoxon rank sum test.
DISCUSSION
A rapidly increasing number of protein molecules are being identified as potential biomarkers for human disease by investigating affected tissues using for example immunohistochemistry, mass spectrometry, or other proteomic approaches or on the basis of the expression of the corresponding transcripts. This motivates the search for these same proteins in blood as possible reporters of disease processes that may cause their release, but sensitivity of detection often is found to be insufficient (15). Therefore, methods are needed that offer high sensitivity and high specificity as required when dealing with complex biologic material (16, 17). The aim of this study was to establish a general and straightforward method for detecting proteins in complex biological samples at levels too low to be detected by standard means. To do so, we combined the advantages of the PLA technique with those of sandwich immunoassays (11, 16, 17) to create sensitive and specific immunoassays. We report herein the development of SP-PLA, a microparticle-based PLA, in which the solid support allows concentration of target molecules and removal of excess probes as well as potentially interfering agents from complex biological samples followed by amplification of ligated reporter DNA strands for sensitive protein detection. The two different solid supports used to perform SP-PLA performed equally well in singleplex protein detection performed in buffer. We have preliminary data suggesting that the microparticle-based protocol presents advantages in multiplex protein analysis where different antibodies are immobilized on separate microparticles.2 Furthermore, the microparticle-based protocol can be used to analyze larger sample volumes if necessary.
SP-PLA consistently exhibited a very low limit of detection in the low fm range, a broad linear dynamic range of up to 6 orders of magnitude, and CV% between 7 and 25%. In addition, only 5-μl sample aliquots were used in the standard protocol, a feature that is of great importance in examining valuable and limited specimens such as biobank material. The assays performed equally well for a variety of target proteins, and we observed a very low LOD for nine different analytes, ranging from small cytokine molecules to larger proteins.
Compared with sandwich ELISA, SP-PLA offered around 100-fold greater sensitivity. Similar levels of sensitivity have been reported for immuno-PCR assays in which a single oligonucleotide-antibody conjugate is used for detection of target molecules captured via another antibody (18). We expect that the requirement for recognition of three epitopes in SP-PLA can serve to further enhance the detection specificity by eliminating background signals due to nonspecifically bound detectable reagents or cross-reactive detection of related target molecules. The requirement for multiple binding also facilitates analysis of more complex target structures such as proteins with posttranslational modification and binary or ternary protein interactions. Moreover, the requirement for specific ligation of the attached DNA strands has the important advantage of allowing detection reactions to be restricted to cognate sets of antibodies among large numbers of reagents added to an assay. Thereby, the assays can avoid the rapidly increasing risks of cross-reactions between non-cognate antibody pairs observed when regular sandwich assays are multiplexed (19). Accordingly, the PLA approach is promising for assays where large numbers of analytes are simultaneously interrogated in one sample. The SP-PLA was also compared with published data for homogenous-phase PLA by Fredriksson et al. (9). Although the assays described in this publication were performed in multiplex, their performance is similar to that of separate singleplex assays. When compared with homogenous-phase dual (9) or triple (8) recognition PLA, SP-PLA performs equally well for detection of VEGF. Homogenous-phase PLA presents several attractive features. The assay uses only 1-μl samples compared with 5 μl of sample for SP-PLA, making it highly suitable in applications where sample amounts are limited; even lower amounts of antibodies are used compared with SP-PLA; and the assay is performed without washing steps, thereby simplifying the procedure and avoiding sources of variability. The distinct advantages of SP-PLA lies in its suitability for analysis of complex biologic material such as undiluted plasma or serum and even whole blood, which could be inhibitory in the case of homogeneous PLA. In addition, SP-PLA has proven robust and easy to set up, and new assays can be established well within a working day starting from a suitable biotinylated polyclonal antibody preparation. The requirement for target recognition by three antibody molecules can be conveniently achieved with a single antiserum raised against the whole target protein to ensure specific binding of several epitopes on the target protein. Alternatively, assays can be performed using proper combinations of sets of three monoclonal antibodies as described previously (8). We have so far established SP-PLA tests for detection of over 46 different proteins with excellent results.
SP-PLA was successfully used to estimate levels of a marker for coronary disease in clinical plasma samples. The assay exhibited very good reproducibility, low intra-assay CV%, and a satisfactory interassay CV%. Furthermore, assays performed on the same samples on different occasions correlated very well with one another and with levels measured for the same samples using ELISA. Nevertheless, further development will be required for SP-PLA to be routinely applied in clinical contexts.
There is room for further improvement of the assay as presented here, for example to decrease assay time while retaining sensitivity and to allow the use of considerably larger volumes of samples when target molecules must be detected at even lower concentrations. The use of internal controls will serve to improve reproducibility. Furthermore, as discussed herein, the SP-PLA approach is promising for high multiplex immunoassays where large sets of target molecules are simultaneously interrogated in minimal sample aliquots for diagnostic and research purposes.
In conclusion, we have shown that SP-PLA is a robust and highly sensitive protein detection technique. It is suitable for clinical applications where proteins need to be quantified at low concentrations and for analyzing biobanked samples in minute sample aliquots. In addition, the SP-PLA mechanism exhibits excellent potential for development of highly multiplexed immunoassays.
Supplementary Material
Acknowledgments
We thank Katerina Pardali and Mikaela Friedman for critical comments on the manuscript. We also thank Lars Wallentin of the Uppsala University Hospital for advice on this study and for kindly providing clinical samples.
Footnotes
* This work was supported by funding from the Knut and Alice Wallenberg Foundation as well as by the Swedish Research Council, Vinnova/Uppsala Bio, Uppsala Berzelii Centre and the European Community's 6th and 7th Framework Programs.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Fig. S1.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Fig. S1.
2 S. Darmanis and R. Y. Nong, unpublished data.
1 The abbreviations used are:
- PLA
- proximity ligation assay
- SP
- solid-phase
- IL
- interleukin
- PSA
- prostate-specific antigen
- VEGF
- vascular endothelial growth factor
- TNFα
- tumor necrosis factor α
- ICAM-1
- intercellular adhesion molecule-1
- GDF-15
- growth differentiation factor-15
- RT
- room temperature
- q-PCR
- quantitative real time PCR
- LOD
- limit of detection
- Ct
- cycle threshold
- CV
- coefficient of variation.
REFERENCES
- 1.Anderson N. L., Anderson N. G. ( 2002) The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845– 867 [DOI] [PubMed] [Google Scholar]
- 2.Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gústafsdóttir S. M., Ostman A., Landegren U. ( 2002) Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol 20, 473– 477 [DOI] [PubMed] [Google Scholar]
- 3.Gustafsdottir S. M., Nordengrahn A., Fredriksson S., Wallgren P., Rivera E., Schallmeiner E., Merza M., Landegren U. ( 2006) Detection of individual microbial pathogens by proximity ligation. Clin. Chem 52, 1152– 1160 [DOI] [PubMed] [Google Scholar]
- 4.Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. ( 2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995– 1000 [DOI] [PubMed] [Google Scholar]
- 5.Gustafsdottir S. M., Schlingemann J., Rada-Iglesias A., Schallmeiner E., Kamali-Moghaddam M., Wadelius C., Landegren U. ( 2007) In vitro analysis of DNA-protein interactions by proximity ligation. Proc. Natl. Acad. Sci. U.S.A 104, 3067– 3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu L., Koistinen H., Landegren U., Stenman U. H. ( 2009) Proximity ligation measurement of the complex between prostate specific antigen and alpha1-protease inhibitor. Clin. Chem 55, 1665– 1671 [DOI] [PubMed] [Google Scholar]
- 7.Gullberg M., Gústafsdóttir S. M., Schallmeiner E., Jarvius J., Bjarnegård M., Betsholtz C., Landegren U., Fredriksson S. ( 2004) Cytokine detection by antibody-based proximity ligation. Proc. Natl. Acad. Sci. U.S.A 101, 8420– 8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schallmeiner E., Oksanen E., Ericsson O., Spångberg L., Eriksson S., Stenman U. H., Pettersson K., Landegren U. ( 2007) Sensitive protein detection via triple-binder proximity ligation assays. Nat. Methods 4, 135– 137 [DOI] [PubMed] [Google Scholar]
- 9.Fredriksson S., Dixon W., Ji H., Koong A. C., Mindrinos M., Davis R. W. ( 2007) Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat. Methods 4, 327– 329 [DOI] [PubMed] [Google Scholar]
- 10.Fredriksson S., Horecka J., Brustugun O. T., Schlingemann J., Koong A. C., Tibshirani R., Davis R. W. ( 2008) Multiplexed proximity ligation assays to profile putative plasma biomarkers relevant to pancreatic and ovarian cancer. Clin. Chem 54, 582– 589 [DOI] [PubMed] [Google Scholar]
- 11.Meza M. B. ( 2000) Bead-based HTS applications in drug discovery. Drug Discov. Today 5, 38 [Google Scholar]
- 12.Verpoorte E. ( 2003) Beads and chips: new recipes for analysis. Lab Chip 3, 60N– 68N [DOI] [PubMed] [Google Scholar]
- 13.Ericsson O., Jarvius J., Schallmeiner E., Howell M., Nong R. Y., Reuter H., Hahn M., Stenberg J., Nilsson M., Landegren U. ( 2008) A dual-tag microarray platform for high-performance nucleic acid and protein analyses. Nucleic Acids Res 36, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggers K. M., Kempf T., Allhoff T., Lindahl B., Wallentin L., Wollert K. C. ( 2008) Growth-differentiation factor-15 for early risk stratification in patients with acute chest pain. Eur. Heart J 29, 2327– 2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanash S. M., Pitteri S. J., Faca V. M. ( 2008) Mining the plasma proteome for cancer biomarkers. Nature 452, 571– 579 [DOI] [PubMed] [Google Scholar]
- 16.de Jager W., Rijkers G. T. ( 2006) Solid-phase and bead-based cytokine immunoassay: a comparison. Methods 38, 294– 303 [DOI] [PubMed] [Google Scholar]
- 17.Elshal M. F., McCoy J. P. ( 2006) Multiplex bead array assays: performance evaluation and comparison of sensitivity to ELISA. Methods 38, 317– 323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sims P. W., Vasser M., Wong W. L., Williams P. M., Meng Y. G. ( 2000) Immunopolymerase chain reaction using real-time polymerase chain reaction for detection. Anal. Biochem 281, 230– 232 [DOI] [PubMed] [Google Scholar]
- 19.Kingsmore S. F. ( 2006) Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat. Rev. Drug Discov 5, 310– 320 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.