Abstract
Compartmentalization of biological processes and the associated cellular components is crucial for cell function. Typically, the location of a component is revealed through a co-localization and/or co-purification with an organelle marker. Therefore, the identification of reliable markers is critical for a thorough understanding of cellular function and dysfunction. We fractionated macrophage-like RAW264.7 cells, both in the resting and endotoxin-activated states, into six fractions representing the major organelles/compartments: nuclei, mitochondria, cytoplasm, endoplasmic reticulum, and plasma membrane as well as an additional dense microsomal fraction. The identity of the first five of these fractions was confirmed via the distribution of conventional enzymatic markers. Through a quantitative liquid chromatography/mass spectrometry-based proteomics analysis of the fractions, we identified 50-member ensembles of marker proteins (“marker ensembles”) specific for each of the corresponding organelles/compartments. Our analysis attributed 206 of the 250 marker proteins (∼82%) to organelles that are consistent with the location annotations in the public domain (obtained using DAVID 2008, EntrezGene, Swiss-Prot, and references therein). Moreover, we were able to correct locations for a subset of the remaining proteins, thus proving the superior power of analysis using multiple organelles as compared with an analysis using one specific organelle. The marker ensembles were used to calculate the organelle composition of the six above mentioned subcellular fractions. Knowledge of the precise composition of these fractions can be used to calculate the levels of metabolites in the pure organelles. As a proof of principle, we applied these calculations to known mitochondria-specific lipids (cardiolipins and ubiquinones) and demonstrated their exclusive mitochondrial location. We speculate that the organelle-specific protein ensembles may be used to systematically redefine originally morphologically defined organelles as biochemical entities.
One of the basic concepts of cell biology is compartmentalization of the cellular processes within subcellular structures, termed organelles. Organelles were originally identified in the 19th century as the morphological entities that are still reflected in their names (e.g. “nucleus” from the Latin “little nut,” “mitochondria” from the Greek “thread” + “grain,” or “reticulum” from the Latin “little net”). Later, the progress of biochemistry made it possible to assign to the various organelles their specific biological functions. Thus, detailed information about the location of biochemical reactions became crucial for the understanding of their roles in cell function or dysfunction. Current technology allows the location of a cell component (a protein or a metabolite) to be linked directly to a morphologically defined organelle (or even a suborganellar compartment) by using electron microscopy. However, more typically, the location of a component is determined on the basis of its co-localization with a known marker for the organelle or subcellular compartment. This co-localization can be either visualized microscopically (imaging approach) to preserve some degree of morphological information or determined through co-purification of the component and the marker in a subcellular fractionation (biochemical approach).
For both the imaging and the biochemical approaches, optimal organelle markers are of the utmost importance. Conventional markers include proteins, DNA (for nucleus), and even physical/chemical parameters (electric potential for mitochondria and acidic pH for lysosomes). Protein markers are assayed using either an interaction with specific antibodies or their enzymatic activities. Unfortunately, the former is typically non-quantitative, whereas the latter, although semiquantitative, is subject to interference from multiple parameters of the environment as well as substrate and product sharing with non-marker proteins. For a biochemical approach, tightness of the anchoring of a marker to the corresponding organelle is also an issue. Moreover, an inherent problem is that most proteins are located in several organelles/compartments, which may result in false localization conclusions.
Our goal was to identify specific, reliable, and universal protein markers for major subcellular organelles/compartments. The following principles were chosen as the basis for our approach. First, the search had to be conducted without a preconceived notion of the nature of the markers (e.g. we did not expect to necessarily confirm conventional markers as optimal). Second, the search had to be conducted in all major organelles/compartments simultaneously. Third, the aim was to identify relatively large panels (ensembles) of markers as opposed to the best single marker. The last two principles allowed us to address the problem of multiple locations of potential marker proteins. Some of them can be eliminated as markers; for others, the impact of multiple locations on further analysis can be negated by averaging of the data for large numbers of proteins (derivation of marker ensembles).
To meet these goals, we performed a complete “quantitative” proteomics analysis of all major subcellular fractions in a single cell type. Numerous reports have focused on the proteomes of specific organelles or interrelated sets of organelles in various cell types (for reviews, see Refs. 1 and 2). However, a need for an integral systematic study in a single cell type has been evident for some time (2), and the present study is the first step aimed at addressing this need.
The marker ensembles that we identified from the proteome data were used to quantify the composition of the subcellular fractions. It is becoming appreciated that a physical association of various organelles makes it next to impossible to completely separate the organelles and obtain pure fractions acceptable for detailed proteomics analysis (e.g. see Ref. 3). Therefore, correlative approaches such as protein correlation profiling (1, 3, 4) and localization of organelle proteins by isotope tagging (5, 6) have been suggested to address this problem. These approaches allowed the assignment of protein locations based on co-localization with known markers in a density gradient (1, 4–6) or in multiple fractions (7). We took this approach a step further and derived a quantitative composition of the fractions based on the distribution of the marker ensembles. Furthermore, this enabled us to calculate levels of various components (lipids and proteins) in pure organelles from experimental data obtained with less than pure fractions.
The choice of a particular cell type for this study was somewhat arbitrary, and the resulting marker ensembles were optimal for the cell type for which they were generated; of course, they may have to be adjusted to be adapted for other cell types. We chose macrophage cells partly because this study was an integral part of a larger subcellular lipidomics/proteomics study under the auspices of the Lipid Metabolites and Pathways Strategy (LIPID MAPS Consortium). The macrophage plays a central role in inflammation and innate and adaptive immunity. The macrophage detects and attacks pathogens and orchestrates a host response by sending signals to other cells and tissues; in this process, the macrophage itself transits from a resting to an activated state. These two states differ vastly in function, morphology, and underlying protein expression profiles, and therefore, we aimed to identify marker ensembles that would be invariant with regard to the activation process.
In the present study, the activation paradigm was treatment with Kdo21-lipid A. This defined, nearly homogeneous reagent is a form of lipopolysaccharide endotoxin that has all the essential biological properties of lipopolysaccharide (8).
EXPERIMENTAL PROCEDURES
Materials
RAW264.7 cells were from ATCC (catalog number TIB-71). Dulbecco's modified Eagle's medium (catalog number 10-013) and Dulbecco's PBS (catalog number 21-031-CV) were from Mediatech. Fetal calf serum with low endotoxin content was from Hyclone (SH30071.03 ANG19242). Kdo2-lipid A was obtained from Avanti Polar Lipids. Iodixanol (OptiPrepTM from Axis-Shield) was obtained through Sigma-Aldrich. The Quant-iTTM DNA assay kit and Vybrant® cytotoxicity assay kit were from Invitrogen. Tris(2-carboxyethyl)phosphine and iodoacetamide were from Fisher (catalog numbers AC36383 and AC12227, respectively). Trypsin was from Roche Applied Science (catalog number 03 708 969 001). Potassium cyanide, EGTA, and magnesium chloride from Fluka were obtained through Sigma. Solvents were chromatography grade and purchased from OmniSolv. All other reagents/kits were from Sigma-Aldrich. All aqueous solutions were prepared using distilled deionized water (catalog number 25-055-CV) from Mediatech. Isolation media were prepared K+- and Na+-free; pH was adjusted by Tris base (Trizma).
Tissue Culture
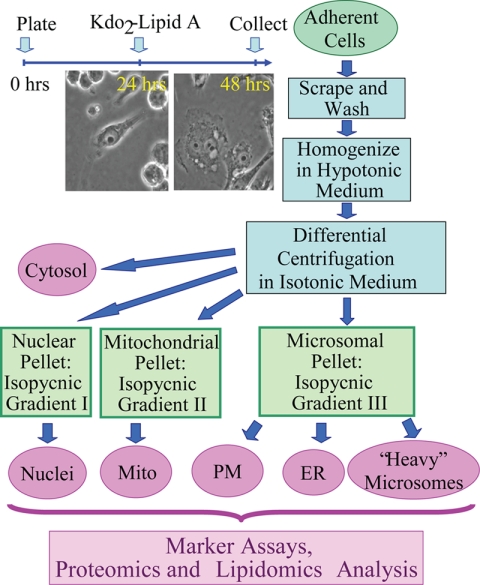
Three separate cultures of both resting and activated macrophages were generated for subsequent proteomics and lipidomics analyses. The replicates were started 1 week apart to reflect biological variability in its entirety as followed from our error analysis of eicosanoid production (data not shown). A schematic outline of our procedure is shown in Fig. 1. RAW264.7 mouse macrophage-derived cells were maintained between passages 4 and 24 at 37 °C and 10% CO2. The medium was composed of high glucose- and l-glutamine-containing Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. For an experiment, five T-150 flasks of the cells were plated at a density of 36 × 106 cells/flask in 24 ml of the same medium. At 24 h after plating, they were treated (or left untreated) with 100 ng/ml Kdo2-lipid A for another 24 h followed by subcellular fractionation.
Fig. 1.
Subcellular fractionation. The cell treatment timeline is shown in the top left. Upon treatment with Kdo2-lipid A, a cell transition from round or bipolar (at 24 h) to an extremely spread morphology (at 48 h), a hallmark of activation, occurs. At 48 h, the cells were harvested and fractionated; an outline of the fractionation procedure is shown on the right (see text for further detail). The six resulting fractions (shown in purple ovals) were all subjected to the same panel of analytical assays (listed on the bottom). Mito, mitochondria; PM, plasma membrane.
Subcellular Fractionation
The cultured medium was removed and used for eicosanoid analysis of Kdo2-lipid A-treated versus control cells as described elsewhere (9) to confirm their activated state. In these experiments, eicosanoid profiles and levels were consistent with the results of whole cell experiments (9) (data not shown).
The cells were harvested by scraping in Dulbecco's PBS (total of 35 ml), pelleted at 200 × g for 7 min, resuspended in 35 ml of the isolation medium (250 mm sucrose, 10 mm HEPES-Tris, pH 7.4, 1 mm EGTA-Tris), and pelleted again to remove salts. For effective homogenization, the cells were subjected to mild osmotic shock by resuspending in 35 ml of slightly hypotonic medium (same as the isolation medium above but with only 100 mm sucrose) and pelleted. The supernatant was set aside; the cell pellet was carefully transferred into a 7-ml glass Dounce homogenizer, homogenized in 10 ml of the supernatant by 40 strokes of the tight fitting pestle, and recombined with the supernatant. The osmotic shock and the details of homogenization are essential for effective cell lysis, organelle separation, and the final yield.
The homogenate was brought to an isotonic state by the addition of 3.2 ml of the hypertonic medium (same as the isolation medium above but with 1.78 m sucrose) and supplemented with 2 mm MgCl2, essential for preservation of the nuclei throughout the preparation. Differential centrifugation parameters were as follows: 200 × g for 10 min to pellet the nuclei/unbroken cells (the initial “nuclear” pellet), 5,000 × g for 10 min to pellet the mitochondria, and 100,000 × g for 1 h to pellet the microsomes. Postnuclear and postmitochondrial supernatants were additionally spun at 300 × g and 5,000 × g for 10 min, respectively, to additionally remove residual nuclei and mitochondria, respectively. The initial nuclear and mitochondrial pellets were additionally washed by resuspending/pelleting in Mg2+-containing and Mg2+-free media, respectively. The supernatant from the 100,000 × g spin was retained as the cytosolic fraction.
The nuclear, mitochondrial, and microsomal pellets were additionally separated in the stepwise gradients of iodixanol in an SW-41 bucket rotor. All gradient media were prepared according to the manufacturer's instructions based on the isolation medium above; the media for the nuclear preparation were supplemented with 5 mm MgCl2.
Nuclei were purified according to the manufacturer-suggested protocol; briefly, the nuclear pellet was brought to 25% iodixanol (12 ml), the iodixanol gradient was built from the bottom up in three 12-ml tubes (4 ml of 10%, 4 ml of nuclei in 25%, 2.5 ml of 30%, and 1.5 ml of 35%) and spun at 10,000 × g for 20 min. Nuclei banded at the 30/35% interface.
The mitochondrial and microsomal pellets were resuspended in the isolation medium, brought to 35% iodixanol (6 ml), and fractionated by flotation for 2 h at 50,000 × g in three 12-ml tubes each. The following iodixanol gradient was used: 2 ml of 10%, 4 ml of 17.5%, 4 ml of 25%, and 2 ml of the corresponding pellet resuspended in the 35% iodixanol. Mitochondria banded at the 17.5/25% interface; plasma membrane and the ER banded at 10/17.5 and 17.5/25% interfaces, respectively. The third fraction originating from the microsomal pellet banded at the most dense 25/35% interface and was termed “dense microsomes.” All samples were frozen and stored at −80 °C.
Proteomics Analysis
Proteomics analysis of each of the three biological replicates was performed in duplicate using the quadruplex or octuplex iTRAQTM (Applied Biosystems, Foster City, CA) approach as follows. For duplicates, the quartets of samples for each iTRAQ run were permuted to enable either direct or indirect calculations of all possible sample-to-sample ratios.
TCA was added to samples to a final concentration of 15% (w/v) to precipitate proteins. Samples were incubated at 4 °C for 2 h and then spun down in a refrigerated centrifuge at 4,000 × g for 15 min. The supernatant was discarded. Protein pellets were solubilized in 1 ml of 0.1% RapiGest (Waters) and 75 mm HEPES buffer, pH 7.0. Cysteines were reduced and alkylated using 1 mm tris(2-carboxyethyl)phosphine at 95 °C for 5 min followed by 2.5 mm iodoacetamide at 37 °C in the dark for 15 min. Proteins were digested with trypsin at an enzyme-to-substrate ratio (w/w) of 1:50 overnight.
For iTRAQ derivatization, an aliquot of each digested sample (100 μg of total protein) was treated with one tube of one of the iTRAQ reagents in 70% isopropanol at pH 7.2 for 2 h at room temperature. Labeled samples were dried down in a vacuum concentrator. 100 μl of water was added to each tube to dissolve the peptides. Samples tagged with four different iTRAQ reagents were pooled together. 1% TFA, pH 1.4 was added to precipitate RapiGest. Samples were incubated at 4 °C overnight and then centrifuged at 16,100 × g for 15 min. Supernatant was collected and centrifuged through a 0.22-μm filter and was used for LC-MS/MS analysis. iTRAQ labeling efficiency was calculated by searching the MS/MS data, specifying four possible iTRAQ modifications: 1) fully labeled, 2) N terminus-labeled only, 3) lysine-labeled only, and 4) non-labeled. Using the above protocol, we obtained higher than 95% iTRAQ labeling efficiency for all data sets.
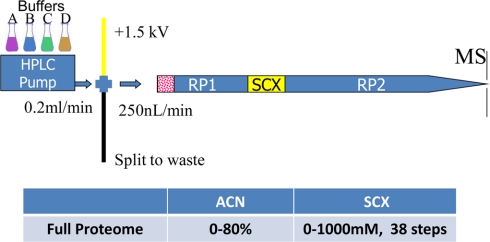
An Agilent 1100 HPLC system (Agilent Technologies, Santa Clara, CA) delivered a flow rate of 300 nl/min to a three-phase capillary chromatography column through a splitter. Using a custom pressure cell, 5-μm Zorbax SB-C18 (Agilent) was packed into fused silica capillary tubing (200-μm inner diameter, 360-μm outer diameter, 20 cm long) to form the first reverse phase column (RP1). A 5-cm-long strong cation exchange (SCX) column packed with 5-μm polysulfoethyl (PolyLC, Inc.) was connected to RP1 using a zero dead volume 1-μm filter (Upchurch, M548) attached to the exit of the RP1 column. A fused silica capillary (100-μm inner diameter, 360-μm outer diameter, 20 cm long) packed with 5-μm Zorbax SB-C18 (Agilent) was connected to the SCX column as the analytical column (the second reverse phase column; Fig. 2). The electrospray tip of the fused silica tubing was pulled to a sharp tip with the inner diameter smaller than 1 μm using a laser puller (Sutter P-2000). The peptide mixtures were loaded onto the RP1 using the custom pressure cell. Columns were not reused. Peptides were first eluted from the RP1 to the SCX column using a 0–80% acetonitrile gradient for 150 min. The peptides were fractionated by the SCX column using a series of salt gradients (from 10 mm to 1 m ammonium acetate for 20 min) followed by high resolution reverse phase separation using an acetonitrile gradient of 0–80% for 120 min (Fig. 2). Typically, it takes 4 days (38 salt fractions) for each full proteome analysis.
Fig. 2.
Schematics of three-phase on-line multidimensional nano-LC system. Peptides are loaded directly onto RP1 and subject to multiple step SCX fractionation followed by high resolution separation on the analytical column (RP2). Typically, it takes 4 days for each full proteome analysis.
Spectra were acquired using an LTQ linear ion trap tandem mass spectrometer (Thermo Electron Corp., San Jose, CA) using automated, data-dependent acquisition. The mass spectrometer was operated in positive ion mode with a source temperature of 150 °C.
The full MS scan range of 400–2,000 m/z was divided into three smaller scan ranges (400–800, 800–1,050, and 1,050–2,000 m/z) to improve the dynamic range. Both CID and pulsed Q dissociation (PQD) scans of the same parent ion were collected for protein identification and quantitation. Each MS scan was followed by four pairs of CID-PQD MS/MS scans of the most intense ions from the parent MS scan. A dynamic exclusion of 1 min was used to improve the duty cycle of MS/MS scans. About 20,000 MS/MS spectra were collected for each salt step fractionation.
The raw data were extracted and searched using Spectrum Mill v3.03 (Agilent). The CID and PQD scans from the same parent ion were merged together. MS/MS spectra with a sequence tag length of 1 or less were considered to be poor spectra and were discarded. The rest of the MS/MS spectra were searched against the International Protein Index (IPI) mouse database (v3.31, 56,555 protein sequences). The enzyme parameter was limited to fully tryptic peptides with a maximum miscleavage of 1. All other search parameters were set to the default settings of Spectrum Mill (carbamidomethylation of cysteines, iTRAQ modification, ±2.5 Da for precursor ions, ±0.7 Da for fragment ions, and a minimum matched peak intensity (scored peak intensity) of 50%). A concatenated forward-reverse database was constructed to calculate the in situ false discovery rate (FDR). The total number of protein sequences in the combined database was 113,110. Cutoff scores were dynamically assigned to each data set to maintain the false discovery rate at less than 1% at the protein level. The resulting spectrum scores/spectrum scored peak intensities were >14/>50%, >12/>50%, and >14/>50% for 1+ peptides, 2+ peptides, and 3+ peptides, respectively. Proteins that share common peptides were grouped to address the database redundancy issue. The proteins within the same group shared the same set or subset of unique peptides.
Protein iTRAQ intensities were calculated by summing the peptide iTRAQ intensities from each protein group. Peptides shared among different protein groups were removed before quantitation. A minimal total iTRAQ intensity of 100 was used to filter out low intensity spectra. Isotope impurities of iTRAQ reagents were corrected using correction factors provided by the manufacturer (Applied Biosystems).
Protein identification information (unique scores, numbers of unique peptides, and percent coverage) is summarized in supplemental Table S1. Semiquantitatively, raw protein abundances were calculated by normalization of the data by the total iTRAQ reporter intensities for each sample (supplemental Table S1). Because the same amount of total protein was used in the analysis of each sample, the latter approach is equivalent to normalization to total protein. In all subsequent analyses, abundances of proteins undetected in particular fractions were regarded as missing data rather than zero amounts. Therefore, the duplicate protein abundances were averaged if protein was detected in both iTRAQ runs; otherwise, the single replicate was used.
To derive protein distributions among six fractions, these raw protein abundances were normalized either to the sum total of all six fractions (supplemental Table S1) or to protein abundance in the main fraction (supplemental Table S2; selected marker proteins only). To assess the biological variability of the protein distributions, means and S.E. of biological triplicates were calculated for each of the 2,642 detected proteins in each of six fractions from the resting and activated cells (supplemental Table S1) for which duplicate/triplicate data had been obtained.
Measurement of Conventional Marker Enzymes/DNA
The purity of the fractions was characterized with regard to the intensities of the conventional markers for each organelle/cell compartment. DNA was measured as the marker for nuclei using a Quant-iT DNA assay kit according to the manufacturer's protocol. Measurements were performed using a FluoroMax-2 spectrofluorometer (Horiba Jobin-Yvon). To ensure reproducibility, the sample aliquots were supplemented with 5% ethanol and frozen-thawed prior to the assay.
Succinate dehydrogenase served as the marker enzyme for mitochondria. The enzyme quantity was assayed using a partial enzymatic reaction, reduction of p-iodonitrotetrazolium violet (INT), according to the method described by Munujos et al. (10) with minor modifications. The assay medium contained 50 mm Tris-HCl, pH 8.1, 1 mm EGTA, 12 mg/ml detergent Cremaphor EL, and 20 mm succinate. All reactions were performed in triplicate in the 96-well plates in the ELx808iu plate reader (BioTek Instruments). Control reactions in the absence of succinate were set up for each sample to account for the background reduction of INT. The reactions were started with the addition of 2 mm INT and followed for 10 min at 490 nm; the succinate-dependent rates were calculated by subtraction.
Cytochrome P450 reductase served as the marker enzyme for the ER. Its quantity was assayed using the NADPH-dependent cytochrome c reductase activity of the enzyme using a cytochrome c reductase (NADPH) assay kit according to the manufacturer's protocol. The measurements were performed in an Uvikon-XL spectrophotometer (BioTek Instruments).
K+-dependent phosphatase reaction served as the marker activity for plasma membrane and was measured as K+-stimulated p-nitrophenylphosphatase according to the method of Kashiwamata et al. (11) with modifications. The assay medium contained 50 mm Tris-HCl, pH 8.1, 2 mm MgCl2, and 25 mm KCl. All reactions were performed in triplicate in the 96-well plates in an ELx808iu plate reader (BioTek Instruments). Control reactions in the absence of KCl were set up for each sample to account for the background phosphatase reaction. The reactions were started with the addition of 10 mm p-nitrophenyl phosphate and followed for 30 min at 410 nm; the K+-dependent rates were calculated by subtraction.
Glucose-6-phospate dehydrogenase served as the marker enzyme for cytoplasm. The enzyme quantity was determined using a Vybrant cytotoxicity assay kit essentially according to the manufacturer's protocol. The measurements were performed using a FluoroMax-2 spectrofluorometer (Horiba Jobin-Yvon). Due to a non-linear dose response, prior to the assay, the samples were diluted to achieve similar reaction rates.
Lipidomics Analysis
Lipids were extracted as described previously (12) and analyzed as follows.
Coenzyme Q
Coenzymes Q9 and Q10 were quantified by LC-MRM experiments performed using a Shimadzu LC system (comprising a solvent degasser, two LC-10A pumps, and an SCL-10A system controller) coupled to a 4000 Q-Trap hybrid triple quadrupole linear ion trap mass spectrometer equipped with a Turbo V ion source (Applied Biosystems). LC was performed at a flow rate of 200 μl/min with a linear gradient as follows: 100% mobile phase A was held isocratically for 2 min and then linearly increased to 100% mobile phase B over 14 min and held at 100% mobile phase B for 4 min. Mobile phase A consisted of methanol, acetonitrile, and aqueous 1 mm ammonium acetate (60:20:20, v/v/v). Mobile phase B consisted of 100% ethanol containing 1 mm ammonium acetate. A Zorbax SB-C8 reverse phase column (5 μm, 2.1 × 50 mm) was obtained from Agilent.
MRM was performed in the positive ion mode with MS settings as follows: curtain gas, 10 p.s.i.; GS1, 20 p.s.i.; GS2, 30 p.s.i.; ion source, +5,000 V; temperature, 350 °C; interface heater, on; declustering potential, +100 V; entrance potential, +10 V; and collision cell exit potential, +5 V. The voltage used for collision-induced dissociation was +55 V. To quantify coenzyme Q9 and coenzyme Q10 in the subcellular fractions of RAW cells, a known quantity of coenzyme Q6 (Sigma) was added as an internal reference. The MRM pairs for coenzyme Q6, coenzyme Q9, and coenzyme Q10 are 608/197, 812/197, and 880/197, respectively. In these MRM pairs, the precursor ions are the [M + NH4]+ ions, and the m/z 197 is the major fragment ion corresponding to a proton adduct of the quinone ring of coenzyme Q.
Cardiolipin
Cardiolipins were quantified by using normal phase LC coupled to a QSTAR XL quadrupole time-of-flight tandem mass spectrometer (Applied Biosystems) equipped with an electrospray source. Normal phase LC using an Ascentis® silica HPLC column (5 μm, 25 cm × 2.1 mm) was performed on an Agilent 1200 Quaternary LC system. Mobile phase A consisted of chloroform/methanol/aqueous ammonium hydroxide (800:195:5, v/v/v). Mobile phase B consisted of chloroform/methanol/water/aqueous ammonium hydroxide (600:340:50:5, v/v/v/v). Mobile phase C consisted of chloroform/methanol/water/aqueous ammonium hydroxide (450:450:95:5, v/v/v/v). The elution program consisted of the following: a linear gradient begun at 100% mobile phase A to 100% mobile phase B over 14 min and held at 100% mobile phase B for 11 min followed by a 5-min linear gradient to 100% mobile phase C and held for 2 min. A 5-min gradient back to 100% mobile phase B was then held for 2 min, returned to 100% mobile phase A over 5 min, and held for an additional 8 min. The total LC flow rate was 300 μl/min. The postcolumn splitter diverted ∼10% of the LC flow to the ESI source of the QSTAR XL mass spectrometer with MS settings as follows: ion source, −4,200 V; curtain gas, 20 p.s.i.; GS1 = 20 p.s.i.; declustering potential, −55 V; and focusing potential, −265 V.
Four synthetic cardiolipin standards (CL57:4, CL61:1, CL80:4, and CL86:4 made by Avanti) were used as internal references. The 57:4 standard is 1′-[1,2-di-(9Z-tetradecenoyl)-sn-glycero-3-phospho, 3′-[1-(9Z-tetradecenoyl), 2-(10Z-pentadecenoyl)-sn-glycero-3-phospho]-sn-glycerol. The 61:1 standard is 1′-[1,2-dipentadecanoyl-sn-glycero-3-phospho], 3′-[1-(pentadecanoyl), 2-(9Z-hexadecenoyl)-sn-glycero-3-phospho]-sn-glycerol. The 80:4 standard is 1′-[1,2-di-(13Z-docosenoyl)-sn-glycero-3-phospho], 3′-[1-(13Z-docosenoyl), 2-(9Z-tetradecenoyl)-sn-glycero-3-phospho]-sn-glycerol. The 86:4 standard is 1′-[1,2-di-(15Z-tetracosenoyl)-sn-glycero-3-phospho], 3′-[1-(15Z-tetracosenoyl), 2-(9Z-tetradecenoyl)-sn-glycero-3-phospho]-sn-glycerol. The exact masses of these standards are 1,245.792, 1,307.902, 1,568.152, and 1,652.247 atomic mass units for CL57:4, CL61:1, CL80:4 and CL86:4, respectively. Data were analyzed as described previously (12).
Databases and Bioinformatics Tools
Results of the proteomics and lipidomics analyses, as well as the respective search tools, are available on line at the web site of the LIPID MAPS Consortium. The proteomics search tool allows the selection of the proteins predominantly located in the specific fractions, their ranking based on the measure of the prevalence, and the effect of cell activation. Here, we present the initial analysis of these data.
RESULTS
Characterization of Subcellular Preparations Using Conventional Markers
Macrophage-like RAW264.7 cells were grown both in the resting and endotoxin-activated states and subjected to subcellular fractionation. The combination of differential centrifugation and isopycnic gradients used (see “Experimental Procedures”) separated organelles based on their mass and density. On the basis of the separation procedure (and therefore these physical properties), the resulting fractions could be identified as the nuclear, mitochondrial, cytosolic, endoplasmic reticulum, and plasma membrane fractions plus an unidentified dense microsomes (as we defined under “Experimental Procedures”) fraction.
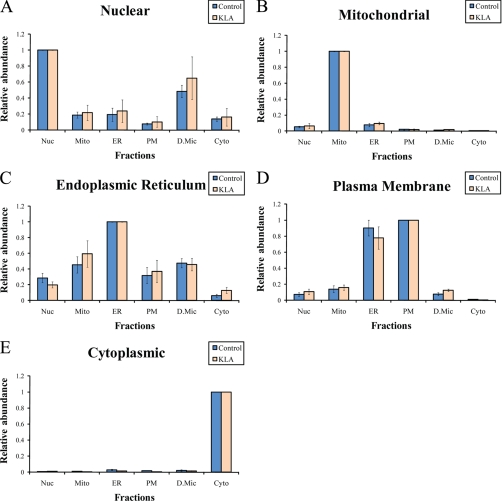
A panel of five markers, one for each target organelle/compartment,2 was used to initially characterize composition of these fractions. All five markers were detected in each of the fractions, and their distributions are shown in Fig. 3. These distributions demonstrate, at the biochemical level, that the fractions were identified correctly and contain predominant levels of the anticipated (title) organelles.
Fig. 3.
Distribution of conventional markers between fractions (A–E). Intensities of the markers were calculated on a per protein basis followed by normalization to the main fraction. Nuc, nuclei; Mito, mitochondria; PM, plasma membrane; D.Mic, dense microsomes; Cyto, cytosol; KLA, Kdo2-lipid A-activated cells. The data are mean ± S.E.; n = 6.
Initially, we attempted a calculation of fraction composition (as described under “Characterization of Subcellular Preparations Using Proteomic Markers”) for the five fractions for which we had markers. However, this calculation was not productive because the solution included negative marker intensities in pure organelles (result not shown).
Thus, the conventional markers did not allow the assessment of the composition of each fraction. Additionally, these conventional markers have several obvious shortcomings as outlined in the Introduction. For instance, the nuclei marker DNA may be released from the nucleus and nonspecifically bind to other organelles via electrostatic interactions. It is obvious, for example, that cytosol (supernatant after 1-h 100,000 × g spin) cannot be 15% contaminated with intact nuclei (which pellet after 10 min at 5,000 × g) as shown on Fig. 3A. Additionally, we microscopically examined all fractions after staining with the nuclear stain 4′,6-diamidino-2-phenylindole and did not detect any nuclei-like structures in any fractions other than nuclei (data not shown).
Therefore, our next step was to identify sizable panels of potentially novel organelle markers without any a priori assumptions as to their nature. The identification was to be performed on the basis of our semiquantitative proteomics analysis and, therefore, was designed to be relatively free of the shortcomings of the functional enzymatic assays and/or the use of DNA. We further intended to do an a posteriori bioinformatics analysis of the resulting markers to confirm their correct locations and, thus, correct identifications.
Protein Identification
13,190 IPI protein sequences were identified using the filtering criteria described under “Experimental Procedures.” Among them, 12,760 proteins were from the forward database, and 430 were from the reverse database, corresponding to a cumulative FDR of 3.4% at the protein level. 6,705 proteins were identified in all three biological replicates and were used for quantification and downstream analysis. The FDR of those proteins is 0.15% (10 of 6,705); 3,074 protein groups were obtained from the 6,705 protein sequences. The FDR at the protein group level is 0.13% (four reverse protein groups of 3,074 total protein groups).
Proteomic Marker Panels
We started the analysis by preselecting a subset of proteins (or more precisely the entries in the IPI protein database) that were detected in all six fractions for at least one biological replicate. This preselection was necessary given the probability that a protein in a particular fraction may be undetected due to limitations of the mass spectrometry method rather than of low/zero abundance. It should be noted that by doing this preselection we risked missing some of the best markers (i.e. the ones with exclusive localization). Therefore, the resulting ensembles may be considered conservative in the sense that they may overestimate cross-contamination of fractions rather than vice versa.
These preselected protein entries were then each designated as the candidate markers for those organelles in which they were the most abundant; only the protein entries that have the same designation for the control and activated cells and were consistent among replicates were used for further analysis. To evaluate the quality of the candidate markers we introduced the concept of a marker index (M): the ratio of the abundance of a protein (as defined under “Experimental Procedures”) in its main fraction to the sum of its abundances in all other fractions. For example, if the protein E is the most abundant in the nuclear fraction it is a potential nuclear marker with a marker index of MENuc = IE, Nuc/(IE, Mito + IE, PM + IE, ER + IE, D.Mic + IE, Cyto) where IE, Mito, IE, PM, IE, ER, IE, D.Mic, and IE, Cyto are the protein abundances in the mitochondrial, plasma membrane, ER, dense microsomal, and cytosolic fractions, respectively.
The greater the calculated ME, the higher the quality of the marker. Theoretically, for an ideal marker (that is exclusively distributed to one organelle) and pure fractions, the index should approach infinity. An index equal to 1 may be roughly interpreted as an equal distribution between one main fraction and the rest of the fractions in the total.
These preselected protein entries were ranked based on the average of their marker indices for control and activated cells. Finally, the top 50 candidate markers for each organelle were selected to comprise the completed 50-protein marker panels (supplemental Tables S2–S9). The size of the panel was arbitrary chosen; we considered the 50-member panels as fairly large to be representative. Indeed, a fluctuation in one protein intensity would be diluted 50-fold, resulting in a mere 2% bias.
A subsequent bioinformatics analysis has demonstrated that these marker panels were consistent with the legacy information in public databases (Table I and supplemental Tables S3–S7). For this, we matched our empirically observed locations with the location annotations obtained using the Functional Annotation Search Tool from Database for Annotation, Visualization and Integrated Discovery (DAVID) 2008 and, if necessary, by further drilling down into EntrezGene and Swiss-Prot databases; immediate references in those databases were also critically reviewed and taken into account.
Table I. Summary of bioinformatics analysis of marker panels.
50-member marker panels identified for each organelle were divided into three groups according to their cell component annotations (see supplemental Tables S3–S7). For many proteins, databases indicate more than one location. In general, we considered annotated location consistent with the one observed in our proteomics analysis if a list of database locations included the correct location. In special cases, we added markers to this group based on additional information in the databases (see footnotes to this table and the supplemental tables). GO, gene ontology.
| Markers for (organelle) | GO term (cell component 4) |
Annotated location of markers (number of markers) |
||||
|---|---|---|---|---|---|---|
| Top term | Top term p value | Additional terms (rank 2–4) | Consistent with observed location | Alternative to observed locationa | Unknownb | |
| Nuclei | Nucleus | 9.90e−21 | Nuclear part, intracellular membrane-bound organelle, spliceosome, intracellular organelle part | 47 | None | 3 |
| Mitochondria | Mitochondrion | 1.50e−53 | Mitochondrial part, cytoplasmic part, mitochondrial membrane, mitochondrial envelope | 50 | None | None |
| Cytoplasm | Cytoplasm | 3.70e−13 | Intracellular part, cytosol, cytoskeleton, cytoplasmic part | 41 | 1 | 7 |
| Plasma membrane | Plasma membrane | 2.20e−08 | Plasma membrane part, cytoplasm, coated pit, cytoplasmic vesicle | 33c | 6 | 7 |
| ER | Endoplasmic reticulum | 2.20e−25 | Endoplasmic reticulum part, cytoplasmic part, endoplasmic reticulum membrane, nuclear envelope-endoplasmic reticulum network | 35 | 7 | 3 |
a Correctable annotations (one for cytoplasm, four for plasma membrane, and five for ER; see Table II) are excluded.
b Includes annotations as unspecified “membrane” (for plasma membrane and ER markers).
c Includes five proteins annotated as localized to plasma membrane-associated structures (G protein-coupled receptors, ruffle, lamellipodium, cell projection membrane, and apical part of the cell).
For example, 47 of the 50 nuclear markers (Table I and supplemental Table S3) have annotations consistent with the nuclear location. Three remaining proteins have no annotated locations.
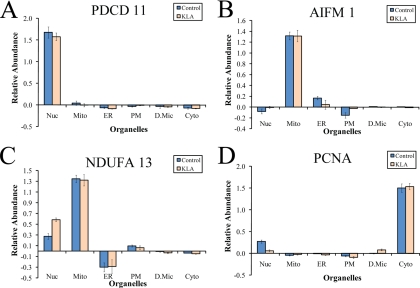
Programmed cell death protein 11 (PDCD 11) was not mapped to the nucleus cell component by DAVID 2008 (among a few other proteins), but manual inspection of EntrezGene revealed a dual nuclear/cytoplasmic annotation. However, we found that the level of PDCD 11 in the cytosolic fraction is ∼ 20 times lower than in the nuclear fraction (supplemental Table S2). We termed this and all similar annotations “correctable.” All annotations that point to a location in one of the six major fractions but do not match our assignment are correctable based on our experimental subcellular profiles (supplemental Table S2). Note that our further analysis showed that in the macrophages PDCD 11 was completely absent from the alternative cytoplasmic location (see “Protein Distribution in Pure Organelles” and Fig. 8A). We regard as consistent with our results all annotations that include a matching location (summarized in Table I) rather than only exclusive matches; this assignment gains additional support when the annotations are correctable and the alternative locations have low levels of the proteins.
Fig. 8.
Distribution of selected protein markers among pure organelles (A–D). Protein content in pure organelles was calculated based on the organellar composition of fractions (Fig. 4) and protein distribution in fractions (measured in the iTRAQ experiment). See text for detail on calculations. Nuc, nuclei; Mito, mitochondria; PM, plasma membrane; D.Mic, dense microsomes; Cyto, cytosol; KLA, Kdo2-lipid A-activated cells. The data are mean ± S.E. for three independent preparations.
Similar analysis was done for all other fractions. In the mitochondrial panel (supplemental Table S4), all 50 markers are annotated as mitochondrial (Table I).
Database annotations for the cytoplasmic marker panel (supplemental Table S5) were less unambiguous. 41 of the 50 proteins have annotations consistent with cytoplasmic location, and seven do not have an explicit organellar location. Two remaining proteins are annotated as having alternative locations. Nuclear location of proliferating cell nuclear antigen (PCNA) is correctable (supplemental Table S5), and indeed, this protein is predominantly cytosolic in comparison with the nuclear fraction (Fig. 4 and Table II). However, to be conservative, we do not positively log it as “consistent” with cytoplasmic location but only exclude this protein from the “alternative” group and, accordingly, from the corresponding column in Table I. The same treatment was applied to nine more proteins (from the ER and the plasma membrane marker panels listed in supplemental Tables S6 and S7) that had correctable annotations pointing exclusively to alternative locations (summarized in Table II).
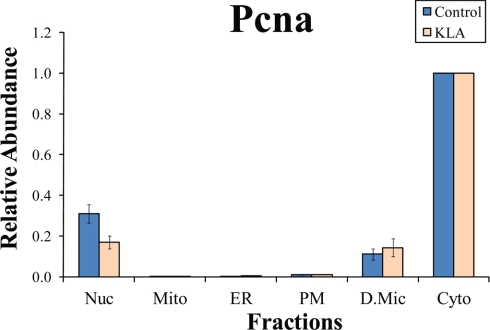
Fig. 4.
Subcellular distribution of PCNA. The y axis shows abundances of PCNA in various fractions relative to the abundance in the main (in this case, nuclear) fraction. Nuc, nuclei; Mito, mitochondria; PM, plasma membrane; D.Mic, dense microsomes; Cyto, cytosol; KLA, Kdo2-lipid A-activated cells. Note the high prevalence of the cytoplasmic, as opposed to reported nuclear, location of PCNA. The data are mean +/- S.E. for three independent preparations.
Table II. Summary of correctable annotations.
Only those markers that are unequivocally annotated as located in alternative organelles are listed. Nuc, nucleus; PM, plasma membrane; Mito, mitochondrion; Cyto, cytoplasm.
| Marker |
Observed location |
Annotated locations |
|||||
|---|---|---|---|---|---|---|---|
| Gene symbol | Accession no. | Protein | Fraction | Relative abundancea | Organelle | Relative abundanceb |
|
| Control | Kdo | ||||||
| Hist1h1e | IPI00223714 | Histone H1.4 | ER | 1 | Nuc | 0.29 ± 0.07, n = 2 | 0.22 ± 0.01, n = 2 |
| Tmem48 | IPI00165794 | Nucleoporin NDC1 | ER | 1 | Nuc | 0.26, n = 1 | 0.18, n = 1 |
| Krt1 | IPI00625729 | Keratin, type II cytoskeletal 1 | ER | 1 | PM | 0.55, n = 1 | 0.22, n = 1 |
| Clptm1 | IPI00121627 | Cleft lip and palate transmembrane protein 1 homolog | ER | 1 | PM | 0.63 ± 0.19, n = 3 | 0.48 ± 0.05, n = 3 |
| Ubxd8 | IPI00265386 | Isoform 1 of Ubiquitin regulatory X (UBX) domain-containing protein 8 | ER | 1 | Cyto | 0.01, n = 1 | 0.03, n = 1 |
| Srgap2 | IPI00652316 | Slit/Robo Rho GTPase-activating protein 2 isoform 10 | PM | 1 | Mito | 0.06, n = 1 | 0.05, n = 1 |
| Preb | IPI00124980 | Prolactin regulatory element-binding protein | PM | 1 | ER | 0.48 ± 0.15, n = 3 | 0.60 ± 0.21, n = 3 |
| Nuc | 0.17, n = 1 | 0.23, n = 1 | |||||
| Coro1b | IPI00124819 | Coronin-1B | PM | 1 | Cyto | 0.15, n = 1 | 0.04, n = 1 |
| Lgals9 | IPI00114396 | Isoform long of Galectin-9 | PM | 1 | Cyto | 0.19 ± 0.05, n = 3 | 0.08 ± 0.05, n = 3 |
| Pcna | IPI00113870 | Proliferating cell nuclear antigen | Cyto | 1 | Nuc | 0.30 ± 0.04, n = 3 | 0.16 ± 0.03, n = 3 |
a Relative abundances reflect normalized marker distributions between locations. Relative abundance in main organelle is 1 by definition (see text for detail).
b Relative abundances observed in locations listed in the databases (annotated locations). Note the relatively low levels of the markers in these locations with the exception of ER proteins in plasma membrane fraction and vice versa. The latter is consistent with the distribution of the ER and plasma membrane marker ensembles as whole (Fig. 5C), resulting from cross-contamination of these two fractions (Fig. 6).
Galectin-1 (LGALS1) remains the only member of the cytoplasmic panel regarded as having an alternative location (Table I). It should be noted, however, that this assignment is also conservative. This protein is secretable and listed in the databases as extracellular, although its cytoplasmic location has been directly demonstrated (Ref. 13 and references therein). Thus, overall, the bioinformatics analysis unequivocally confirms the quality of the cytoplasmic marker panel.
We regard as consistent with the plasma membrane location (Table I and supplemental Table S6) not only the markers that are explicitly annotated in the databases as such but also the proteins that are annotated as a part of G protein-coupled receptor as well as the proteins that are annotated as located in the plasma membrane structures such as ruffle, cell projection, apical part of the cell, etc. Thus, overall, the location information for 33 markers is consistent with the plasma membrane (Table I and supplemental Table S6). Seven proteins have no precise location annotations (they are either not annotated or annotated as unspecified “membrane” proteins). The remaining 10 proteins have alternative annotated locations (in some cases more than one): three as cytoplasmic, two as mitochondrial, two as ER, one as nuclear, three as lysosomal, two as Golgi, one as endosomal, and one as extracellular. However, the annotations to the former four locations are correctable (Table II).
35 markers in the ER marker panel are annotated as ER proteins (Table I and supplemental Table S7). Three proteins have no precise location annotations. 12 other proteins have alternative location annotations (in some cases more than one): three as plasma membrane, two as nuclear, two as cytoplasmic, four as peroxisomal, two as extracellular, and one as Golgi. Again, annotations to the former three locations are correctable; five markers fall in this category (Table II).
It should be noted that plasma membrane and ER may still contain additional minor organelles that are not correctable (not present as a separate fraction in our analysis). For example, the plasma membrane may be the main lysosome- and/or endosome-containing fraction based on the presence of the β-galactosidase precursor (GLB1), glucosylceramidase precursor (GBA), cathepsin D precursor (CTSD), and transmembrane 9 superfamily protein member 2 precursor (TM9SF2), respectively, in the marker panel for this fraction. Similarly, the presence of peroxisomal acyl-coenzyme A oxidase 3 (ACOX3), serine hydrolase-like protein (SERHL), vesicle-associated membrane protein-associated protein B (VAPB), and isoform 1 of fatty acyl-CoA reductase 1 (MLSTD2) in the marker panel for the ER may indicate that peroxisomes are contained mainly in the ER fraction. It should be further noted that, based on the low number of corresponding markers (four in plasma membrane and seven in ER), these additional organelles are indeed minor and generate a very small bias in the marker ensemble as a whole.
Thus, marker panels identified in the present study are in a surprisingly good agreement with the legacy data (>82%). The minimal percentage of correct annotations is 66% (for the ER marker), whereas the maximal percentage of a particular alternative location is less than 10% (four peroxisome-annotated proteins among the ER markers).
Composition of Dense Microsomes
The dense microsomal fraction was prepared without any preconceived notion regarding its composition. Marker analysis allowed us to propose identities of the components comprising the dense microsomal fraction (supplemental Tables S8 and S9). This is most likely a mixture of several components. A functional annotation search using DAVID 2008 (supplemental Table S8) revealed that relevant cell components were ribosomes (second from the top) and related components (ribonucleoprotein complex, intracellular non-membrane-bound organelle, etc.), COPI-coated vesicle (and related terms), and Golgi. It should be noted that although the most relevant term is cytoplasm it is defined according to gene ontology as the whole cell except for the nucleus and the plasma membrane (including the three components mentioned above). Results of more detailed manual examination of EntrezGene and Swiss-Prot databases confirmed that the dense microsomal fraction is mainly a mixture of ribosomes with other minor cytosolic vesicles (see supplemental Table S9). 23 of 50 marker proteins are annotated as ribosomal (or components of ribonucleoprotein complexes), and four are related to COPI-coated vesicles. Other locations include Golgi apparatus, P-body, and fragments of plasma membrane (including cell junction). All these components are protein- and nucleic acid-rich and consequently dense, which is consistent with their banding at the high density interface (25/35% of iodixanol). It should also be noted that some of the proteins have more than one potential location because of intracellular trafficking. For example, COPI vesicles are known to transverse from Golgi to ER, and it is impossible to predict whether it is located in one, the other, or both in the particular case of RAW264.7 macrophages in either their basal or activated state. Because this fraction is an insoluble fraction, we interpreted cytoplasmic annotations as association with (unspecified) vesicles within the cytoplasm (nine members; supplemental Table S9).
Surprisingly, DNA-directed RNA polymerase I, subunit RPA1 (annotated as nuclear protein) was detected in this fraction. This is one correctable annotation in the 50-member panel, and our data show with great confidence (supplemental Table S2; conclusion based on biological triplicates) that this protein localizes to dense microsomes with 5-fold higher abundance than to the nucleus. Either this protein is simply misannotated, or it is located in a specific subnuclear compartment (similar to the ribosomes of ER or coated pits of the plasma membrane) that dissociates from the major organelle and co-purifies with the dense microsomes. For eight proteins, location is either not specified or specified as membrane.
Characterization of Subcellular Preparations Using Proteomic Markers
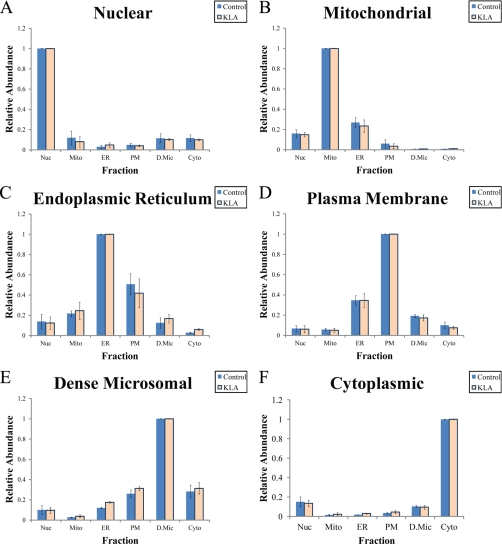
Theoretically, the distribution of an ideal organelle marker among subcellular fractions would reflect a distribution of the corresponding organelle in the fractions. To simulate the distribution of an ideal marker using experimental data, the 50-member marker panels for each organelle were averaged to derive an integrated marker ensemble. Because we use normalized protein intensities (supplemental Table S2), each protein contributes to the normalized marker ensemble intensities equally regardless of its general abundance, thus allowing us to avoid a potential bias from the high abundance proteins.
A marker ensemble may be treated as a virtual “protein”; in particular, it may be characterized by its subcellular distribution (Fig. 5) and by a marker index. For each of the six prepared fractions, we obtained intensities (I) of six marker ensembles, one for each of the organelles (see data in Fig. 5). These data may be organized in a 6 × 6 matrix [Ii,j] where i is an organelle and j is a fraction.
Fig. 5.
Distribution of marker ensembles between fractions (A–F). Nuc, nuclei; Mito, mitochondria; PM, plasma membrane; D.Mic, dense microsomes; Cyto, cytosol; KLA, Kdo2-lipid A-activated cells. Distributions of ensembles are averages of all 50 markers for each organelle. The data are mean ± S.E. for three independent preparations.
The intensity of the marker ensemble Ii,j may be used to calculate a percentage of the organelle i in the fraction j (Pi,j). Indeed, if the so far unknown value of the intensity of the marker ensemble for a pure organelle i (Iiorg) were known, we could calculate the Pi,j simply by dividing the measured intensity of the marker by the intensity in a pure organelle (and expressing it as a percentage) as follows.
Equation 1 may be rearranged to linear form by introducing a reciprocal of Iiorg (Xiorg) to give
where Xiorg = 1/Iiorg. For each fraction j, the sum total of all six organelles is 100%.
 |
or
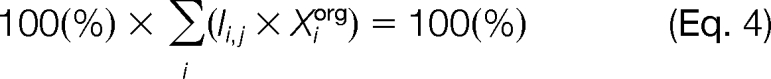 |
or
 |
A system of six such equations (one for each fraction j) may be presented in the matrix form,
where [Xiorg] is a vector of the reciprocal marker intensities for six pure organelles and [1] is a unit vector representing the composition total for the six fractions.
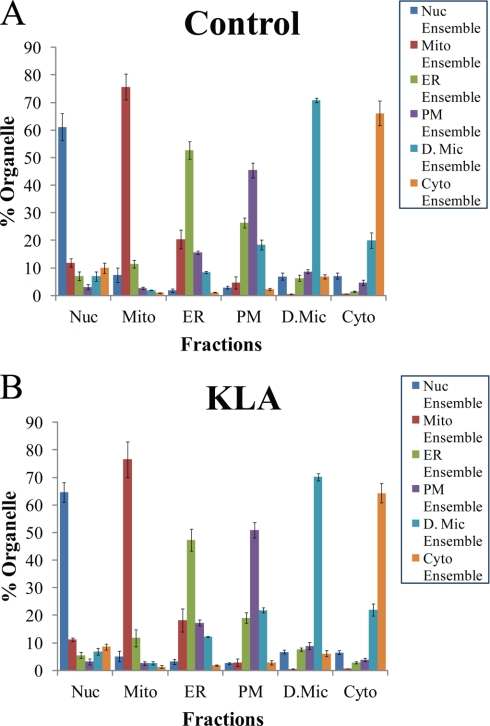
By solving Equation 6, we found the values of all six Xiorg. Using Equation 2 for every i and j, we calculated all Pi,j and, thus, generated a matrix of the organellar composition of the fractions [Pi,j]. This composition is presented in graphic form in Fig. 6. The composition matrix [Pi,j] was further used to calculate the distribution of lipids and proteins among the pure organelles.
Fig. 6.
Fraction composition based on proteomic markers (A and B). The percentage of different organelles in each fraction was calculated from the distribution of the organelle-specific marker ensembles among the fractions (see “Results” for details). Nuc, nuclei; Mito, mitochondria; PM, plasma membrane; D.Mic, dense microsomes; Cyto, cytosol; KLA, Kdo2-lipid A-activated cells. The data are mean ± S.E. for three independent preparations.
Distribution of Lipid Markers
Knowing the organellar composition of the fractions and a distribution of a lipid among them, it is possible to calculate a distribution of the lipid among the pure organelles. Indeed, the content of lipid k in the fraction j (Lj,k) is a linear combination of its content in each organelle (Li,korg) multiplied by the content of this organelle in the fraction (Pi,j/100%).
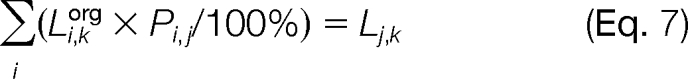 |
In the matrix format, the system of six Equations 7 (one for each j) may be presented as
where [Li,korg] is an unknown vector of the content of lipid k in six organelles, [Lj,k] is a vector of the observed content of this lipid in the corresponding fractions (e.g. see Fig. 7A), and [Pi,j/100%] is a matrix of organellar compositions of these fractions (see Fig. 6).
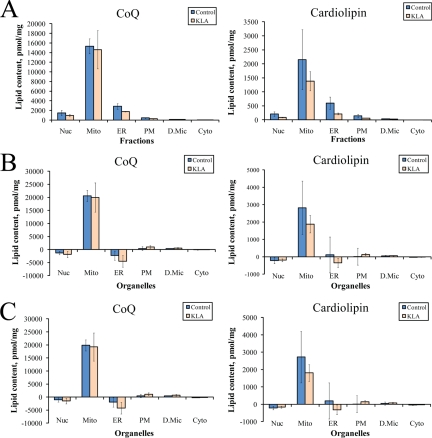
Fig. 7.
Distribution of lipid markers between subcellular organelles. A, measured levels of mitochondrial lipids in the subcellular fractions. The lipid content for all fractions is the sum total of all detected species of each subclass. B, calculated levels of mitochondrial lipids in pure organelles based on marker ensembles. C, calculated levels of mitochondrial lipids in pure organelles based on the six-marker panel (Table III). See text for detail on calculations. Nuc, nuclei; Mito, mitochondria; PM, plasma membrane; D.Mic, dense microsomes; Cyto, cytosol; Cardiolipin, total cardiolipin; CoQ, total coenzyme Q; KLA, Kdo2-lipid A-activated cells. The data are mean ± S.E. for three independent preparations.
By solving Equation 8, it is possible to obtain the originally unknown values of organellar lipid content Li,korg. This procedure may be repeated for each lipid k to generate a complete subcellular lipidome (to be reported elsewhere).
We applied this algorithm to the lipids that may be considered organellar lipid markers. Cardiolipin and coenzyme Q are known to be exclusively mitochondrial lipids. For each fraction, we calculated sum totals of all detected species of these lipid subclasses (16 cardiolipins and two ubiquinones, coenzyme Q9, and coenzyme Q10; Fig. 7A) and used them to build the [Lj,k] vectors. By solving Equation 8 for each of these lipid markers, we obtained the content of the total cardiolipin and coenzyme Q in the pure organelles (Fig. 7B).
Six-marker Panel
Because measuring levels of 50 proteins per fraction may be sometimes impractical for future studies, a question arises whether a single marker can be used as a surrogate. We selected a panel of six organelle markers (one per organelle), distributions of which closely conform to the marker ensembles (Table III). This panel was used instead of marker ensembles to derive organellar lipid levels (Fig. 7C) and gave a result similar to the one obtained with marker ensembles (Fig. 7B).
Table III. Six-marker panel.
For selection of this panel, unidentified and putative proteins were excluded. Distributions of individual proteins among six fractions with and without Kdo2-lipid A treatment were compared with distributions of corresponding ensembles, and the proteins with the least maximal deviation were selected. Linear regression parameters for these proteins versus corresponding ensembles (A, slope; B, intersect; and r2) are given in the table.
| Organelle | Marker |
Single marker vs. ensemble regression parameters |
||||
|---|---|---|---|---|---|---|
| Gene symbol | Accession no. | Protein | A | B | r2 | |
| Nuclei | Ddx21 | IPI00652987 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 | 0.988351 | 0.009351 | 0.990617 |
| Mitochondria | Etfb | IPI00121440 | Electron transfer flavoprotein subunit β | 1.002542 | −0.00527 | 0.999439 |
| ER | Snx22 | IPI00135686 | PPIB, peptidylprolyl isomerase B | 0.960789 | 0.040539 | 0.981165 |
| Plasma membrane | Rhog | IPI00116558 | Rho-related GTP-binding protein RhoG precursor | 1.04213 | −0.06685 | 0.984108 |
| Dense microsomes | Rpl15 | IPI00273803 | 60 S ribosomal protein L15 | 0.995402 | −0.00666 | 0.981378 |
| Cytoplasm | Anxa1 | IPI00230395 | Annexin A1 | 0.995659 | 0.004414 | 0.995243 |
Protein Distribution in Pure Organelles
The same algorithm as described above can be applied to any cellular component, including proteins, to derive organellar proteomes. In particular, it may be used to assess whether alternative location annotations of a protein in the databases likely result from the actual distribution between two or more organelles or from a contamination with one organelle that is an exclusive location of the protein. This may also be applied to any protein from the 50-member marker panel. A marker ensemble reflects an average of all 50 markers and is, therefore, resistant to bias from any single marker. Thus, it is conceivable that within the marker panels there may be proteins with a strong deviation from the marker ensemble (i.e. from the corresponding organelle). This would indicate that the protein has a location in more than one organelle.
A few examples of such analysis are presented in Fig. 8. The above mentioned PDCD 11 demonstrates exclusive location in nucleus despite dual (nuclear and cytosolic) annotation in EntrezGene (Fig. 8A). It is claimed that AIF is released from mitochondria upon the activation of an apoptotic program and redistributes to the cytoplasm (14) or to the nucleus (15). We did not detect cytoplasmic AIF (Fig. 8B), consistent with the lack of the apoptotic cell morphology (data not shown). According to the literature, a subunit of the respiratory complex I, GRIM-19 (NDUFA13), may translocate to the nucleus (16), and our data confirm this for both control and activated macrophages (Fig. 8C). Finally, PCNA, which is annotated as nuclear, was found both in the cytoplasm and the nucleus (Fig. 8D). PCNA abundance in the nucleus is much lower than in the cytoplasm, which, given that the cytoplasm constitutes the main portion of the total cell protein, makes the nuclear PCNA a very minor component.
DISCUSSION
Marker Ensembles
In the present study, a search for marker proteins undertaken without any preconception as to their identities resulted in the identification of 50-member marker panels for each of the major subcellular organelles/compartments. These marker panels were in fairly good agreement with the legacy data (annotations in the public databases). Moreover, we were able to suggest corrections to the location annotations for a number of proteins.
One reason for the significant number of incorrect annotations may be a typical contamination of an organellar preparation with other organelles. This problem is well appreciated in the proteomics field (1, 3–7). As an extreme example of the basis for incorrect annotation, the location of leucine-rich pentatricopeptide repeat motif-containing protein (LRPPRC) and prohibitin (IPI00321718, PHB2) was deduced in part from a very crude separation of cell homogenate into only two fractions (17, 18). Although these fractions were called “nuclear” and “cytosolic” in those studies, it is obvious that all other organelles are distributed between these two fractions. As a result, the two mitochondrial proteins (supplemental Table S2) were erroneously assigned both nuclear and cytoplasmic locations in EntrezGene.
Even when a study attempts to address specifically a preparation of high quality organellar fractions (e.g. mitochondria (19, 20)), the possibility of contamination cannot be excluded. The more superior the proteomics analysis, the greater is the probability to include into the proteome some minor components arising from such contaminations (3). When only a single fraction is analyzed, there is no way to assess whether a minor protein is a true component of the target organelle or whether it originates from a minor contaminant organelle with a high abundance of that protein. Correlative approaches such as protein correlation profiling (1, 3, 4) and localization of organelle proteins by isotope tagging (5, 6) that use continuous gradients (or multiple fractions) may be used to identify proteins localized to specific organelles. However, this identification is typically based on co-localization with a protein marker that is assumed to be organelle-specific. Our approach was (i) to avoid such a priori assumptions and (ii) to base our analysis on a large (50-member) panel of markers as opposed to a single “best” marker. As a result, we were able to suggest corrections of location annotations for 14 proteins (Table II); additionally, it is possible to derive an actual organellar distribution of a large set of proteins that have multiple location annotations (e.g. Fig. 8). Moreover, we took this approach a step further to quantify organellar compositions of subcellular fractions.
The 50-member marker panels were used to derive the marker ensembles that represent all 50 proteins as a whole. Organellar compositions of the fractions were calculated based on the distribution of the marker ensembles among them. Furthermore, we demonstrated that this organellar composition could be used to calculate the true distribution of cellular components between pure organelles from their distribution between mixed fractions (Figs. 7 and 8).
The composition of the marker panels demonstrates that isolated organelles are relatively intact. Indeed, the nuclear panel includes both chromatin- and nuclear envelope-associated proteins (e.g. histone HIST1h4f and nuclear pore complex components NUP 35, NUP 43, NUP 93, and NUP 155, respectively). The mitochondrial panel includes inner membrane proteins (e.g. NADH dehydrogenase subunit NDUFA13, cytochrome c oxidase subunit mt-CO2, and ATP synthase subunits ATP5b, ATP5f1, and ATP5a1), outer membrane proteins (porin VDAC2), intermembrane space proteins (AIFM1), and matrix proteins (citrate synthase and isocitrate dehydrogenase IDH2).
Six-marker Panel
Certainly, a direct application of marker ensembles based on 50-member panels may be a resource-consuming endeavor; however, it is possible to perform a complete analysis based on a limited number of (or even a single) protein(s) from each panel (Fig. 7C) that may be quantified using ELISA or similar methods. It should be noted that the best candidates for these abridged panels should be selected from the proteins most closely conforming to the distribution of the marker ensembles (e.g. Table III). Additionally, it should be noted that depending on the cell type the subcellular distribution of a protein may change. It is our opinion that a reliable marker identification and assessment of the fraction composition may be made only when based on a distribution of several markers. Indeed, any given protein may either have an alternative location or be poorly expressed, if at all, in a particular cell type. On the other hand, this cannot be expected for a large set of proteins. This reasoning is supported by the results shown in Fig. 7. We have made every effort to select the best possible six-marker panel (Table III); however, in this case, the deconvoluted distribution of mitochondrial lipids (Fig. 7C) does not show the same clear mitochondrial localization as with the use of marker ensembles (Fig. 7B).
The composition of the six-marker panel reemphasizes the legitimacy of our approaches. All members of the panel are bona fide components of corresponding organelles.
The concept of the current study was to relate cellular components (such as proteins, lipids, etc.) to ensembles of proteins (the marker ensembles) as the representatives of various organelles/subcellular compartments. Therefore, this study contributes to redefining the organelles as biochemical rather than historically prevailing morphological entities.
Conclusion
Indeed, subcellular fractionation was pioneered by De Duve (21) and co-workers in the 1950s using liver tissue. Much progress has been made in the techniques in the following years to obtain purer subcellular fractions. However, it is now clear that by the very nature it may not be practical to obtain all of the organelles in a pure and defined form for a given cell type. Our marker ensemble approach now provides a means to determine the organellar composition of less than fully purified fractions so that one can fully analyze and determine the protein and metabolite composition of each organelle, a valuable step forward for cell biology and metabolomic profiling.
Supplementary Material
Acknowledgments
We are grateful to Drs. Anatoly Starkov and Richard Harkewicz for helpful discussion and critical reading of the manuscript.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grant GM069338, a Lipid Metabolites and Pathways Strategy (LIPID MAPS) large scale collaborative grant.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Tables S1–S9.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Tables S1–S9.
2 Strictly speaking, cytosol and plasma membrane are not organelles, but for brevity, we use the term “organelles” as a substitute for “organelles and other subcellular compartments” to include these two important subcellular locations.
1 The abbreviations used are:
- Kdo2
- (3-deoxy-d-manno-octulosonic acid)2
- ER
- endoplasmic reticulum
- INT
- p-iodonitrotetrazolium violet
- CL
- cardiolipin
- SCX
- strong cation exchange
- RP1
- the first reverse phase column
- iTRAQ
- isobaric tag for relative and absolute quantitation
- MRM
- multiple reaction monitoring
- PQD
- pulsed Q dissociation
- IPI
- International Protein Index
- FDR
- false discovery rate
- PDCD
- programmed cell death protein
- PCNA
- proliferating cell nuclear antigen
- COPI
- coat protein complex I
- AIF
- apoptosis-inducing factor.
REFERENCES
- 1.Andersen J. S., Mann M. ( 2006) Organellar proteomics: turning inventories into insights. EMBO Rep 7, 874– 879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yates J. R., 3rd, Gilchrist A., Howell K. E., Bergeron J. J. ( 2005) Proteomics of organelles and large cellular structures. Nat. Rev. Mol. Cell Biol 6, 702– 714 [DOI] [PubMed] [Google Scholar]
- 3.Forner F., Foster L. J., Campanaro S., Valle G., Mann M. ( 2006) Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol. Cell. Proteomics 5, 608– 619 [DOI] [PubMed] [Google Scholar]
- 4.Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. ( 2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570– 574 [DOI] [PubMed] [Google Scholar]
- 5.Dunkley T. P., Watson R., Griffin J. L., Dupree P., Lilley K. S. ( 2004) Localization of organelle proteins by isotope tagging (LOPIT). Mol. Cell. Proteomics 3, 1128– 1134 [DOI] [PubMed] [Google Scholar]
- 6.Sadowski P. G., Dunkley T. P., Shadforth I. P., Dupree P., Bessant C., Griffin J. L., Lilley K. S. ( 2006) Quantitative proteomic approach to study subcellular localization of membrane proteins. Nat. Protoc 1, 1778– 1789 [DOI] [PubMed] [Google Scholar]
- 7.Gilchrist A., Au C. E., Hiding J., Bell A. W., Fernandez-Rodriguez J., Lesimple S., Nagaya H., Roy L., Gosline S. J., Hallett M., Paiement J., Kearney R. E., Nilsson T., Bergeron J. J. ( 2006) Quantitative proteomics analysis of the secretory pathway. Cell 127, 1265– 1281 [DOI] [PubMed] [Google Scholar]
- 8.Raetz C. R., Garrett T. A., Reynolds C. M., Shaw W. A., Moore J. D., Smith D. C., Jr., Ribeiro A. A., Murphy R. C., Ulevitch R. J., Fearns C., Reichart D., Glass C. K., Benner C., Subramaniam S., Harkewicz R., Bowers-Gentry R. C., Buczynski M. W., Cooper J. A., Deems R. A., Dennis E. A. ( 2006) Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J. Lipid Res 47, 1097– 1111 [DOI] [PubMed] [Google Scholar]
- 9.Buczynski M. W., Stephens D. L., Bowers-Gentry R. C., Grkovich A., Deems R. A., Dennis E. A.( 2007) TLR-4 and sustained calcium agonists synergistically produce eicosanoids independent of protein synthesis in RAW264.7 cells. J. Biol. Chem 282, 22834– 22847 [DOI] [PubMed] [Google Scholar]
- 10.Munujos P., Coll-Cantí J., González-Sastre F., Gella F. J. ( 1993) Assay of succinate dehydrogenase activity by a colorimetric-continuous method using iodonitrotetrazolium chloride as electron acceptor. Anal. Biochem 212, 506– 509 [DOI] [PubMed] [Google Scholar]
- 11.Kashiwamata S., Goto S., Semba R. K., Suzuki F. N. ( 1979) Inhibition by bilirubin of (Na+ + K+)-activated adenosine triphosphatase and K+-activated p-nitrophenylphosphatase activities of NaI-treated microsomes from young rat cerebrum. J. Biol. Chem 254, 4577– 4584 [PubMed] [Google Scholar]
- 12.Garrett T. A., Guan Z., Raetz C. R. ( 2007) Analysis of ubiquinones, dolichols, and dolichol diphosphate-oligosaccharides by liquid chromatography-electrospray ionization-mass spectrometry. Methods Enzymol 432, 117– 143 [DOI] [PubMed] [Google Scholar]
- 13.Goldring K., Jones G. E., Thiagarajah R., Watt D. J. ( 2002) The effect of galectin-1 on the differentiation of fibroblasts and myoblasts in vitro. J. Cell Sci 115, 355– 366 [DOI] [PubMed] [Google Scholar]
- 14.Daugas E., Nochy D., Ravagnan L., Loeffler M., Susin S. A., Zamzami N., Kroemer G. ( 2000) Apoptosis-inducing factor (AIF): a ubiquitous mitochondrial oxidoreductase involved in apoptosis. FEBS Lett 476, 118– 123 [DOI] [PubMed] [Google Scholar]
- 15.Lakhani S. A., Masud A., Kuida K., Porter G. A., Jr., Booth C. J., Mehal W. Z., Inayat I., Flavell R. A. ( 2006) Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science 311, 847– 851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angell J. E., Lindner D. J., Shapiro P. S., Hofmann E. R., Kalvakolanu D. V. ( 2000) Identification of GRIM-19, a novel cell death-regulatory gene induced by the interferon-beta and retinoic acid combination, using a genetic approach. J. Biol. Chem 275, 33416– 33426 [DOI] [PubMed] [Google Scholar]
- 17.Tsuchiya N., Fukuda H., Sugimura T., Nagao M., Nakagama H.( 2002) LRP130, a protein containing nine pentatricopeptide repeat motifs, interacts with a single-stranded cytosine-rich sequence of mouse hypervariable minisatellite Pc-1. Eur. J. Biochem 269, 2927– 2933 [DOI] [PubMed] [Google Scholar]
- 18.Kurtev V., Margueron R., Kroboth K., Ogris E., Cavailles V., Seiser C. ( 2004) Transcriptional regulation by the repressor of estrogen receptor activity via recruitment of histone deacetylases. J. Biol. Chem 279, 24834– 24843 [DOI] [PubMed] [Google Scholar]
- 19.Mootha V. K., Bunkenborg J., Olsen J. V., Hjerrild M., Wisniewski J. R., Stahl E., Bolouri M. S., Ray H. N., Sihag S., Kamal M., Patterson N., Lander E. S., Mann M. ( 2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115, 629– 640 [DOI] [PubMed] [Google Scholar]
- 20.Taylor S. W., Fahy E., Zhang B., Glenn G. M., Warnock D. E., Wiley S., Murphy A. N., Gaucher S. P., Capaldi R. A., Gibson B. W., Ghosh S. S. ( 2003) Characterization of the human heart mitochondrial proteome. Nat. Biotechnol 21, 281– 286 [DOI] [PubMed] [Google Scholar]
- 21.De Duve C. ( 1965) The separation and characterization of subcellular particles. Harvey Lect 59, 49– 87 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.