Abstract
Lanthanide-based luminescent ligand binding assays are superior to traditional radiolabel assays due to improved sensitivity and affordability in high throughput screening while eliminating the use of radioactivity. Despite significant progress using lanthanide(III)-coordinated chelators such as DTPA derivatives, dissociation-enhanced lanthanide fluoroimmunoassays (DELFIA) have not yet been successfully used with more stable chelators, e.g. DOTA derivatives, due to the incomplete release of lanthanide(III) ions from the complex. Here, a modified and an optimized DELFIA procedure incorporating an acid treatment protocol is introduced for use with Eu(III)-DOTA labeled peptides. Complete release of Eu(III) ions from DOTA labeled ligands was observed using hydrochloric acid (2.0 M) prior to the luminescent enhancement step. NDP-α-MSH labeled with Eu(III)-DOTA was synthesized and the binding affinity to cells overexpressing the human melanocortin-4 receptors (hMC4R) was evaluated using the modified protocol. Binding data indicate that the Eu(III)-DOTA linked peptide bound to these cells with an affinity similar to its DTPA analogue. The modified DELFIA procedure was further used to monitor the binding of an Eu(III)-DOTA labeled heterobivalent peptide to the cells expressing both hMC4R and CCK-2 (Cholecystokinin) receptors. The modified assay provides superior results and is appropriate for high-throughput screening of ligand libraries.
Keywords: DELFIA, Lanthanide, DOTA derivatives, Time-resolved luminescence, Binding assay, Solid-phase peptide synthesis, Melanocortin receptor
Introduction
Evaluation of ligand-receptor interactions is considered to be a fundamental screening assay in drug discovery [1]. Lanthanide-based luminescence assays provide an attractive alternative to the traditional radiolabel assays used for monitoring ligand-receptor interactions in terms of sensitivity, appropriateness for high throughput screening, and radioactive biohazards. The superiority of lanthanide-based luminescence is due to the narrow emission bands, large Stokes shifts, limited photo-bleaching, and long luminescent lifetimes which allow time-resolved fluorometric assays with high sensitivity [2–5]. Work in our lab has shown that the detection limit for this technology is 0.2 amole per 100 μL [6]. Among the various lanthanide-based assay platforms, DELFIA technology (Wallac/PerkinElmer Lifesciences) has been widely employed as a bio-analytical tool in both clinical and biomedical applications [7–15].
In DELFIA-based ligand binding assays, a ligand is labeled with a lanthanide(III) ion [such as Eu(III)] chelate and is competed off with test ligands or directly used for saturation binding studies [16]. The amount of specifically bound lanthanide-labeled ligand is then analyzed by adding an “enhancement solution” following wash steps to remove the unbound compounds. The acidic (pH=3.5–4) enhancement solution is capable of completely releasing Eu(III) ions from relatively weak chelators such as DTPA and their derivatives. The enhancement solution is comprised of sensitizer chelators (NTA and TOPO) that form highly luminescent coordination complexes with the released lanthanide(III) ions [17, 18]. This process enhances the lanthanide-centered luminescence by up to 107-fold. The signal at 615 nm is then detected using TRL with a delay time of up to 400 μs following 340 nm excitation.
DELFIA methods have been widely employed by us and others to study receptor binding of ligands labeled with derivatives of DTPA or DTTA chelates of Eu(III) [6, 14, 16, 19]. For example, we previously demonstrated the use of Eu(III)-DTPA constructs to characterize and optimize ligands binding to human melanocortin-4 and δ–opioid receptors. However, the eventual in vivo use of DTPA or DTTA chelates with metal ion coordination is limited due to their relative low affinity constants for metal chelation. Heterocyclic chelators, such as DOTA, are much stronger chelators, however, DOTA derivatives have not yet been used in DELFIA approaches due to the incomplete release of lanthanide(III) ions from the chelate under the traditional conditions [19]]. Lanthanide(III) complexes formed with DOTA and their derivatives have received intense scrutiny in a wide range of biomedical applications including MRI, PARACEST MRI and positron emission tomography [20–23]. DOTA and its derivatives offer the highest thermodynamic and kinetic stability with a wide selection of metal ions among the commonly used polyaminocarboxylic acid chelators (Stability constants of Gd(III)-DOTA and Gd(III)-DTPA, 25.3 and 22.4, respectively) [23–25]. DOTA complexes are more suitable for in vivo radiopharmaceutical applications under conditions where the clearance of the contrast agent is relatively slow [26]. As a part of our ongoing research, we are designing and developing DOTA-labeled ligands for biomedical applications with Gd(III) and In(III)-based chelation chemistries for MRI and gamma ray detection, respectively. This article discusses the development and evaluation of a modified DELFIA technology based on an acid treatment protocol for the use of lanthanide(III)-DOTA labeled ligands in whole cell ligand-receptor binding studies.
We have chosen to evaluate the binding of Eu(III)-DOTA and DTPA labeled NDP-α-MSH [27] to Hek293 cells expressing the hMC4R [28] as a model system where one of the carboxylic acid groups of DOTA or DTPA is used for ligand coupling. DTPA labeled compounds were used for comparison with the traditional DELFIA. Furthermore the modified DELFIA protocol was used to demonstrate the targeting of a heterobivalent ligand to two different combinations of cell-surface receptors as a proof-of-study where heteromultivalency is used to crosslink multiple receptors [29]. The Eu(III)-DOTA labeled heterobivalent ligand was constructed from analogues of NDP-α-MSH peptide and the CCK-6 peptide targeted to cells expressing both hMC4R and CCK-2 receptors.
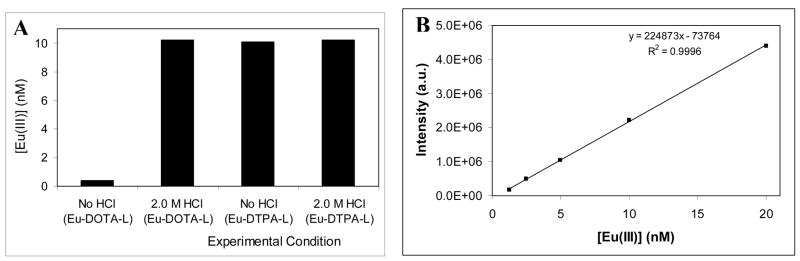
The requirement of an acid treatment procedure for a complete release of lanthanide(III) ions from DOTA derivative is shown in Fig. 1 by comparing the photo-physical characteristics of Eu(III)-DOTA and –DTPA labeled NDP-α-MSH ligands. No significant change in the Eu(III)-signal of DTPA-labeled ligand was observed with the acid treatment according to Fig. 1A, confirming that the enhancement solution is capable of completely releasing lanthanide(III) ions from DTPA. However data in Fig. 1B indicate that DOTA bound Eu(III) ions can not be quantitatively released by simply adding the enhancement solution. However, pretreatment of Eu(III)-DOTA labeled ligands with 2M HCl completely released Eu(III) ions from the chelator, which was detected following neutralization.
Fig. 1.
Excitation and emission spectra of (A) Eu(III)-DTPA-PEG9-NDP-α-MSH and (B) Eu(III)-DOTA-PEG9-NDP-α-MSH (each 10 nM) using DELFIA. The compounds were excited at 340 nm. Acidification: Samples (20 μL) were incubated with 2.0 M HCl (50 μL) for 5 hours, neutralized with 2.0 M NaOH (55 μL), treated with the enhancement solution (115 μL), and incubated for 30 min before reading.
Materials and Methods
General chemistry methods
Fmoc-protected amino acids, HBTU, HOBt, and bifunctional polyethyleneglycol linker PEGO were purchased from SynPep (Dublin, CA) or from Novabiochem (San Diego, CA). Rink amide Tentagel S resin was acquired from Rapp Polymere (Tubingen, Germany). DOTA-NHS ester was purchased from Macrocyclics (Dallas, TX, USA). DTPA dianhydride and Fmoc-PEG9, were purchased from Aldrich and Peptides International, respectively. For the N-α-Fmoc-protected amino acids, the following side chain protecting groups were used: Arg(Ng-Pbf); Glu(O-tBu); Asp(O-tBu); His(Nim-Trt); Ser(tBu), Trp(Ni-Boc), Lys(Nε-Aloc), Lys(Nε-Boc) and Nα-Boc-Tyr(tBu). Reagent grade solvents for HPLC and other reagents were acquired from VWR (West Chester, PA) or Aldrich-Sigma (Milwaukee, WI), and were used without further purification unless otherwise noted. Solid-phase synthesis was performed in fritted syringes using a Domino manual synthesizer obtained from Torviq (Niles, MI).
Purification of the compounds was achieved using a Hewlett-Packard 1100 series HPLC instrument with a reverse-phase column (Vydac, 10 mm × 220 mm, 10 μm, 300 Å) or size exclusion chromatography [borosilicate glass column (2.6 · 250 mm, Sigma, St. Louis, MO, USA) filled with medium-size Sephadex G25]. Separations were monitored at 230 and 280 nm with Hewlett-Packard 110 series UV detector and integrated with Hewlett-Packard 3396 series III integrator (flow rate = 5 mL/min). Purity of the peptides was ensured using analytical HPLC (Waters Alliance 2695 separation model with a dual wavelength detector Waters 2487) with a reverse-phase column (Waters Symmetry, 4.6 · 75 mm, 3.5 μm; flow rate = 0.3 mL/min). (Conditions: HPLC, linear gradient from 10 to 90% B over 30 min, where A is 0.1% TFA and B is acetonitrile). Structures were characterized by ESI (Finnigan, Thermoquest LCQ ion trap instrument), MALDI-TOF or FT-ICR mass spectrometry. An appropriate mixture of standard peptides was used for internal calibrations.
Ligand Synthesis
The ligands were synthesized manually as previously described using Nα-Fmoc/tBu solid-phase peptide synthesis strategy and standard DIC/HOBt or DIEA/HBTU activations on Rink amide Tentagel resin (0.23 mmol/g) [6, 29]. The Rink resin was swollen in DMF for an hour. The resin was washed with DMF, and Nα-Fmoc protecting groups were removed with 50% piperidine in DMF (1 × 2 min and 1 × 20 min). The resin was washed again with DMF, 1.0 M HOBt in DMF, DMF, and the next Nα-Fmoc amino acid was coupled using preactivated 0.3 M HOBt ester in THF (3 equiv. of Nα-Fmoc amino acid, 3 equiv. of HOBt, and 3 equiv. of DIC). The resin slurry was stirred for 2 hours or until the Kaiser test became negative. If the test failed, the resin was washed with DMF and the amino acid was coupled again with HBTU/DIEA procedure (0.3 M solution of 3 equiv. of Nα-Fmoc amino acid, 3 equiv. of HBTU, and 6 equiv. of DIEA in DMF) for 3 hours. If the second coupling did not result in a negative Kaiser test, the resin was washed with DMF, and the rest of amino groups were capped with 50% acetic anhydride in pyridine for 10 min. When the coupling reaction was finished, the resin was washed with DMF, and the same procedure was repeated for the next amino acid until all the amino acids in the sequence were attached. Attachment of PEG9 or PEGO linkers was performed using DIC/HOBt activation (3 equiv. of Nα-Fmoc-PEG9 or Fmoc-PEGO, 3 equiv. of HOBt, and 3 equiv. of DIC).
Aloc cleavage
The orthogonal protecting Aloc group of lysine was cleaved as following for the attachment of PEG9 linker in heterobivalent ligand. The resin was washed with DCM then flushed with argon for 10 min. A cleavage mixture of dimethylbarbituric acid (5 equiv.), Pd(TPP)4 (0.2 equiv.) in DCM (0.5 M solution) was flushed with argon and injected into the syringe. The reaction mixture was stirred for 30 min then repeated. The resin is washed with DMF, 10%DIEA in DMF, DMF, 2% sodium diethyldithiocarbamate trihydride, 10%DIEA in DMF, and DMF.
DTPA and DOTA attachment
The DTPA chelator was attached to the N-terminus of resin bound PEG9-NDP-α-MSH-resin. After Fmoc removal, the free-amine resin was washed with DMSO. DTPA dianhydride (10 equiv.) and HOBt (30 equiv.) were dissolved in 1 mL of dry DMSO at 50 °C and stirred for 20 min at room temperature. The reagent mixture was then injected into the syringe reactor. The resin was stirred overnight then washed with DMSO, THF, 20% aqueous THF, THF, 5% DIEA in THF (5 minutes) and THF. DOTA was attached to resin–bound PEG9-NDP-α-MSH-resin and Nε-amine lysine side chain of Ac-MSH(7)-PEGO-[ProGly]6-Lys(PEG9)-PEGO-CCK(6)-resin. DIEA (10 equiv.) and DOTA-NHS ester (3 equiv.) in DMF were injected into the free-amine resin and stirred overnight. The resin was washed with DMF, THF, and DCM.
Cleavage of ligands from the resin
The resin was washed with DMF, THF, and DCM. A cleavage mixture (10 mL per 1 g of the resin) consists of TFA (91%), water (3%), 1,2-ethandithiol (3%), and thioanisole (3%) was injected into the resin and then stirred for 4 hours at room temperature. The solution was filtered off, and the resin was washed with TFA (2×3 min). Collected filtrates were concentrated by a stream of nitrogen and the product was precipitated by cold ether. The peptide pellets were washed three times with cold ether, dried, dissolved in 1.0 M acetic acid, and lyophilized.
Eu(III) labeling of peptides
The lyophilized peptide was dissolved in 0.1 M ammonium acetate (pH was adjusted to 8 with aqueous 0.1 M NH4OH) and treated with 3 equiv. of EuCl3.6H2O in water. The reaction mixture was stirred at room temperature overnight. The excess of Eu(III) chloride and ammonium salts were removed using a SEPAC C-18 reverse-phase column with repetitive washing (20 mL of HPLC grade water). The final product was eluted using 50 % aqueous acetonitrile (4 mL) and lyophilized.
Cell culture
Hek293 cells were engineered to overexpress hMC4R as previously described [28]. The coding region of hMC4R gene was expressed in pcDNA3.1 (Invitrogen, V790-20). Hek293 cells expressing both hMC4R and CCK receptors were used for heterobivalent binding studies. All cells were maintained under standard conditions (37 C, 5% CO2) and were grown in DMEM supplemented with 10% FBS. The dual expressing cells maintained were under selection in the above media containing Zeocin (100mg/mL) and Geneticin (0.4 mg/mL). At 90% cell confluency (verified by microscopy), cells were harvested for plating in 96-well plates for performing the receptor-ligand binding assays.
Eu(III)-based ligand binding assays
Cells were plated in black CoStar 96-well plates (#3603) at a density of 20,000 cells/well and were allowed to grow for 3 days. The HEK293 cells adhere to the plastic reasonably well, however if weakly attaching cells are used, the wells can be coated with extracellular matricies or polylysine to improve cell retention during the assay incubation period. On the day of the experiment, media was aspirated from all wells prior to the addition of the ligands to be tested. Ligands were diluted in binding media (DMEM, 1mM 1,10-Phenanthroline, 200mg/L Bacitracin, 0.5mg/L Leupeptin, 0.3% BSA) and samples were tested in quadruplicate, unless otherwise noted. Saturation binding assays were carried out as follows: increasing amounts of Eu(III)-labeled peptides were added to cells and incubated for 1 hour at 37 °C. Following the incubation, cells were washed three times with Wash Buffer (50 mM Tris-HCl, 0.2%BSA, 30 mM NaCl). For our experiments, the media was carefully replaced manually using a vacuum manifold. It is important to make certain that cells are not released during the washing maneuver. Microscopic analysis of the aspirate can provide a simple evaluation of cell release in control experiments. After the cell wash period, no additional removal of media from the wells can occur, since addition of both the enhancement solutions and acid treatment are cytotoxic. When testing Eu(III)-DTPA-labeled ligands, the enhancement solution (Perkin Elmer; 1244-105) was added directly to the wells (100 μL/well) after washing and the plates were incubated for at least 30 min at 37°C prior to reading. The plates were read on a Wallac VICTOR3 instrument using the standard Eu(III) TRL measurement (340 nm excitation, 400 μsec delay, and emission collection for 400 μsec at 615 nm).
For Eu(III)-DOTA-labeled ligands, 2.0 M HCl (50 μL/well) was added after the wash step and the plates were incubated at 37 °C for 2 hours. The samples were then neutralized using 2.0 M NaOH (55 μL/well) followed by the enhancement solution (115 μL/well). Analysis procedure was carried out as described for Eu(III) TRL measurements. Competitive binding assays were carried out using variable concentrations of non-labeled ligand (50 μL/well) and a fixed concentration of Eu(III)-labeled ligand (10 nM, 50 μL/well) and using otherwise identical conditions provided above. Data were analyzed with GraphPad Prism Software using appropriate non-linear regression analyses.
Results
Synthesis of Eu(III)-labeled ligands
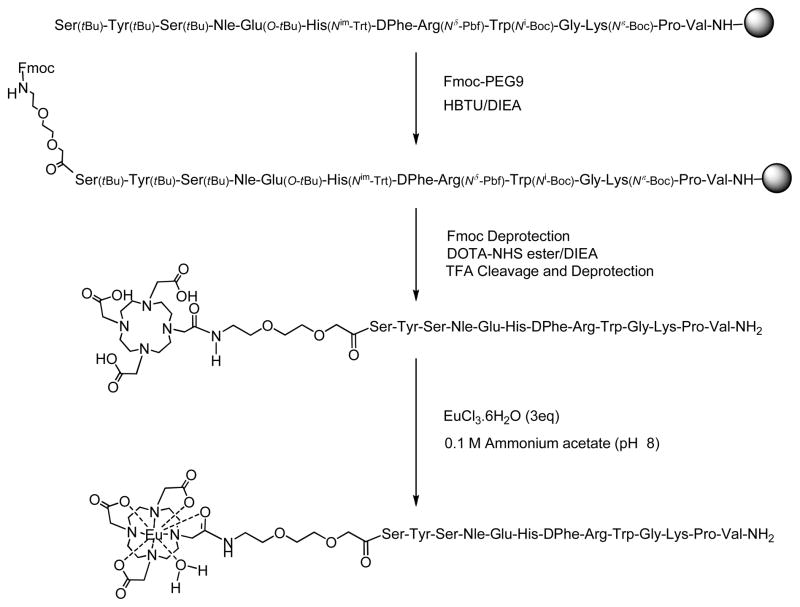
All peptides were synthesized using standard solid-phase technology whereas Eu(III) labeling was performed in the solution phase. A representative synthetic scheme of Eu(III)-DOTA-PEG9-NDP-α-MSH is shown in Fig. 2. Eu(III) chelators (DTPA and DOTA) were attached through a PEG9 semi rigid linker to the α-amino terminal of serine (in NDP-α-MSH) or to the ε-amino group of lysine (in the heterobivalent ligand). A previously reported procedure was adapted for the attachment of DTPA to NDP-α-MSH wherein an in situ di-HOBt ester of DTPA was formed before the coupling with the free amino group of the resin-bound ligands [6]. Activated DOTA-NHS ester was used for the coupling of DOTA chelator to the PEG9 linker.
Fig. 2.
Synthetic scheme of Eu(III)-DOTA-PEG9-NDP-α-MSH.
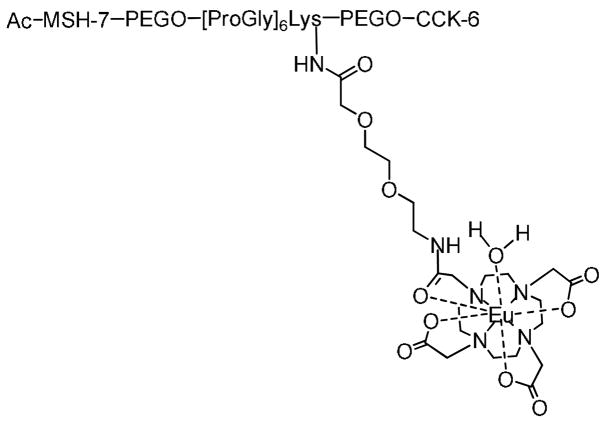
MSH-7, a heptapeptide fragment of NDP-α-MSH and CCK-6, a hexapeptide fragment of [Nle28, Nle31]CCK-6, respectively, were conjoined with PEGO-[Pro-Gly]6-Lys-PEGO linker to construct the heterobivalent ligand with an appropriate distance for favorable binding to the cells with dual receptor expression. This distance was optimized in the previous study using linkers of varying rigidity, and lengths, and evaluating the binding of these heterobivalent constructs to cells that expressed one or both complementary receptors by competing against Eu(III)-labeled monovalent ligands (Eu(III)-DTPA-NDP-α-MSH and Eu(III)-DTPA-CCK-8) [29]. A similar DOTA labeled, heterobivalent ligand was used in the present study as shown in Fig. 3. All compounds were purified by HPLC (see supporting information). Eu(III) labeling was performed under pH=8 buffer conditions. Basic conditions facilitate the coordination of Lewis acidic Eu(III) ions to Lewis basic oxygen bearing carboxylate moieties. All the ligands were characterized by high-resolution mass spectroscopy. Our initial results indicate that the MALDI method removes Eu(III) ions from the chelator during the analysis. However FT-ICR mass spectrometry with ESI was found to be non-destructive towards Eu(III)-DOTA labeled ligands. Therefore Eu(III)-based isotope patterns of the DOTA-linked ligands were analyzed using the FT-ICR method (see supporting information).
Fig. 3.
Structure of Eu(III)-DOTA-labeled heterobivalent ligand. MSH(7)=Ser-Nle-Glu-His-DPhe-Arg-Trp; CCK(6)=Nle-Gly-Trp-Nle-Asp-Phe-NH2.
Optimization of the modified DELFIA protocol
We have quantitatively evaluated the Eu(III) ion concentration of both DOTA- and DTPA-labeled NDP-α-MSH as shown in Fig. 4. Without the acid treatment, we were only able to detect 0.4 nM of Eu(III) ions in a pre-determined solution of 10 nM Eu(III)-DOTA labeled NDP-α-MSH. In contrast, 10 nM of Eu(III) was detected after acid treatment. Complementary experiments performed with or without acid treatment gave 10 nM of Eu(III) ions in DTPA-labeled ligands, which agreed with our qualitative assessment shown in Fig. 1.
Fig. 4.
(A) Measured Eu(III) ion concentrations of Eu(III)-DOTA and DTPA labeled NDP-α-MSH ligands (L=PEG9-NDP-α-MSH). 10 nM ligand solutions predetermined using tryptophan assay were used for the analysis. (B) Calibration curve based on EuCl3.6H2O.
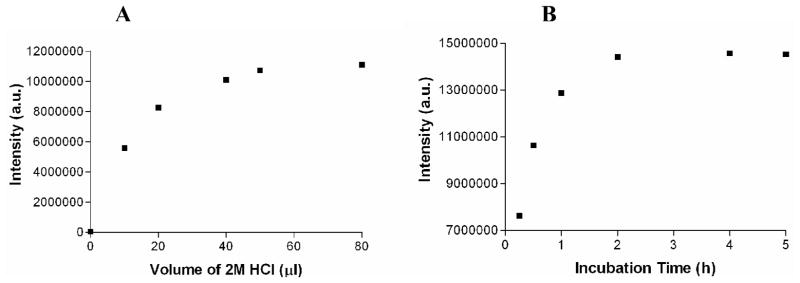
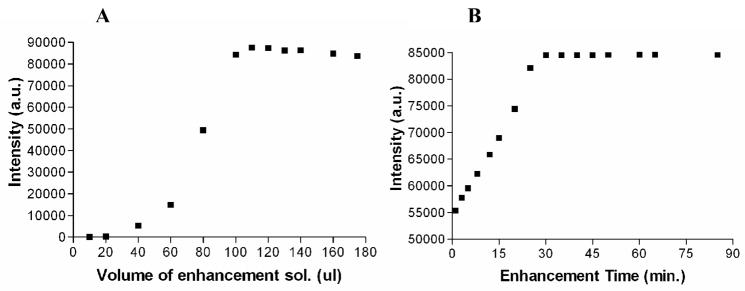
The modified DELFIA protocol consists of additional acidification and neutralization steps prior to the addition of the enhancement solution. HCl was chosen to acidify the Eu(III)-DOTA-labeled ligands because excess Cl− ions do not interfere with the formation of Eu(NTA)3(TPPO)2 complex or the luminescent signal. The conditions (concentrations, time, and temperature) were optimized for the complete release of Eu(III) ions (Fig. 5). By increasing the acid strength or the incubation time, Eu(III) ions are released from DOTA according to a mechanism similar to first order kinetics (Supporting Information). By carefully analyzing the acid strengths we decided to use 2.0 M HCl acid for the acidification step. The optimized volume of 2.0 M HCl and the incubation time with the acid are 50 μL and 2 hours, respectively (Fig. 5).
Fig. 5.
Optimization of acidification step for release of Eu(III). (A) Effect of volume of 2.0 M HCl on Eu(III) release. (B) Effect of incubation time with 2.0 M HCl (50 μL) on Eu(III) release.
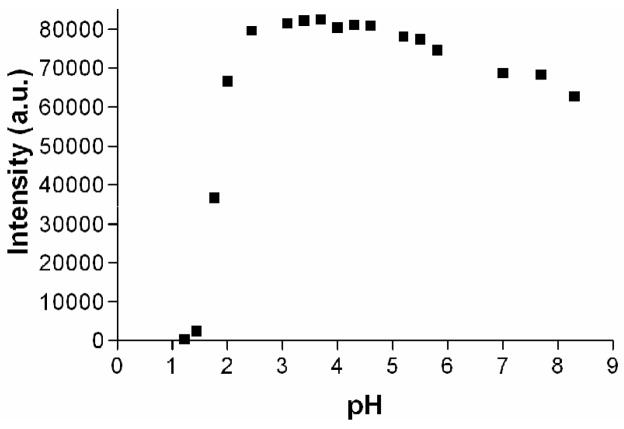
The formation of Eu(NTA)3(TPPO)2 complex depends on the pH of the medium. This has previously been shown in the presence of different percentages of HClO3 or HNO3 acid and EuCl3 [6]. Here we have performed a systematic study using HCl acid. Fig. 6 shows the measured Eu(III)-centered luminescent intensity at different pH values. The intensity drops dramatically below pH=2. This may be due to the instability of the Eu(NTA)3(TPPO)2 complex and the micellar structures under strong acidic conditions. Specially the formation of lanthanide(III)-β-diketonates are not favorable under extremely acidic conditions. The maximum luminescent intensity was observed in the pH range of 3–5 and a slight but noticeable decrement of the intensity was evident at higher pH values. At higher pH conditions, formation of lanthanide-oxo and –hydroxo species are favorable with lower intensities due to the quenching of Eu(III)-based luminescent in the presence of O-H type bonds. Therefore, the final pH of the samples was adjusted back to around 4 using 2.0 M aqueous NaOH before adding the enhancement solution.
Fig. 6.
Optimal pH for DELFIA assay. The luminescent signal shows strong pH dependence. The intensity drops significantly at pH lower than 2. The optimal range for DELFIA assay is pH 3–5. Increasing amounts of HCl were added to 1×10−13 mol of Eu(III) ions dissolved in distilled water. Enhancement solution (100 μL) was added to each sample before reading.
We have revisited the concentration- and volume-dependence of the enhancement step due to the differences in the matrix used for Eu(III)-DOTA-labeled ligands compared to that of DTPA. Fig. 7 indicates enhancement volume- and time-dependence in the presence of Eu(III)-DOTA-NDP-α-MSH with 2.0 M HCl and 2.0 M NaOH. According to the Figure 7B, the samples should be incubated for at least 30 min to fully develop the luminescent intensity. This incubation time is consistent with our prior observations using Eu(III)-DTPA-labeled peptides [6]. In the case of DTPA, the enhancement solution is used both to release Eu(III) ions from the DTPA chelator and to provide photo-active chelators, NTA. On the other hand, HCl is additionally used to release Eu(III) ions from DOTA chelator prior to the addition of the enhancement solution. Therefore, the observed similar enhancement times indicate that the overall formation of the photo-active Eu(III) complexes with the micellar structure is the rate determining step in lanthanide-based dissociation enhanced assays. A significant level of the luminescence was observed within minutes after the addition of the enhancement solution. However the optimal signal was achieved after the incubation of the plates for 30 minutes. Part of this has to be due to the diffusion control of the enhancement solution and the formation of the micellar structures in the presence of TOPO. Addition of 115 μL of the enhancement solution was sufficient to detect the highest luminescent signal intensity in Eu(III)-DOTA-labeled ligands. The value was higher than that observed for DTPA-based ligands (100 μL) due to the presence of NaOH.
Fig. 7.
(A) Enhancement solution volume-dependence on Eu(III)-based luminescence. Luminescence was measured using 1×10−13 mol of Eu-DOTA-PEG9-NDP-α-MSH. Samples were incubated for 1.5 hrs after adding 50 μL of 2.0 M HCl, 55 μL of 2.0 M NaOH, and increasing amount of the enhancement solution. (B) Time-dependence of the Eu(III)-based luminescence. Luminescence measured using 1×10−13 mol of Eu(III) with the addition of 50 μL of 2.0 M HCl, 55 μL of 2.0 M NaOH, and 115 μL of the enhancement solution.
Cell binding assays using Eu(III)-DOTA-labeled NDP-α-MSH
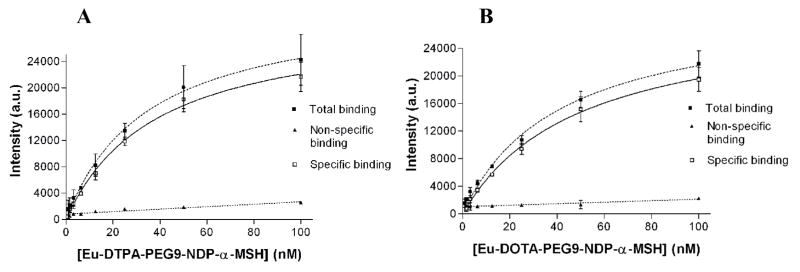
The modified DELFIA protocol was used to evaluate the binding of Eu(III)-DOTA-labeled NDP-α-MSH to HEK293 cells overexpressing hMC4R. As a comparison, traditional DELFIA protocol was also carried out using Eu(III)-DTPA-labeled analogue on the same cell line using an otherwise identical cell preparation and washing conditions. The saturation binding curves using Eu(III)-DTPA-PEG9-NDP-α-MSH and Eu(III)-DOTA-PEG9-NDP-α-MSH are shown in Fig. 8. Non-specific binding was determined in the presence of 100 μM NDP-α-MSH. Kd values determined for Eu(III)-DTPA and Eu(III)-DOTA-labeled ligands are within the same range (38 and 49 nM, respectively). Therefore, the modified DELFIA protocol (using Eu(III)-DOTA-labeled ligand) produced identical results to the traditional DELFIA (using DTPA analogue) in a whole cell binding assay using 96-well plates. The observed Kd value of the Eu(III)-DTPA-labeled NDP-α-MSH through a PEG9 linker (in this study) was slightly higher than that previously observed using ε-aminocaproic acid as the linker (Kd=18.8 nM) [6]. This might be due to the interference of different linker structures on ligand-receptor binding.
Fig. 8.
Saturation binding curves of (A) Eu(III)-DTPA-PEG9-NDP-α-MSH (Kd = 38 nM); and (B) Eu(III)-DOTA-PEG9-NDP-α-MSH (Kd = 49 nM) binding to Hek293 cells overexpressing hMC4R. Non-linear regression data fitting; R2=0.95.
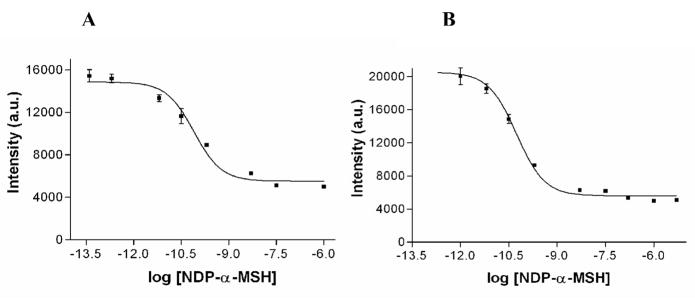
Competitive binding assays against NDP-α-MSH were performed for both Eu(III)-DTPA and -DOTA labeled ligands (Fig. 9). The IC50 values of both Eu(III)-DTPA and –DOTA-labeled ligands were similar (0.079 nM and 0.055 nM, respectively). Hence, the presence of the Eu-DOTA chelate with NDP-α-MSH did not significantly alter its binding affinity compared to that of DTPA.
Fig. 9.
Competitive binding assay curves using (A) Eu(III)-DTPA-PEG9-NDP-α-MSH (IC50 = 0.079 nM); and (B) Eu(III)-DOTA-PEG9-NDP-α-MSH (IC50 = 0.055 nM). Non-linear regression data fitting; R2=0.98
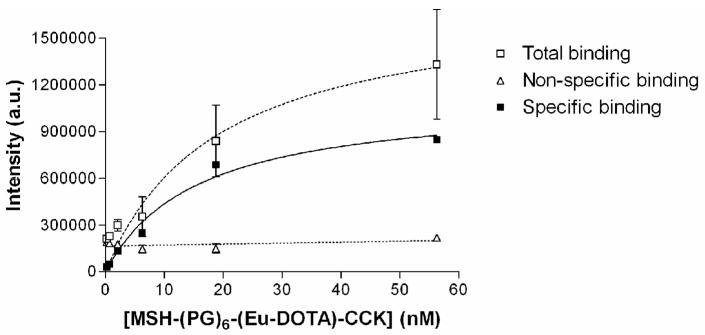
Saturation binding of Eu(III)-DOTA-labeled MSH/CCK heterobivalent ligand
To further verify the use of the modified DELFIA, the Eu(III)-DOTA-labeled heterobivalent ligand containing both MSH and CCK was evaluated in a saturation binding assay. We have previously shown that the heterobivalent ligands can bind to multiple receptors leading to enhanced avidity and selectivity [29]. Fig. 10 shows the saturation binding curve of Eu(III)-DOTA-labeled heterobivalent ligand to Hek293 cells overexpressing both hMC4R and CCK-2 receptors. Non-specific binding was determined in the presence of 10 μM NDP-α-MSH and CCK(8). The Kd value of the labeled ligand was found to be 10 nM. This value is consistent with the Kd value obtained in a single cell level fluorescence binding assay carried out using Cy5 labeled analogue (Data not shown).
Fig. 10.
Saturation binding curve of Eu(III)-DOTA-labeled heterobivalent ligand binding to Hek293 cells overexpressing both hMC4R and CCK-2 receptors (Kd = 10 nM). Non-linear regression data fitting; R2=0.95.
Evaluation of assay quality and validity of the modified DELFIA
The modified DELFIA assay quality was assessed using both a Z′-factor and signal-to-noise ratio analysis (S:N). S:N was calculated by taking into account variation of both the control and the responding sample population using the following equation (see ref. [30] for description and testing). A S:N of 2.5 is generally accepted as indicating excellent assay sensitivity.
| (1) |
where μ = mean signal, σ = standard deviation, p = positive control, and n = negative control.
This parameter was evaluated for Eu(III)-DOTA and Eu(III)-DTPA labeled NDP-α-MSH ligands in their competitive and saturation binding assay platforms (supporting information). The calculated S:N values of DOTA and DTPA labeled ligands near their IC50 values are 15.4 and 8.9, respectively. Consequently DOTA derivatives, using our modified DELFIA protocol, can be utilized in receptor-ligand binding assays with a relatively higher S:N.
The Z′-factor is a standard approach to evaluate the sensitivity of a small preliminary assay for its potential use for a screening assay involving a large number of comparisons such as gene or protein arrays [31]. Z′-factor was calculated according to:
| (2) |
The calculated Z′-factor values of DOTA and DTPA labeled ligands near their IC50 values are 0.79 and 0.64, respectively (supporting information) with a Z′-factor greater than 0.5 indicating adequate assay sensitivity.
We have evaluated the detection limits of Eu(III)-DOTA and Eu(III)-DTPA labeled NDP-α-MSH using two different approaches and their S:N was evaluated (supporting information). Our results confirm that the modified DELFIA can be used to detect attomoles of Eu(III) ions per well in a 96-well plate using Eu(III)-DOTA NDP-α-MSH.
Discussion
Traditional DELFIA technology has its limitations for use with more stable derivatives of lanthanide chelators such as DOTA due to the incomplete release of Eu(III) ions required for detection. To overcome this problem, we have introduced a modified and an optimized DELFIA protocol where Eu(III)-DOTA-labeled ligands were used to evaluate the ligand-receptor interactions. In traditional DELFIA protocol, the essential steps involve incubation of the cells with the labeled-ligands, wash steps to remove the unbound ligands, incubation with the enhancement solution, and TRL measurements. The modified protocol consists of additional acidification and neutralization steps prior to the enhancement step.
Strong acidic conditions with pH less than 2 are necessary for a complete release of Eu(III) ions from DOTA derivative. 2.0 M HCl was chosen for the acidification step and the volume of the acid and the incubation time were optimized. Subsequent neutralization of the medium was essential as the Eu(III)-centered luminescence was not observed under strong acidic conditions and the enhancement solution by itself is not capable of raising the pH to its optimal value (3–5) to detect the maximum signal. Therefore the final pH of the samples was adjusted back to neutral pH or higher using 2.0 M NaOH before the enhancement step. Thus the enhancement solution can now re-adjust the pH of the samples to their optimal value for maximum signal detection.
Our attempts to use moderately volatile acids (such as trifluoroacetic acid) were unsuccessful. A HCl acid concentration of 2.0 M was chosen to successfully release Eu(III) ions within hours and considering the practicability of adjusting the pH back to the optimal value. Higher acid concentrations such as 5.0 M can be used to release Eu(III) ions within minutes but the re-adjustment of the pH was more variable. Many strong acids can serve our purpose. However, an acid must be selected such that the counterions will not interfere with the formation of the lanthanide(III) luminescent complex and its micellar structure. Lastly the neutralization must be tested before the enhancement step for higher sensitivity.
The use of lanthanide-based binding assays has greatly improved the sensitivity of receptor-ligand measurements. With the advent of synthetic procedures for making potent lanthanide-labeled ligands, the need for radiolabeled ligands is diminishing. Moreover, the use of TRL measurements with lanthanides provides a method for substantially decreasing background noise, and thereby improving signal-to-noise ratio and assay sensitivity into the atomolar range. Thus, the modified DELFIA provides a platform to explore novel ligands with highly stable lanthanide chelators such as DOTA for evaluating ligand-receptor interactions. Importantly, the developed protocol is highly reproducible, easy to set up, and is amenable to high-throughput screening.
Supplementary Material
Acknowledgments
The authors thank Professor Eugene Mash and Dr. Jatinder Josan (Department of Chemistry, University of Arizona) for useful discussions and Craig Weber and Ashley Dyett (Department of Physiology, University of Arizona) for technical assistance. We thank Merry Warner and Margie Colie (University of Arizona) for administrative assistance. This research was supported by grants from the U. S. Public Health Service, NIH/National Cancer Institute (RO1-CA123547 and RO1-CA97360) and the Arizona Biomedical Research Commission (# 0707).
Abbreviations used
- DELFIA
dissociation-enhanced lanthanide fluoroimmunoassay
- Eu
europium
- DTPA
diethylenetriaminepentaacetic acid
- NTA
2-naphthoyl trifluoroacetone
- TOPO
trioctylphosphineoxide
- TRL
time-resolved luminescence
- DTTA
diethylenetriaminetetraacetic acid
- DOTA
tetraazacyclododecyltetraacetic acid
- MRI
magnetic resonance imaging
- PARACEST
paramagnetic chemical exchange saturation transfer
- Gd
gadolinium
- In
indium
- α-MSH
α-melanocyte stimulating hormone
- NDP-α-MSH
[Nle4, D-Phe7]-α-MSH
- hMC4R
human melanocortin 4 receptor
- CCK
cholecystokinin
- Fmoc
N-α-9-fluorenylmethoxycarbonyl
- Boc
t-butoxycarbonyl
- tBu
t-butoxy
- Aloc
allyloxycarbonyl
- Pbf
2,2,4,6,7-pentamethyl-dihydrobenzofurane-5-sulfonyl
- Trt
trityl
- HOBt
1-hydroxybenzotriazole
- HBTU
2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexafluorophosphate
- PEGO
19-amino-5-oxo-3,10,13,16-tetraoxa-6-azanonadecan-1-oic acid
- PEG9
8-amino-3,6-dioxaoctanoic acid
- DOTA-NHS ester
DOTA mono N-hydroxysuccinimide ester
- TFA
trifluoroacetic acid
- ESI
electrospray ionization
- MALDI-TOF
matrix-assisted laser desorption ionization time-of-flight
- FT-ICR
Fourier transform ion cyclotron resonance
- tBu
tert-butyl
- DIC
diisopropylcarbodiimide
- TPP
triphenylphosphine
- DMF
dimethylformamide
- THF
tetrahydrofuran
- DCM
dichloromethane
- DIEA
diisopropylethylamine
- DMSO
dimethylsulfoxide
- DMEM
Dulbecco’s modified eagle medium
- FBS
fetal bovine serum
- BSA
bovine serum albumin
Footnotes
Supplementary materials associated with this article can be found, in the online version, at
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jones DS, Silverman AP, Cochran JR. Developing therapeutic proteins by engineering ligand-receptor interactions. Trends in Biotech. 2008;26:498–505. doi: 10.1016/j.tibtech.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Bunzli JCG, Piguet C. Taking advantage of luminescent lanthanide ions. Chem Soc Rev. 2005;34:1048–1077. doi: 10.1039/b406082m. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Parker D, Pal R, Poole RA, Cann MJ. A europium complex that selectively stains nucleoli of cells. J Am Chem Soc. 2009;128:2294–2299. doi: 10.1021/ja056303g. [DOI] [PubMed] [Google Scholar]
- 4.Moore EG, Samuel APS, Raymond KN. From antenna to assay: Lessons learned in lanthanide luminescence. Acc Chem Res. 2009;42:542–552. doi: 10.1021/ar800211j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comby S, Imbert D, Vandevyver C, Bunzli JCG. A novel strategy for the design of 8-hydroxyquinolinate-based lanthanide bioprobes that emit in the near infrared range. Chem Eur J. 2007;13:936–944. doi: 10.1002/chem.200600964. [DOI] [PubMed] [Google Scholar]
- 6.Handl HL, Vagner J, Yamamura HI, Hruby VJ, Gillies RJ. Lanthanide-based time-resolved fluorescence of in cyto ligand-receptor interactions. Anal Biochem. 2004;330:242–250. doi: 10.1016/j.ab.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Hemmila I, Dakubu S, Mukkala VM, Siitari H, Lovgren T. Europium as a label in time-resolved immunofluorometric assays. Anal Biochem. 1984;137:335–343. doi: 10.1016/0003-2697(84)90095-2. [DOI] [PubMed] [Google Scholar]
- 8.Wu FB, Han SQ, Xu T, He YF. Sensitive time-resolved fluoroimmunoassay for simultaneous detection of serum thyroid-stimulating hormone and total thyroxin with Eu and Sm as labels. Anal Biochem. 2003;314:87. doi: 10.1016/s0003-2697(02)00624-3. [DOI] [PubMed] [Google Scholar]
- 9.Schoket B, Doty WA, Vincze I, Strickland PT, Ferri GM, Assennato G, Poirier MC. Increased sensitivity for determination of polycyclic aromatic hydrocarbon-DNA adducts in human DNA samples by dissociation-enhanced lanthanide fluoroimmunoassay (DELFIA) Cancer Epidem Biomark Preven. 1993;2:349–353. [PubMed] [Google Scholar]
- 10.Kashem MA, Nelson RM, Yingling JD, Pullen SS, Prokopowicz AS, Jones JW, Wolak JP, Rogers GR, Morelock MM, Snow RJ, Homon CA, Jakes S. Three mechanistically distinct kinase assays compared: Measurement of intrinsic atpase activity identified the most comprehensive set of ITK inhibitors. J Biomol Screening. 2007;12:70–83. doi: 10.1177/1087057106296047. [DOI] [PubMed] [Google Scholar]
- 11.Elliott CT, Francis KS, McCaughey WJ. Investigation of dissociation enhanced lanthanide fluoroimmunoassay as an alternative screening test for veterinary drug residues. Analyst. 1994;119:2565–2569. doi: 10.1039/an9941902565. [DOI] [PubMed] [Google Scholar]
- 12.Waddleton D, Ramachandran C, Wang Q. Development of a time-resolved fluorescent assay for measuring tyrosine-phosphorylated proteins in cells. Anal Biochem. 2002;309:150–157. doi: 10.1016/s0003-2697(02)00292-0. [DOI] [PubMed] [Google Scholar]
- 13.Butcher H, Kennette W, Collins O, Demoor J, Koropatnick J. A sensitive time-resolved fluorescent immunoassay for metallothionein protein. J Immun Methods. 2003;272:247–256. doi: 10.1016/s0022-1759(02)00441-6. [DOI] [PubMed] [Google Scholar]
- 14.Deng SJ, Liu W, Simmons CA, Moore JT, Tian G. Identifying substrates for endothelium-specific TIE-2 receptor tyrosine kinase from phage-displayed peptide libraries for high throughput screening. Combin Chem High Throughput Screen. 2001;4:525–533. doi: 10.2174/088800199276958. [DOI] [PubMed] [Google Scholar]
- 15.Franz KJ, Nitz M, Imperiali B. Lanthanide-binding tags as versatile protein coexpression probes. Chembiochem. 2003;4:265–271. doi: 10.1002/cbic.200390046. [DOI] [PubMed] [Google Scholar]
- 16.Handl HL, Gillies RJ. Lanthanide-based luminescent assays for ligand-receptor interactions. Life Sciences. 2005;77:361–371. doi: 10.1016/j.lfs.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Nishioka T, Fukui K, Matsumoto K. Lanthanide chelates as luminescent labels in biomedical analyses. Handbook on the physics and chemistry of rare earths. 2007;37:171–216. [Google Scholar]
- 18.Wilkinson D. A one-step fluorescent detection method for lipid fingerprints; Eu(TTA)3.2TOPO. Forensic Sci Int. 1999;99:5–23. doi: 10.1016/s0379-0738(98)00176-5. [DOI] [PubMed] [Google Scholar]
- 19.Handl HL, Vagner J, Yamamura HI, Hruby VJ, Gillies RJ. Development of a lanthanide-based assay for detection of receptor-ligand interactions at the δ-opioid receptor. Anal Biochem. 2005;343:299–307. doi: 10.1016/j.ab.2005.05.040. [DOI] [PubMed] [Google Scholar]
- 20.Bari LD, Pescitelli G, Sherry AD, Woods M. Structural and chiroptical properties of the two coordination isomers of YbDOTA-type complexes. Inorg Chem. 2005;44:8391–8398. doi: 10.1021/ic0511118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo B, Pagel MD. A PARACEST MRI contrast agent to detect enzyme activity. J Am Chem Soc. 2006;128:14032–14033. doi: 10.1021/ja063874f. [DOI] [PubMed] [Google Scholar]
- 22.De Leon-Rodriguez LM, Lubag AJM, Malloy CR, Martinez GV, Gillies RJ, Sherry AD. Responsive MRI agents for sensing metabolism in vivo. Acc Chem Res. 2009 doi: 10.1021/ar800237f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka K, Fukase K. PET (positron emission tomography) imaging of biomolecules using metal DOTA complexes: A new collaborative challenge by chemists, biologists, and physicians for future diagnostics and exploration of in vivo dynamics. Org Biomol Chem. 2008;6:815–828. doi: 10.1039/b718157b. [DOI] [PubMed] [Google Scholar]
- 24.Caravan P, Ellison JJ, Mcmurry TJ, Lauffer RB. Gadolinium(III) chelates as MRI contrast agents: Structure, dynamics, and applications. Chem Rev. 1999;99:2293–2352. doi: 10.1021/cr980440x. [DOI] [PubMed] [Google Scholar]
- 25.Kovacs Z, De Leon-Rodriguez LM. Conjugation of 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetracetic acid (DOTA) and its derivatives to peptides: Synthesis, applications and future prospects. Mini-Reviews in Org Chem. 2007;4:281–291. [Google Scholar]
- 26.De Leon-Rodriguez LM, Kovacs Z. The synthesis and chelation chemistry of DOTA-peptide conjugates. Bioconjug Chem. 2008;19:391–402. doi: 10.1021/bc700328s. [DOI] [PubMed] [Google Scholar]
- 27.Sawyer TK, Sanfilippo PJ, Hruby VJ, Engel MH, Heward CB, Burnett JB, Hadley ME. 4-norleucine, 7-D-phenylalanine-alpha-melanocyte-stimullating hormone: A highly potent alpha-melanotropin with ultralong biological activity. PNAS USA. 1980;77:5754–5758. doi: 10.1073/pnas.77.10.5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grieco P, Lavecchia A, Cai MY, Trivedi D, Weinberg D, MacNeil T, Van der Ploeg LHT, Hruby VJ. Structure-activity studies of the melanocortin peptides: Discovery of potent and selective affinity antagonists for the hMC3 and hMC4 receptors. J Med Chem. 2002;45:5287–5294. doi: 10.1021/jm0202526. [DOI] [PubMed] [Google Scholar]
- 29.Vagner J, Xu L, Handl L, Josan JS, Morse DL, Mash EA, Gillies RJ, Hruby VJ. Heterobivalent ligands crosslink multiple cell-surface receptors: The human melanocortin-4 and δ-opioid receptors. Angew Chem Int Ed. 2008;47:1685–1688. doi: 10.1002/anie.200702770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang XD. A pair of new statistical parameters for quality control in RNA interference high-throughput screening assays. Genomics. 2007;89:552–561. doi: 10.1016/j.ygeno.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Ji-Hu, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.