Abstract
The benzylisoquinoline alkaloids (BIAs) are a diverse class of metabolites that exhibit a broad range of pharmacological activities and are synthesized through plant biosynthetic pathways comprised of complex enzyme activities and regulatory strategies. We have engineered yeast to produce the key intermediate reticuline and downstream BIA metabolites from a commercially available substrate. An enzyme tuning strategy was implemented that identified activity differences between variants from different plants and determined optimal expression levels. By synthesizing both stereoisomer forms of reticuline and integrating enzyme activities from three plant sources and humans, we demonstrated the synthesis of metabolites in the sanguinarine/berberine and morphinan branches. We also demonstrated that a human P450 enzyme exhibits a novel activity in the conversion of (R)-reticuline to the morphinan alkaloid salutaridine. Our engineered microbial hosts offer access to a rich group of BIA molecules and associated activities that will be further expanded through synthetic chemistry and biology approaches.
The engineering of biological systems holds much promise for the synthesis of new chemicals, materials and products that will address pressing human needs in energy, the environment, sustainability and human health. Synthetic biology is advancing foundational technologies in support of engineering biological systems1 and furthering the complexity of systems that can be forward engineered2. Such capabilities are enabling new approaches to biosynthesis that integrate diverse activities from natural sources into heterologous hosts and can often provide new insights into nature's own biosynthetic strategies. Several recent efforts directed toward the construction of metabolic pathways in microbial hosts have resulted in new biological systems that produce a variety of disparate natural products3–5.
The BIAs are a large and structurally diverse family of plant secondary metabolites derived from tyrosine (1) that exhibit a wide range of pharmacological activities. For example, the key intermediate reticuline (2) has been shown to (i) inhibit Ca2+ transport, resulting in hypotensive and antispasmodic effects6, (ii) act as a central nervous system depressant7 and (iii) accelerate hair growth8. The antimicrobial agents sanguinarine (3) and berberine (4) have also shown potential as anticancer therapeutics9. Other BIAs have been studied for their antioxidative properties10, evaluated as ligands for neuronal nicotinic acetylcholine receptors11 and identified as anti-HIV therapeutics12. More complex molecules, such as the bisbenzylisoquinoline alkaloid tetrandrine (5), have been used to treat autoimmune disorders13 and hypertension14, whereas other BIAs demonstrate similar vasorelaxing properties15. Among the most widely used BIAs are the analgesics morphine (6) and codeine (7) and the muscle relaxant (+)-tubocurarine (8)16,17.
Although certain alkaloids such as morphine and berberine can reach relatively high levels in plants, there is currently no general source for obtaining significant quantities of many of the BIA molecules. The molecular and functional diversity of BIAs accessible from natural sources is limited, as many of the native plant hosts display distinct alkaloid profiles with accumulation of only a select few BIA products18. In addition, synthetic chemistry approaches have not been effective at producing many of the BIAs owing to their molecular complexity17. Efforts have been directed toward elucidating the associated biosynthetic pathways in plants and cloning and characterizing many of the related enzyme activities17. However, metabolic engineering efforts directed at increasing the accumulation of BIA metabolites in the native plant hosts18–20 have been largely limited by the lack of tools for forward engineering biosynthetic pathways in plants21 and the complexity of the BIA pathway and its regulation22.
The reconstitution of a BIA biosynthetic pathway in an engineered microbial host presents several advantages over traditional efforts directed toward the native plant hosts, including the isolated production of key intermediate molecules, rapid biomass accumulation, ease in purification and the availability of genetic tools for forward engineering and pathway optimization. Recent work demonstrated reticuline production from dopamine (9) at ~10 mg l–1 in Escherichia coli expressing three pathway enzymes from Coptis japonica and a bacterial monoamine oxidase23. Yeast strains expressing one or two pathway enzymes from C. japonica were added in a co-culture system along with the engineered bacteria to demonstrate additional conversion steps on (S)-reticuline (2a). Though co-culture systems can be used to synthesize BIA metabolites in microbial hosts, these systems face limitations in metabolite transport across the cell membranes of the two microorganisms and challenges in fermentation scale-up. In addition, given that many of the BIA pathway enzymes are available from diverse plant hosts, it is of interest to construct synthetic pathways comprised of different enzyme variants to examine relative activities and synergistic interactions in an engineered host.
We have engineered yeast strains expressing combinations of enzymes from three plant sources and humans as microbial hosts for the production of a wide array of BIA metabolites. In particular, we examined the ability of different combinations of three recombinant enzymes from Thalictrum flavum and Papaver somniferum to produce the early BIA metabolite reticuline from norlaudanosoline (10). In addition, we describe a new enzyme tuning strategy that can be generally applied to determine optimal enzyme expression levels to conserve cellular resources and improve growth and production rates without compromising pathway flux. These studies demonstrate that reticuline production levels vary from ~10 to 150 mg l–1 depending on the combination of enzyme variants used, which highlights the differences in activities and interactions among pathway variants in the engineered host. We also engineered yeast strains that produce BIA metabolites along two of the major branches from reticuline: the sanguinarine and berberine branch and the morphinan branch. Expression of three downstream enzymes from T. flavum and P. somniferum and a reductase partner from Arabidopsis thaliana resulted in the synthesis of (S)-scoulerine (11), (S)-tetrahydrocolumbamine (12) and (S)-tetrahydroberberine (13) from (S)-reticuline. In addition, expression of a human P450 enzyme and its native reductase partner resulted in the synthesis of salutaridine (14) from (R)-reticuline (2b), thereby demonstrating a novel activity for this P450 enzyme. As the enzymatic activities leading to salutaridine have not yet been cloned and characterized from plant hosts24, our synthetic pathway highlights the diversity of BIA products that can be synthesized in microbial hosts by combining activities from diverse sources.
RESULTS
Synthesis of reticuline from norlaudanosoline in yeast
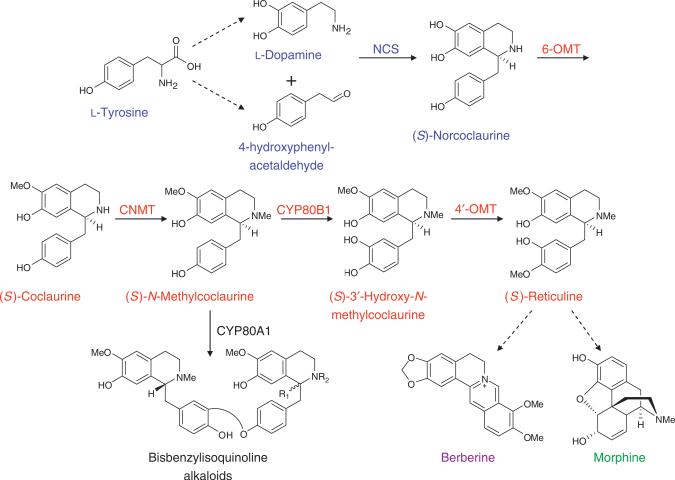
In native plant hosts such as T. flavum and P. somniferum, the first committed step in BIA biosynthesis is the condensation of dopamine and 4-hydroxyphenylacetaldehyde (4-HPA; 15) catalyzed by norcoclaurine synthase (NCS) to produce (S)-norcoclaurine (16) (Scheme 1). This natural intermediate undergoes a series of methylation reactions catalyzed by norcoclaurine 6-O-methyltransferase (6-OMT), coclaurine-N-methyltransferase (CNMT) and 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase (4′-OMT) and a hydroxylation reaction catalyzed by cytochrome P450 80B1 (CYP80B1) to produce the major branch point intermediate (S)-reticuline. (S)-Reticuline is converted to downstream metabolites along various branches, ultimately resulting in the synthesis of pharmacologically relevant molecules such as morphine and berberine.
Scheme 1.
The native BIA pathway. Color schemes for the metabolites and enzymes are as follows: upstream portion of the pathway to the first BIA backbone molecule norcoclaurine, blue; middle portion of the pathway from norcoclaurine to the branch point metabolite reticuline, red; sanguinarine/berberine branch, purple; morphine branch, green. Conversion steps for which the entire set of enzymes has not been completely elucidated or cloned are indicated by dashed arrows.
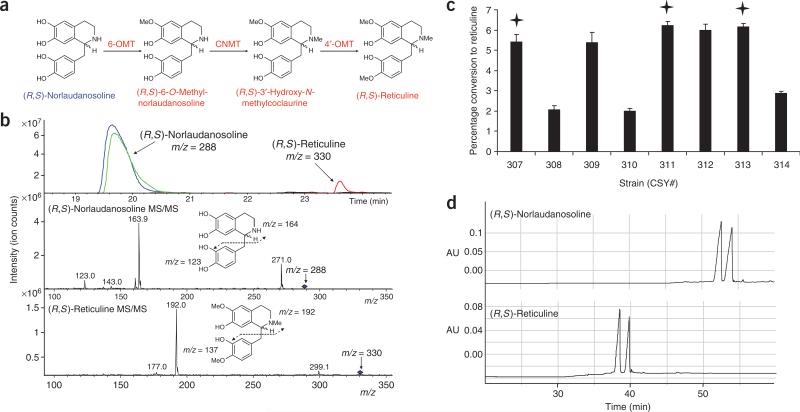
The total synthesis of the BIA backbone from tyrosine represents a substantial engineering challenge, as many of the plant enzymes that perform the early conversion reactions have not yet been isolated and cloned. As an alternative route, we have constructed a synthetic BIA pathway in S. cerevisiae that converts the commercially available substrate (R,S)-norlaudanosoline to (R,S)-reticuline (Fig. 1a). Norlaudanosoline differs from the natural substrate norcoclaurine by the presence of a 3′-OH group that is added to the BIA backbone by CYP80B1 in the native pathway. Therefore, our synthetic BIA pathway is comprised of three heterologous AdoMet-dependent methyltransferase enzymes (6-OMT, CNMT and 4′-OMT) that convert norlaudanosoline to reticuline. Norlaudanosoline was preferred over dopamine as the starting substrate in this work as the initial conversion step from dopamine proved to be extremely inefficient in yeast cells expressing either the E. coli or human monoamine oxidase enzyme variant with or without the T. flavum NCS, requiring fed dopamine concentrations of ~100 mM (data not shown).
Figure 1.
Microbial production of (R,S)-reticuline. (a) The engineered BIA pathway for the synthesis of (R,S)-reticuline from (R,S)-norlaudanosoline. The fed substrate, (R,S)-norlaudanosoline, is indicated in blue. Color schemes follow that defined in Scheme 1. (b) LC-MS/MS analysis of the growth medium of engineered yeast strains supplemented with 1 mM norlaudanosoline and grown for 48 h confirms reticuline production. Extracted ion chromatograms are shown for norlaudanosoline in the wild-type (blue) and CSY307 (green) strains and for reticuline in the wild-type (black, no visible peak) and CSY307 (red) strains. Reticuline is identified as the m/z = 330 ion eluting at 23.6 min showing the expected fragments m/z = 192 and 137 produced by cultures of CSY307 and similar engineered strains. Control experiments in which strains were missing any one of the three required enzymes or grown in the absence of substrate did not accumulate the metabolite peak identified as reticuline. (c) Reticuline production is dependent on the combination of enzyme variants expressed by the engineered yeast strains. Reticuline production is reported as percentage substrate conversion from engineered yeast strains harboring BIA expression constructs for combinatorial expression of different 6-OMT, CNMT and 4′-OMT enzyme variants supplemented with 1 mM norlaudanosoline and grown for 48 h. Stars indicate enzyme combinations used in strains for stable expression. Data are reported as mean ± s.d. from at least three independent experiments. (d) Chiral analysis of (R,S)-norlaudanosoline and (R,S)-reticuline (converted by CSY288). Separation of stereoisomers was performed through capillary electrophoresis on reticuline fractions collected from the LC column.
Yeast strains were engineered to express one or more of the heterologous BIA pathway enzymes from P. somniferum and T. flavum. We constructed yeast BIA expression vectors, where each construct enabled the expression of one or two enzymes from constitutive TEF1 promoters (Supplementary Fig. 1 online). We also constructed single-gene expression plasmids to characterize each enzyme variant individually. The resulting engineered yeast strains were grown in the presence of norlaudanosoline (or appropriate substrate) and assayed for the expected products. We tested the 6-OMT and 4′-OMT activities using norlaudanosoline and laudanosoline (17) as substrates and the CNMT activities using norlaudanosoline and 6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline (18) as substrates (Supplementary Fig. 2 online). As standards are not commercially available for the metabolites of interest, after separation by HPLC, positive identification of BIAs was confirmed using selective reaction monitoring and tandem mass spectrometry (LC-MS/MS), in which the resulting ion fragments are characteristic for a specific molecular structure. Enzymes from both plant species performed the expected methylation reactions on the provided substrates when expressed individually, and yeast strains lacking the heterologous coding sequences were not able to methylate the examined substrates. No differences were observed in the relative activities of the 6-OMT and the 4′-OMT variants, which methylated both norlaudanosoline (giving 19 and 20, respectively) and laudanosoline (giving 21 and 22, respectively) in the expected positions. However, the P. somniferum CNMT variant methylated substantially more of the norlaudanosoline substrate, accumulating over six times the amount of laudanosoline relative to the T. flavum variant. Conversely, the T. flavum variant produced ~20 times the amount of N-methylated 6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline (23), which indicates the different substrate preferences of these orthologous proteins.
We confirmed full-length expression of all methyltransferase variants by western blot analysis using C-terminally tagged constructs (Supplementary Fig. 3 online). Protein expression levels from high-copy plasmids were within two- to three-fold of that observed from a highly expressed yeast-enhanced green fluorescent protein (GFP) variant, which indicates sufficient expression of the methyltransferase enzymes in the microbial host. In addition, the enzyme variants were present at similar levels such that any observed activity differences between variants cannot be attributed to differences in expression but rather must be due to some inherent property of the enzymes. The results also suggest that both CNMT variants were translated more efficiently and/or have a longer half-life in yeast relative to the 6-OMT and 4′-OMT enzymes.
We cotransformed yeast cells with all possible combinations of the 6-OMT, CNMT and 4′-OMT enzymes from T. flavum and P. somniferum (Supplementary Table 1 online). All strains harboring different combinations of the three methyltransferase enzyme variants demonstrated production of reticuline from norlaudanosoline as verified through LC-MS/MS (Fig. 1b). Certain enzyme combinations exhibited greater reticuline accumulation as estimated by percent conversion or by the ratio of the extracted ion chromatogram peak areas of reticuline to norlaudanosoline (Fig. 1c). Notably, all but one strain containing the T. flavum CNMT variant produced ~60% less reticuline than the strains with the P. somniferum CNMT variant, which indicates that the latter variant demonstrated higher activity in this synthetic pathway. This activity difference can be attributed to differences in the substrate affinities of the CNMT variants observed in the single enzyme studies, as the P. somniferum CNMT exhibited higher activity on the pathway substrate norlaudanosoline (Supplementary Fig. 2). Measurements of 5–10% conversion of norlaudanosoline agreed with other estimates of reticuline concentration of ~100 μM (from 1 mM norlaudanosoline) based on comparative peak area analysis of structurally similar standards. Although BIA metabolites up to reticuline exist only in the S conformation in plants, our studies demonstrated that the three methyltransferases accept both stereoisomers as substrates equally in vivo to yield a racemic mixture of (R,S)-reticuline from (R,S)-norlaudanosoline25,26 (Fig. 1d).
Estimation of BIA metabolite levels in the growth medium and cell extract indicated that the norlaudanosoline concentration drop across the membrane was ~10- to 30-fold and that the reticuline-to-norlaudanosoline ratio was slightly greater inside the cell relative to the growth medium (Supplementary Fig. 4 online). The results supported a passive diffusion transport mechanism in the microbial host and confirmed that the substrate was accessible to the intracellular BIA enzymes and that the synthesized metabolites accumulated in the growth medium. This property greatly simplifies metabolite profiling, as production levels can be estimated by direct analysis of the growth medium without rigorous extraction or purification steps. However, the transport of BIA substrates across the cell membrane is somewhat limiting and highlights the importance of reconstructing this pathway in a single microbial host to avoid inefficiencies due to transport of metabolites across two cell membranes.
BIA production in the yeast strains is substrate limited
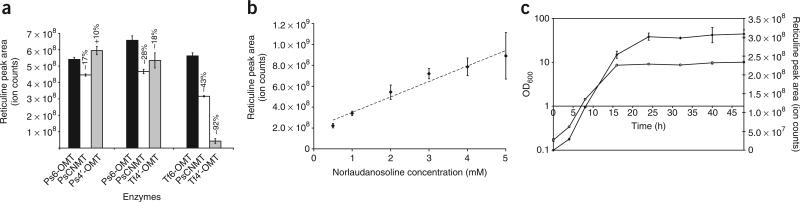
We selected three combinations of the methyltransferase enzyme variants that demonstrated high conversion of norlaudanosoline to reticuline for stable expression in yeast (CSY288, CSY334, CSY456; Supplementary Table 1). Expression of heterologous enzymes from the chromosome is anticipated to result in more consistent and stable expression over time, to facilitate expansion of the synthetic pathway and to enable cultures to be grown in rich medium without selective pressure. However, expression levels from chromosomal integrations are expected to be lower than those from the high-copy plasmid system due to the reduction in DNA copy number. Reticuline production levels compared favorably in all stable expression strains with only one combination of enzymes showing a decrease in production greater than 30% upon integration of the constructs (Fig. 2a). The results indicated that the P. somniferum 6-OMT variant outperformed the T. flavum variant at lower expression levels and exhibited higher specific activity in the synthetic pathway.
Figure 2.
Effects of enzyme levels, substrate levels and culture time on reticuline production levels. (a) Reticuline production is not significantly impacted at lower enzyme expression levels. Reticuline production as measured by LC-MS peak area from three strains engineered to stably express the indicated enzyme variants from the TEF1 promoter (CSY288, CSY334, CSY456; white) or the mutant TEF7 promoter (CSY448, CSY449, CSY458; gray) were compared to strains expressing the enzymes from plasmid-based constructs (CSY307, CSY311, CSY313; black). The percentage change in production observed from the stable strains after 48 h growth in medium supplemented with 2 mM norlaudanosoline is indicated. (b) Reticuline production increases with substrate concentration. Reticuline production was measured in the growth medium of CSY288 supplemented with a range of norlaudanosoline concentrations after 48 h growth. (c) Reticuline production increases with the optical density at 600 nm (OD600) of the culture. Reticuline production (solid diamonds) and OD600 (open circles) were measured at the indicated time points in the growth medium of CSY288 supplemented with 2 mM norlaudanosoline added at t = 0. All data are reported as mean ± s.d. from at least three independent experiments.
The results from the analysis of reticuline production in the stable expression strains suggested that substrate conversion was not severely hindered by enzyme expression levels. We examined the effects of substrate concentration and growth phase on reticuline production levels from cultures of CSY288. Reticuline accumulation increased roughly in proportion to the initial concentration of norlaudanosoline in the medium between 0.5 to 5 mM such that percent conversion was between 7% and 10% in all samples, which supports a substrate limitation model (Fig. 2b). Substrate limitation in our synthetic BIA pathway was not unexpected because in addition to potential transport issues, norlaudanosoline and 6-O-methylnorlaudanosoline are not the natural substrates for 6-OMT and CNMT, respectively. For example, kinetic characterization studies on the 6-OMT variant from C. japonica determined a Km value of 2.23 mM for (R,S)-norlaudonosoline, with a relative activity of 76% compared with (S)-norcoclaurine25. Although the 6-OMT variant from P. somniferum used here has a reported Km value of 10 μM for norcoclaurine26, the value may be higher for norlaudanosoline. We also examined the timescale of production under conditions that model a batch fermentation run and observed substantial reticuline production shortly after substrate addition to a shake flask culture seeded with CSY288. The production rate slowed over time, but reticuline continued to accumulate as cells entered the stationary growth phase (Fig. 2c).
Tuning enzyme levels with a new titration strategy
Pathway optimization strategies often require an analysis of enzyme expression levels and their effects on metabolite accumulation and strain growth rate. Systems that enable heterologous enzymes to be expressed at a minimum level while maintaining maximum pathway flux and product accumulation are desired to avoid wasting cellular resources synthesizing excess proteins. Such strategies become more critical as an engineered pathway is extended to include additional enzymes that will further tax cellular resources. Optimization of pathway enzyme levels should result in an improved strain growth rate and a markedly reduced production time.
To optimize BIA enzyme expression levels, we developed a system that allows each enzyme to be titrated independently. We replaced the constitutive TEF1 promoter with the GAL1-10 galactose-inducible promoter for one of the three BIA enzyme coding sequences in our stable strains to allow tunable expression of one heterologous enzyme while holding the other two constant. In addition, we deleted the GAL2 permease from each strain to enable titratable control over enzyme levels. Deletion of the GAL2 permease has been previously shown to result in a more homogenous, linear induction response from the GAL network relative to the wild-type all-or-none switch-like response27. Our system design resulted in a series of engineered strains stably expressing the BIA enzymes in which levels of one of the three methyltransferases were precisely regulated according to galactose (24) concentration (CSY325–CSY329; Supplementary Table 1).
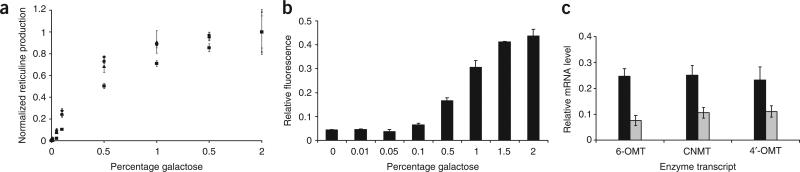
We used the tunable strains to determine relationships between galactose concentration, enzyme expression levels and reticuline production. The titratable yeast strains were grown in 1 mM norlaudanosoline and varying galactose concentrations, and reticuline production was analyzed at 24 h after substrate addition. The enzyme titration experiments demonstrated that in nearly all strains at ~0.5% galactose, conversion of norlaudanosoline to reticuline reached over 70% of the levels attained from fully induced conditions at 2% galactose (Fig. 3a). All strains demonstrated production comparable to the parent strains at maximum induction levels. Only the strain containing the GAL-inducible T. flavum 6-OMT (CSY329; Supplementary Table 1) did not show changes in production corresponding to galactose concentration, as reticuline production was similarly low at all points; this suggests that higher expression levels were necessary to observe activity of this variant in our system (data not shown).
Figure 3.
A new strategy for tuning enzyme expression levels. (a) Reticuline production as a function of galactose concentration in engineered reticuline-producing strains expressing one heterologous enzyme from the titratable GAL expression system; production is normalized to each strain in 2% galactose. CSY325, diamonds; CSY326, squares; CSY327, circles; CSY328, triangles. Cells were grown overnight in noninducing-nonrepressing medium and backdiluted into medium containing the indicated galactose concentration. Following 4 h of induction, 1 mM norlaudanosoline was added and supernatants were analyzed for reticuline production after 24 h. (b) GFP reporter strains were used to guide the determination of relative protein expression levels for the enzyme titration studies. Relative fluorescence data are reported for CSY429 normalized to CSY428. Cells were grown overnight in noninducing-nonrepressing medium and backdiluted into medium containing the indicated galactose concentration. Fluorescence measurements were normalized to OD600 for each sample and taken during the exponential growth phase. (c) qRT-PCR analysis confirms trends in relative transcript levels from the TEF1 and TEF7 integrated expression systems compared with the high-copy plasmid-based systems. Transcript levels in representative stable strains using the TEF1 promoter (CSY288; black) and the TEF7 promoter (CSY448; gray) are normalized to levels from a high-copy plasmid expression system (CSY307). All data are reported as mean ± s.d. from at least two independent experiments.
The relationship between galactose concentration and relative expression levels was obtained indirectly using an analogous fluorescent reporter system. We constructed integration cassettes containing a green fluorescent protein (yEGFP3) under the control of the constitutive TEF1 promoter or the inducible GAL1-10 promoter in a GAL2Δ background to make strains CSY428 and CSY429 (Supplementary Table 1). We compared the relative fluorescence levels from the GAL1-10 promoter at various galactose concentrations to fluorescence levels observed from the TEF1 promoter. The reporter protein titration experiments demonstrated that the expression from the tunable GAL promoter system at 0.5% galactose translated to ~16% expression relative to the native TEF1 promoter (Fig. 3b). The results indicated that the transcriptional activity of the heterologous enzyme promoters could be substantially reduced while maintaining maximal substrate conversion to reticuline.
We optimized BIA enzyme expression levels based on our titration assay results by designing integration cassettes in which the level of expression from the promoter system was minimized without compromising production. We used a mutated TEF promoter library28 in which TEF1 promoter variants with altered levels of gene expression had been generated and characterized. Our titration assays indicated that optimal expression levels for each of the BIA enzymes tested, with the possible exception of P. somniferum 4′-OMT, correspond to ~16% of the native TEF1 promoter, approximating that of the TEF7 mutant. We replaced the TEF1 promoter with the TEF7 promoter in our chromosomal integration cassettes and constructed optimized reticuline-producing strains. TEF7-substituted versions of CSY288 and CSY334 (CSY448, CSY449; Supplementary Table 1) showed comparable production levels (Fig. 2a). However, CSY458 showed greatly reduced production compared with CSY456 and other TEF7-substituted strains. Since the only difference between CSY458 and CSY449 is the 6-OMT variant, the data further supported that this step can become limiting at low expression levels and that the P. somniferum 6-OMT exhibited higher specific activity than the T. flavum variant in the synthetic pathway. We verified trends in relative transcript levels between the TEF1 and TEF7 integrated expression systems and the high-copy plasmid-based system by real-time quantitative reverse transcription PCR (qRT-PCR) (Fig. 3c). Growth rates were not significantly and reproducibly different between the TEF1 and TEF7 strains; however, this optimization strategy, which resulted in reduced metabolic load on the cell without compromising reticuline production levels, will likely prove more important upon further extensions of the pathway. In addition, this tuning strategy can be generally applied to other recombinant pathways in yeast to determine and set the minimal expression level of heterologous enzymes for a desired product yield.
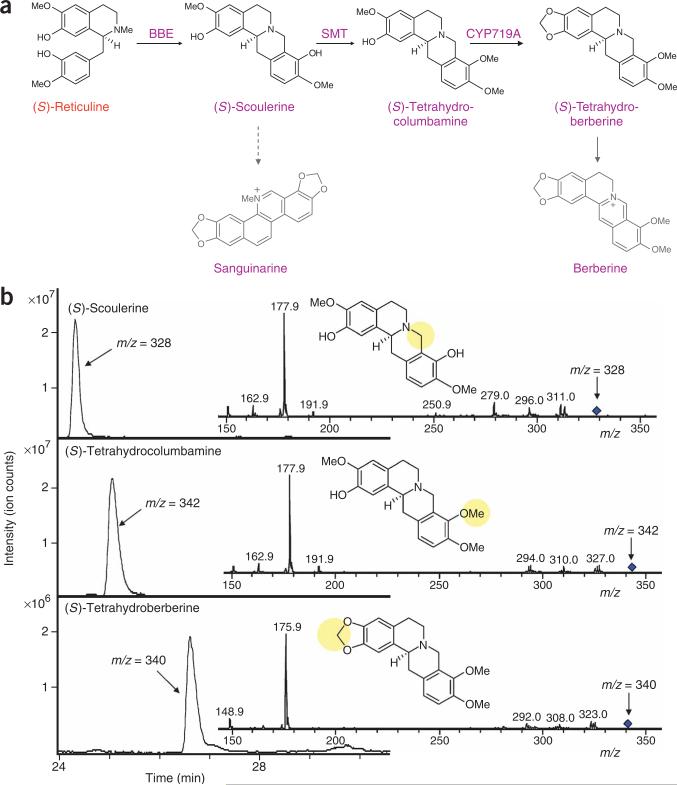
Synthesis of sanguinarine and berberine intermediates
Reticuline represents a major branch point intermediate in plant secondary metabolism from which a wide variety of BIA metabolites can be derived, including sanguinarine, berberine, protopine (25), magnoflorine (26) and morphinan alkaloids. The enzymes in these downstream BIA branches are of particular interest, as many exhibit complex biosynthetic activities. We examined the synthesis of meta-bolites along a major branch, the sanguinarine and berberine branch, by expressing additional enzymes from the native plant pathways as a demonstration of the diverse natural products and activities that can be produced in our engineered yeast strains. This family of benzophenanthridine alkaloids has generated interest for their pharmacological activities, most notably as antimicrobial agents.
The first conversion step in the sanguinarine and berberine branch of the BIA pathway is performed by the berberine bridge enzyme (BBE), which catalyzes the oxidative cyclization of the N-methyl moiety of (S)-reticuline into the berberine bridge carbon of (S)-scoulerine29 (Fig. 4a). This unique reaction forms the protoberberine carbon skeleton via a methylene iminium ion intermediate and cannot be replicated through synthetic chemistry approaches. We constructed a plasmid expressing the P. somniferum BBE complementary DNA for transformation into our reticuline-producing yeast strains CSY288 and CSY334 (CSY336, CSY338; Supplementary Table 1). LC-MS/MS analysis demonstrated that the engineered strains produce (S)-scoulerine (Fig. 4b). The relative peak areas of the appropriate ions indicated ~40% conversion of (R,S)-reticuline to (S)-scoulerine with an effective conversion of ~80% based on an equal mixture of stereoisomers. In addition, we also tested enzyme truncations of the first 25 and 41 amino acids to remove the N-terminal signal sequence30. Neither truncated variant demonstrated increased production of (S)-scoulerine, and the Δ41 truncation showed slightly compromised production (data not shown).
Figure 4.
Microbial production of BIA metabolites along the sanguinarine and berberine branches. (a) Pathway for the synthesis of sanguinarine and berberine metabolites from the key intermediate (S)-reticuline. Color schemes follow that defined in Scheme 1. Shaded portions of the pathway indicate steps not reconstructed in the engineered microbial host. Dashed arrows represent multiple steps, and solid arrows where no enzyme is listed indicate a single step for which the enzyme has not been cloned from plant hosts. (b) LC-MS/MS analysis of the growth medium of engineered yeast strains supplemented with 4 mM norlaudanosoline and grown for 48 h confirms (S)-scoulerine, (S)-tetrahydrocolumbamine and (S)-tetrahydroberberine production. Data are shown for CSY336, CSY337 and CSY410, respectively, and are representative of analogous strains. The fragmentation pattern of the 328 ion corresponding to (S)-scoulerine differs considerably from that of (S)-reticuline, as the formation of the berberine bridge stabilizes (S)-scoulerine so that it does not fragment into the benzyl and isoquinoline moieties, but instead loses methyl and hydroxyl groups. The fragmentation pattern of the 342 ion identified as (S)-tetrahydrocolumbamine is consistent with that of the parent molecule (S)-scoulerine; the observed fragments exhibit an increase (m/z = 14) attributable to the additional methyl group. (S)-Tetrahydroberberine is identified by its major ion (m/z = 340) and exhibits a fragmentation pattern similar to the parent molecule (S)-tetrahydrocolumbamine; the observed fragments show a decrease (m/z = 2) consistent with the formation of the methylenedioxy bridge. Growth medium of strains lacking the required enzyme coding sequences did not display the identified metabolite peaks.
(S)-Scoulerine represents a second important branch point metabolite as it can be converted to BIA metabolites along either the sanguinarine or berberine branch (Fig. 4a). The oxidation of (S)-scoulerine to (S)-cheilanthifoline (27), an intermediate metabolite along the sanguinarine branch, is catalyzed by a cytochrome P450 enzyme that has not yet been cloned from native plant hosts. Alternatively, the methylation of (S)-scoulerine in the 9-OH position by (S)-scoulerine 9-O-methyltransferase (SMT) results in the synthesis of (S)-tetrahydrocolumbamine, an intermediate metabolite along the berberine branch. We constructed a plasmid coexpressing the P. somniferum BBE and T. flavum SMT cDNAs for transformation into our reticuline-producing yeast strains (CSY337, CSY339; Supplementary Table 1). LC-MS/MS analysis demonstrated that the heterologous SMT enzyme performed the expected methylation reaction to produce (S)-tetrahydrocolumbamine (Fig. 4b). Yeast strains producing (S)-tetrahydrocolumbamine from (R,S)-norlaudanosoline exhibited little or no accumulation of (S)-scoulerine, which indicates efficient conversion of the substrate, yielding ~60 mg l–1 (S)-tetrahydrocolumbamine from 4 mM (R,S)-norlaudanosoline.
The next and penultimate metabolite along the berberine branch is (S)-tetrahydroberberine, or (S)-canadine, which is produced by a methylenedioxy bridge-forming reaction from (S)-tetrahydrocolumbamine (Fig. 4a). This reaction is catalyzed by canadine synthase, a cytochrome P450 enzyme (CYP719A1) that has been cloned from multiple berberine-producing plants31. Previous characterization studies on the C. japonica CYP719A1 have been performed in a yeast strain that coexpresses the A. thaliana P450 reductase ATR1 (ref. 31). We therefore constructed a dual-expression plasmid containing the T. flavum CYP719A1 and SMT cDNAs for cotransformation with plasmids containing the A. thaliana ATR1 and P. somniferum BBE cDNAs into our reticuline-producing yeast strains (CSY399, CSY400; Supplementary Table 1). LC-MS/MS analysis demonstrated low levels of (S)-tetrahydroberberine production (<<5 mg l–1) from a starting substrate concentration of 4 mM norlaudanosoline. Similar analysis of engineered strains expressing endogenous levels of the yeast P450-NADPH reductase or additional copies of the yeast or human reductase did not demonstrate (S)-tetrahydroberberine production, which suggests that these CPR1 variants were not suitable reductase partners for CYP719A1 (data not shown). We also constructed a strain in which the A. thaliana ATR1 reductase coding sequence was integrated into the chromosome of CSY288 and transformed with the plasmids expressing BBE, SMT and CYP719A (CSY410; Supplementary Table 1). LC-MS/MS analysis demonstrated that (S)-tetrahydroberberine accumulation was ~10-fold greater in CSY410 than in strains with plasmid-based ATR1 expression (Fig. 4b). We estimated (S)-tetrahydroberberine production to be ~30 mg l–1 from a substrate concentration of 4 mM, or ~1–2% total conversion from norlaudanosoline or laudanosoline before optimization of this heterologous seven-enzyme pathway. The accumulation of several intermediates in the synthetic pathway highlighted remaining flux limitations in our system (Supplementary Fig. 5 online).
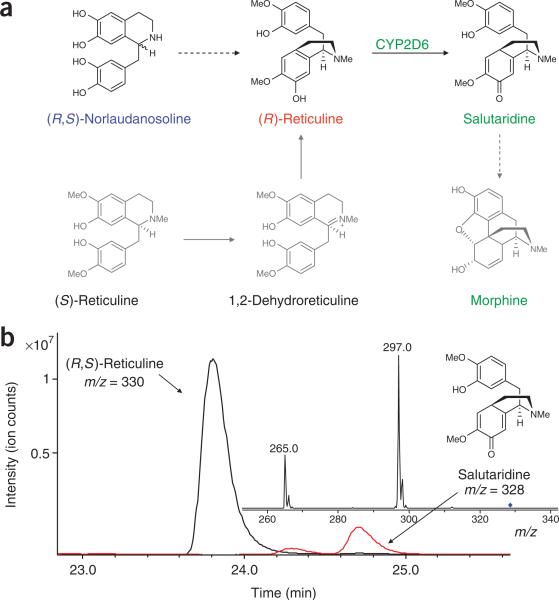
Synthesis of the morphinan intermediate salutaridine
The morphinan alkaloids comprise another major family of BIA metabolites derived from the intermediate reticuline. This family of alkaloids, which includes morphine, codeine and thebaine (28), has received the widest pharmacological use to date and has generated interest mainly for their analgesic properties. However, several of the enzymes that perform the early conversion steps in the morphinan branch have not been cloned and characterized, thereby precluding engineering efforts relying on the native enzymes. To further demonstrate the diversity of small-molecule products that can be synthesized and the power of reconstructing metabolic pathways in engineered hosts, we used an enzyme unrelated to the plant BIA pathway to synthesize an early metabolite in the morphinan branch.
Metabolites in the morphinan branch are synthesized from (S)-reticuline via (R)-reticuline following a two-step isomerization process (Fig. 5a). In the first step, (S)-reticuline is transformed to 1,2-dehydroreticuline (29) by 1,2-dehydroreticuline synthase, an enzyme that has only been partially purified and characterized32. The second step is a reduction of 1,2-dehydroreticuline in the presence of NADPH to (R)-reticuline, catalyzed by an unidentified enzyme. With the lack of available cDNAs for enzymes performing the early isomerization steps, engineering a recombinant host to produce morphinan alkaloids is not possible if only (S)-reticuline is produced. However, using (R,S)-norlaudanosoline as the substrate for our engineered BIA pathway resulted in the synthesis of both forms of reticuline, allowing access to metabolites along the branch extending toward morphine production.
Figure 5.
Microbial production of morphinan alkaloids. (a) Native and engineered pathways for the synthesis of morphinan metabolites. Color schemes and notation follow that defined in Figure 4. In the engineered pathway, (R)-reticuline is produced from (R)-norlaudanosoline. A heterologous human cytochrome P450 (CYP2D6) enzyme can convert (R)-reticuline to the first morphinan alkaloid salutaridine. In the native plant pathway, only (S)-reticuline is produced, and two additional enzymes are required to produce (R)-reticuline, which is used in the synthesis of the morphinan alkaloids. (b) LC-MS/MS analysis of the growth medium of engineered yeast strains supplemented with 4 mM norlaudanosoline and grown for 48 h confirms salutaridine production. Extracted ion chromatograms are shown for reticuline (black) and salutaridine (red) produced by CSY489 and are representative of analogous strains. The salutaridine 328 ion elutes at the same time as (S)-scoulerine, but exhibits a distinctly different fragmentation pattern. The major ions (m/z = 297 and 265) are consistent with the expected fragments reported in the literature for salutaridine44. Strains lacking the CYP2D6 coding sequence did not produce the metabolite peak identified as salutaridine.
The first morphinan alkaloid synthesized from (R)-reticuline is salutaridine, which is synthesized via a complex carbon-carbon phenol coupling reaction catalyzed by salutaridine synthase, which has not been cloned and characterized from plants. However, a recent study demonstrating that humans are capable of synthesizing small amounts of morphine suggests the presence of analogous enzymes in other organisms. The human cytochrome P450 CYP2D6 has been implicated in this pathway, specifically catalyzing the hydroxylation of tyramine (30) to dopamine and the demethylation of codeine to morphine33. Furthermore, it was implied that an unidentified P450 enzyme, possibly CYP2D6, may accept reticuline as a substrate, although experimental evidence has not been shown. We examined the ability of CYP2D6 to convert (R)-reticuline to salutaridine in our engineered yeast strains (Fig. 5b). We constructed plasmids expressing the CYP2D6 cDNA and various reductase partners for coexpression in our reticuline-producing yeast strain backgrounds (CSY463, CSY465, CSY466; Supplementary Table 1).
LC-MS/MS analysis demonstrated that CYP2D6 catalyzes the conversion of (R)-reticuline to salutaridine in strains producing (R,S)-reticuline, thereby highlighting a previously uncharacterized activity for this enzyme (Fig. 5b). Observed levels of salutaridine were consistent with 6–8% conversion of (R,S)-reticuline. Similar to the CYP719A cofactor expression system, chromosomal integration of the gene encoding the human CPR1 reductase (CSY489; Supplementary Table 1) resulted in ~two-fold greater conversion of reticuline to salutaridine compared with strains expressing a plasmid-based P450 reductase or containing only endogenous reductase levels, yielding ~20 mg l–1 from 4 mM norlaudanosoline. However, the activity of CYP2D6 on (R)-reticuline remained relatively low. Codon optimization to improve translational efficiency as well as construction of a fusion protein between CYP2D6 and CPR1 did not improve conversion to salutaridine, which indicates that expression levels and cofactor availability are not limiting (CSY424, CSY425, CSY464, CSY490; Supplementary Table 1). As human P450 enzymes are notoriously promiscuous, the affinity is likely low for (R)-reticuline and may require additional protein engineering or evolution to improve the specificity and activity towards this substrate. In addition, as enzymes that exhibit this activity are cloned and characterized from plant hosts, they can be tested in our system for increased production of salutaridine34.
DISCUSSION
The implementation of synthetic BIA pathways in a microbial host represents a substantial challenge and a rich area for future research, in part due to the complex enzyme activities and regulatory strategies present in the plant hosts that may not translate directly to the engineered host by simply expressing the cloned enzyme activities. For example, a major factor that may generally limit enzymatic activity and conversion is the lack of subcellular compartmentalization in yeast. In particular, BBE and other enzymes are believed to be associated with endomembranes, forming ‘vesicles’ or ‘metabolons’ that facilitate channeling of intermediates and sequestration of toxic metabolites from the cytosol35. Future engineering efforts may enable the assembly of synthetic enzyme complexes in heterologous hosts. A second factor that may limit conversion is the prevalence of cytochrome P450s in the native pathway, which typically exhibit low to no activity in microorganisms. Both CYP719A1 and CYP2D6 exhibited substantially improved activities when coupled to the appropriate reductase partner stably integrated into the host genome as opposed to plasmid-based expression. However, both P450 enzymes did not fully consume their substrates, thereby highlighting the potential for yield improvements at these steps through protein engineering strategies. A third factor that may limit conversion is the lack of an active transport system to facilitate passage of BIA metabolites across microbial cell membranes and to allow for higher intracellular substrate concentrations. Such limitations highlight the importance for reconstruction of these pathways in a single microbial host and may represent a target for future engineering efforts to improve uptake of synthetic substrates.
As a heterologous production host for diverse BIA molecules, our engineered strains will be useful tools in furthering the characterization of a ubiquitous plant secondary metabolic pathway. First, our strains can be used as tools to characterize enzymes and probe regulatory strategies for the BIA pathway in a synthetic host. For example, we used our strains to probe the relative activities of enzyme variants in the engineered pathway. We further described the application of a general tool for titrating individual pathway enzymes to determine optimal expression levels. Such strategies indicated that the 6-OMT and CNMT variants from P. somniferum exhibited superior properties in the engineered pathways relative to the T. flavum variants. Second, our strains can be used as functional genomics tools to further the elucidation of the BIA pathway and characterize new enzyme activities from native plant hosts or non-native sources as demonstrated here. The engineered strains can be used to screen cDNA libraries from BIA-producing plants in high-throughput assays for activities that modify BIA metabolites. Such a tool is particularly powerful in light of the scarce availability of many BIA intermediates and the challenges in plant metabolic engineering.
We have constructed unique production hosts for a diverse set of BIAs offering access to a large family of pharmacologically relevant molecules previously unattainable from other natural sources and synthetic chemistry approaches. Our production hosts offer advantages in directing the production of specific BIA molecules, ease of purification from a GRAS organism, well-established fermentation schemes and rapid biomass accumulation. The microbial BIA production levels reported here reached ~150 mg l–1, similar to previously reported yields from initial engineering efforts on microbial strains to produce other natural plant products3. The application of industrial optimization strategies is expected to substantially increase production levels. More importantly, the engineered yeast strains offer the potential to produce an even broader spectrum of BIA metabolites through extensions of this synthetic pathway. For example, as new enzyme activities are cloned from the native plant hosts, the cDNAs can be expressed in our engineered strains to produce berberine and other BIA metabolites in the sanguinarine, morphinan and bisbenzylisoquinoline branches. As demonstrated here with a human P450 enzyme, there is also the exciting potential to express enzymes that are unrelated to the native BIA pathway but that accept BIA metabolites as substrates in our synthetic hosts to access a wider pool of intermediates and derivatives. Furthermore, strategies that involve recombining native and new enzyme activities and feeding alternative substrates can be used to produce non-natural BIA molecules, thus expanding molecular diversity. Finally, synthetic methods can be used in conjunction with in vivo biosynthesis to attach various functional groups to the molecular backbones, thereby creating a rich population of alkaloids and potentially tapping into new and enhanced pharmacological activities.
METHODS
Plasmid and yeast strain construction
We obtained restriction enzymes, T4 DNA ligase and other cloning enzymes from New England Biolabs. We performed PCR amplifications using Expand High Fidelity PCR system (Roche). Oligonucleotide synthesis was performed by Integrated DNA Technologies. A list of strains and primer sequences for chromosomal integrations and qRT-PCR is provided (Supplementary Tables 1 and 2 online).
We used standard molecular biology techniques to construct the BIA expression vectors36. BIA expression constructs contained the 2μ high-copy yeast origin of replication along with appropriate yeast selection markers and ampicillin resistance. Recombinant enzymes were expressed from the yeast TEF1 promoter and flanked by a CYC1 terminator sequence. We constructed shuttle vectors for subcloning of 1 or 2 cDNA sequences in this fashion (Supplementary Fig. 1 and Supplementary Methods online). Coding sequences for the enzymes of interest, with the exception of human CYP2D6, were donated as cDNAs from P. Facchini (University of Calgary) in plasmids typically suited for expression in E. coli. The human CYP2D6 cDNA was provided by F. Peter Guengerich (Vanderbilt University) as pCW/DB6 (ref. 37), and the yeast codon-optimized version of this gene was synthesized by DNA 2.0. We PCR amplified the endogenous yeast gene encoding the P450 reductase (CPR1) from W303 genomic DNA, the A. thaliana ATR1 gene from WAT11 genomic DNA38 and the Homo sapiens CPR1 gene from pH2E1red (ref. 39).
We transformed ligation reactions into an electrocompetent E. coli strain, DH10B (Invitrogen; F-mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ (ara, leu)7697 galU galK λ-rpsL nupG), using a Gene Pulser Xcell system (BioRAD) according to the manufacturer's instructions. We conducted plasmid isolation using the Wizard Plus SV Minipreps DNA purification system (Promega) according to the manufacturer's instructions. Subcloning was confirmed by restriction analysis and sequence verification (Laragen, Inc.). We transformed plasmids into the appropriate S. cerevisiae strains using a standard lithium acetate protocol40. All yeast strains used in this work were based on the haploid yeast strain W303α (MATα his3-11,15 trp1-1 leu2-3 ura3-1 ade2-1)41. E. coli cells were grown on Luria-Bertani medium (BD Diagnostics) with 100 μg ml–1 ampicillin (EMD Chemicals) for plasmid maintenance, and S. cerevisiae cells were grown in synthetic complete medium (BD Diagnostics) supplemented with the appropriate dropout solution for plasmid maintenance (Calbiochem).
We performed chromosomal integrations of DNA fragments through homologous recombination using a standard lithium acetate transformation protocol to construct strains that stably express combinations of the BIA enzymes. We built gene insertion cassettes harboring the appropriate BIA enzyme expression construct and associated selection marker flanked by loxP sites to allow removal of the selection marker following integration with a Cre-loxP system42.
Growth conditions
For BIA metabolite production assays, with the exception of enzyme titration studies (Supplementary Methods), engineered yeast strains were grown in test tube cultures in volumes ranging from 2 ml to 10 ml at 30 °C and 200 r.p.m. in the appropriate dropout medium with 2% dextrose (w/v) as a sugar source. We diluted overnight cultures 1:20 in fresh medium supplemented with the appropriate pathway substrate as reported, typically norlaudanosoline (CHEMOS GmbH, ACROS Organics) or laudanosoline (ACROS Organics) at concentrations between 0.1 mM and 5 mM diluted from a 10 or 20 mM stock solution in water. With the exception of timecourse experiments, we assayed cultures at 24 or 48 h following substrate addition to observe maximum accumulation as cells reached the stationary growth phase.
Analysis of metabolite production
We evaluated BIA metabolite levels by LC-MS/MS analysis of cell extracts and growth medium. At appropriate time points, aliquots of yeast cultures were centrifuged to recover cells as pellets and allow collection of the growth medium. We analyzed the growth medium or an appropriate dilution directly by LC-MS/MS. Samples were run on an Agilent ZORBAX SB-Aq 3 × 250 mm, 5 μm column with 0.1% acetic acid as solvent A and methanol as solvent B. We used a gradient elution to separate the metabolites of interest as follows: 0–10 min at 100% A, 10–30 min 0–90% B, 30–35 min 90–100% B, followed by a 5 min equilibration at 100% A between samples. Following LC separation, metabolites were injected into an Agilent 6320 ion trap mass spectrometer for detection and identification. We used selective reaction monitoring to isolate ions of interest for MS/MS to verify the molecular structure of each metabolite. We verified chromatogram data including fragmentation patterns through at least three independent experiments and from multiple strains where appropriate. Additional data on the characterization of synthesized metabolites is provided (Supplementary Methods). Quantification of metabolites was based on the integrated area of the extracted ion chromatogram peaks43 calculated using DataAnalysis for 6300 Series Ion Trap LC/MS version 3.4 (Bruker Daltonik GmbH) and reported as the mean ± s.d. When appropriate, we normalized the measured levels to a metabolite peak of known concentration in the growth medium, typically the substrate (norlaudanosoline) peak. We also generated standard curves for norlaudanosoline and laudanosoline to relate peak area to metabolite concentration in our samples. Although slight differences in ionization efficiencies were observed between these available standards, this method further validated our estimates of metabolite concentrations based on percentage substrate conversion. A summary of the average yields for each of the synthesized BIA compounds is provided (Supplementary Table 3 online).
Other methods
Additional methods are available in the Supplementary Methods.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Facchini (University of Calgary), F.P. Guengerich (Vanderbilt University) and D. Pompon (Centre de Génétique Moléculaire, CNRS) for generously providing cDNAs and yeast strains used in this work. This work was supported by the Center for Biological Circuit Design at Caltech (fellowship to K.M.H.) and the US National Institutes of Health (grant to C.D.S.).
Footnotes
Note: Supplementary information and chemical compound information is available on the Nature Chemical Biology website.
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturechemicalbiology/.
Published online at http://www.nature.com/naturechemicalbiology/
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 2.McDaniel R, Weiss R. Advances in synthetic biology: on the path from prototypes to applications. Curr. Opin. Biotechnol. 2005;16:476–483. doi: 10.1016/j.copbio.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Ro DK, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 4.Kealey JT, Liu L, Santi DV, Betlach MC, Barr PJ. Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc. Natl. Acad. Sci. USA. 1998;95:505–509. doi: 10.1073/pnas.95.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szczebara FM, et al. Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nat. Biotechnol. 2003;21:143–149. doi: 10.1038/nbt775. [DOI] [PubMed] [Google Scholar]
- 6.Martin ML, et al. Antispasmodic activity of benzylisoquinoline alkaloids analogous to papaverine. Planta Med. 1993;59:63–67. doi: 10.1055/s-2006-959606. [DOI] [PubMed] [Google Scholar]
- 7.Morais LC, Barbosa-Filho JM, Almeida RN. Central depressant effects of reticuline extracted from Ocotea duckei in rats and mice. J. Ethnopharmacol. 1998;62:57–61. doi: 10.1016/s0378-8741(98)00044-0. [DOI] [PubMed] [Google Scholar]
- 8.Nakaoji K, Nayeshiro H, Tanahashi T. Norreticuline and reticuline as possible new agents for hair growth acceleration. Biol. Pharm. Bull. 1997;20:586–588. doi: 10.1248/bpb.20.586. [DOI] [PubMed] [Google Scholar]
- 9.Kemeny-Beke A, et al. Apoptotic response of uveal melanoma cells upon treatment with chelidonine, sanguinarine and chelerythrine. Cancer Lett. 2006;237:67–75. doi: 10.1016/j.canlet.2005.05.037. [DOI] [PubMed] [Google Scholar]
- 10.Cassels BK, et al. Structure-antioxidative activity relationships in benzylisoquinoline alkaloids. Pharmacol. Res. 1995;31:103–107. doi: 10.1016/1043-6618(95)80054-9. [DOI] [PubMed] [Google Scholar]
- 11.Exley R, et al. Evaluation of benzyltetrahydroisoquinolines as ligands for neuronal nicotinic acetylcholine receptors. Br. J. Pharmacol. 2005;146:15–24. doi: 10.1038/sj.bjp.0706307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kashiwada Y, et al. Anti-HIV benzylisoquinoline alkaloids and flavonoids from the leaves of Nelumbo nucifera, and structure-activity correlations with related alkaloids. Bioorg. Med. Chem. 2005;13:443–448. doi: 10.1016/j.bmc.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 13.Lai JH. Immunomodulatory effects and mechanisms of plant alkaloid tetrandrine in autoimmune diseases. Acta Pharmacol. Sin. 2002;23:1093–1101. [PubMed] [Google Scholar]
- 14.Kwan CY, Achike FI. Tetrandrine and related bis-benzylisoquinoline alkaloids from medicinal herbs: cardiovascular effects and mechanisms of action. Acta Pharmacol. Sin. 2002;23:1057–1068. [PubMed] [Google Scholar]
- 15.Chulia S, et al. Relationships between structure and vascular activity in a series of benzylisoquinolines. Br. J. Pharmacol. 1997;122:409–416. doi: 10.1038/sj.bjp.0701410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bentley KW. β-Phenylethylamines and the isoquinoline alkaloids. Nat. Prod. Rep. 2006;23:444–463. doi: 10.1039/b509523a. [DOI] [PubMed] [Google Scholar]
- 17.Facchini PJ. Alkaloid biosynthesis in plants: biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:29–66. doi: 10.1146/annurev.arplant.52.1.29. [DOI] [PubMed] [Google Scholar]
- 18.Sato F, Inui T, Takemura T. Metabolic engineering in isoquinoline alkaloid biosynthesis. Curr. Pharm. Biotechnol. 2007;8:211–218. doi: 10.2174/138920107781387438. [DOI] [PubMed] [Google Scholar]
- 19.Allen RS, et al. Metabolic engineering of morphinan alkaloids by over-expression and RNAi suppression of salutaridinol 7-O-acetyltransferase in opium poppy. Plant Biotechnol. J. 2008;6:22–30. doi: 10.1111/j.1467-7652.2007.00293.x. [DOI] [PubMed] [Google Scholar]
- 20.Allen RS, et al. RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat. Biotechnol. 2004;22:1559–1566. doi: 10.1038/nbt1033. [DOI] [PubMed] [Google Scholar]
- 21.Sato F, et al. Metabolic engineering of plant alkaloid biosynthesis. Proc. Natl. Acad. Sci. USA. 2001;98:367–372. doi: 10.1073/pnas.011526398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zulak KG, et al. Gene transcript and metabolite profiling of elicitor-induced opium poppy cell cultures reveals the coordinate regulation of primary and secondary metabolism. Planta. 2007;225:1085–1106. doi: 10.1007/s00425-006-0419-5. [DOI] [PubMed] [Google Scholar]
- 23.Minami H, et al. Microbial production of plant benzylisoquinoline alkaloids. Proc. Natl. Acad. Sci. USA. 2008;105:7393–7398. doi: 10.1073/pnas.0802981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ziegler J, et al. Comparative transcript and alkaloid profiling in Papaver species identifies a short chain dehydrogenase/reductase involved in morphine biosynthesis. Plant J. 2006;48:177–192. doi: 10.1111/j.1365-313X.2006.02860.x. [DOI] [PubMed] [Google Scholar]
- 25.Sato F, Tsujita T, Katagiri Y, Yoshida S, Yamada Y. Purification and characterization of S-adenosyl-L-methionine: norcoclaurine 6-O-methyltransferase from cultured Coptis japonica cells. Eur. J. Biochem. 1994;225:125–131. doi: 10.1111/j.1432-1033.1994.00125.x. [DOI] [PubMed] [Google Scholar]
- 26.Ounaroon A, Decker G, Schmidt J, Lottspeich F, Kutchan TM. (R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum - cDNA cloning and characterization of methyl transfer enzymes of alkaloid biosynthesis in opium poppy. Plant J. 2003;36:808–819. doi: 10.1046/j.1365-313x.2003.01928.x. [DOI] [PubMed] [Google Scholar]
- 27.Hawkins KM, Smolke CD. The regulatory roles of the galactose permease and kinase in the induction response of the GAL network in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:13485–13492. doi: 10.1074/jbc.M512317200. [DOI] [PubMed] [Google Scholar]
- 28.Nevoigt E, et al. Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2006;72:5266–5273. doi: 10.1128/AEM.00530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kutchan TM, Dittrich H. Characterization and mechanism of the berberine bridge enzyme, a covalently flavinylated oxidase of benzophenanthridine alkaloid biosynthesis in plants. J. Biol. Chem. 1995;270:24475–24481. doi: 10.1074/jbc.270.41.24475. [DOI] [PubMed] [Google Scholar]
- 30.Bird DA, Facchini PJ. Berberine bridge enzyme, a key branch-point enzyme in benzylisoquinoline alkaloid biosynthesis, contains a vacuolar sorting determinant. Planta. 2001;213:888–897. doi: 10.1007/s004250100582. [DOI] [PubMed] [Google Scholar]
- 31.Ikezawa N, et al. Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel P450 family, from cultured Coptis japonica cells. J. Biol. Chem. 2003;278:38557–38565. doi: 10.1074/jbc.M302470200. [DOI] [PubMed] [Google Scholar]
- 32.Hirata K, Poeaknapo C, Schmidt J, Zenk MH. 1,2-Dehydroreticuline synthase, the branch point enzyme opening the morphinan biosynthetic pathway. Phytochemistry. 2004;65:1039–1046. doi: 10.1016/j.phytochem.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 33.Zhu W, Cadet P, Baggerman G, Mantione KJ, Stefano GB. Human white blood cells synthesize morphine: CYP2D6 modulation. J. Immunol. 2005;175:7357–7362. doi: 10.4049/jimmunol.175.11.7357. [DOI] [PubMed] [Google Scholar]
- 34.Liscombe DK, Facchini PJ. Evolutionary and cellular webs in benzylisoquinoline alkaloid biosynthesis. Curr. Opin. Biotechnol. 2008;19:173–180. doi: 10.1016/j.copbio.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 35.Winkel BS. Metabolic channeling in plants. Annu. Rev. Plant Biol. 2004;55:85–107. doi: 10.1146/annurev.arplant.55.031903.141714. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Russell DW. Molecular Cloning. 3rd edn. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York, USA: 2001. [Google Scholar]
- 37.Gillam EM, Guo Z, Martin MV, Jenkins CM, Guengerich FP. Expression of cytochrome P450 2D6 in Escherichia coli, purification, and spectral and catalytic characterization. Arch. Biochem. Biophys. 1995;319:540–550. doi: 10.1006/abbi.1995.1329. [DOI] [PubMed] [Google Scholar]
- 38.Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J. Biol. Chem. 1997;272:19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- 39.Mapoles J, Berthou F, Alexander A, Simon F, Menez JF. Mammalian PC-12 cell genetically engineered for human cytochrome P450 2E1 expression. Eur. J. Biochem. 1993;214:735–745. doi: 10.1111/j.1432-1033.1993.tb17975.x. [DOI] [PubMed] [Google Scholar]
- 40.Gietz RD, Woods RA. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol. 2006;313:107–120. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- 41.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 42.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, et al. Analysis of major alkaloids in Rhizoma coptidis by capillary electrophoresis-electrospray-time of flight mass spectrometry with different background electrolytes. Electrophoresis. 2008;29:2135–2147. doi: 10.1002/elps.200700797. [DOI] [PubMed] [Google Scholar]
- 44.Raith K, et al. Electrospray tandem mass spectrometric investigations of morphinans. J. Am. Soc. Mass Spectrom. 2003;14:1262–1269. doi: 10.1016/S1044-0305(03)00539-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.