Abstract
As the primary consumers of oxygen within all aerobic organisms, mitochondria are a major source of cellular reactive oxygen species (ROS) derived from the in vivo chemistry of oxygen metabolism. Mitochondrial ROS have been traditionally implicated in aging and in a variety of pathologies, including cancer, neurodegeneration, and diabetes, but recent studies also link controlled mitochondrial ROS fluxes to cell regulation and signaling events. Progress in the development of mitochondrial-targeted fluorescent small-molecule indicators that detect specific ROS with high selectivity offers a promising approach for interrogating mitochondrial ROS production, trafficking, and downstream biological effects.
Introduction
Reactive oxygen species (ROS) have emerged as prevalent and important components of both physiological and pathological states of living organisms [1–9]. Classically, the presence of ROS in biological systems has been associated predominantly with disease and form the underpinning for the “free-radical theory of aging” (FRTA) [10]. Indeed, oxidative stress is connected to many diseases where age is a risk factor [11], including cancer [12, 13] and neurodegenerative disorders such as Alzheimer’s, Parkinson’s, and Huntington’s diseases [14, 15]. On the other hand, the controlled release and compartmentalization of ROS is also critical to maintaining normal physiology. For example, macrophages engulf invading pathogens into phagocytic vesicles and then produce a variety of ROS inside the vesicles to help neutralize the threat. Moreover, the production of certain ROS, such as hydrogen peroxide (H2O2), has been detected in a variety of tissues, and mounting evidence suggests this ROS is utilized as a signaling molecule for a wide range of healthy physiological events [16–22].
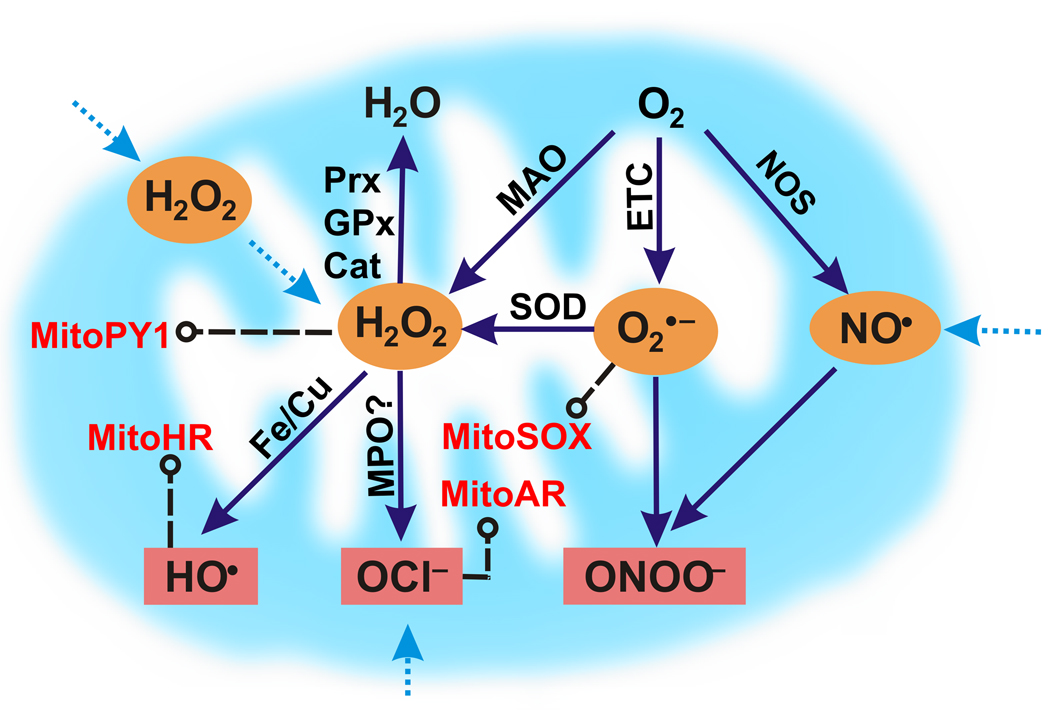
The complex biology of ROS is patently dictated by the chemical properties of each specific oxygen metabolite as well as the localization and trafficking of that metabolite at the cellular level. In this context, mitochondria are the major consumers of cellular oxygen and hence play a central role in ROS biology. Harman’s FRTA focused on mitochondria as the “biologic clocks” of the cell that possess the primary generators and targets of ROS [23]. Figure 1 outlines various potential sources of mitochondrial ROS [24–26], and extensive reviews have been written on this topic [25] [27, 28]. The balance between the production and destruction of ROS in the mitochondria has critical in vivo implications [29], as the overexpression of either superoxide dismutase (SOD) [30] or catalase [31] enzymes that consume O2− and H2O2, respectively, can increase the lifespan of various model organisms. Excess ROS can clearly have aberrant consequences, but the fact that aerobic organisms have conserved the ability to generate a basal level of mitochondrial ROS suggests that a minimum threshold of these oxygen metabolites is beneficial and provides some form of evolutionary fitness.
Figure 1.
A simplified scheme for mitochondrial ROS metabolism. The primary source of ROS in the mitochondria is derived from the one-electron reduction of molecular oxygen (O2) by the electron transport chain (ETC) to form superoxide (O2−). O2− can then be converted to hydrogen peroxide (H2O2) either spontaneously or catalyzed by superoxide dismutases (SOD). H2O2 can also be produced by monoamine oxidases (MAO), which catalyze the oxidative deamination of dietary and neurotransmitter amines. Mitochondrial nitric oxide synthases (NOS) produce nitric oxide (NO) that can potentially react with superoxide to produce peroxynitrite (ONOO−). H2O2 can either be destroyed by peroxiredoxins (PRX), glutathione peroxidases (GPx), or catalases (Cat), converted to a hydroxyl radical (•OH) by iron or copper-mediated Fenton chemistry, or transformed into hypochlorous acid (HOCl) by myeloperoxidase (MPO) catalysis.
The complex interplay between mitochondrial ROS and health, aging, and disease provides motivation for developing new tools to study the chemistry and biology of ROS in living systems with high spatial and temporal resolution. Fluorescence imaging with mitochondrial-targeted reporters for monitoring ROS offers a potentially powerful methodology to achieving this goal, and this review will summarize selected recent examples of small-molecule probes that simultaneously localize to the mitochondria and respond to various ROS molecules. Although outside the scope of this review, we note that protein-based fluorescent reporters provide a complementary approach to targeted mitochondrial ROS imaging and several elegant examples have been published [32–37].
Designing small-molecule probes that target to mitochondria
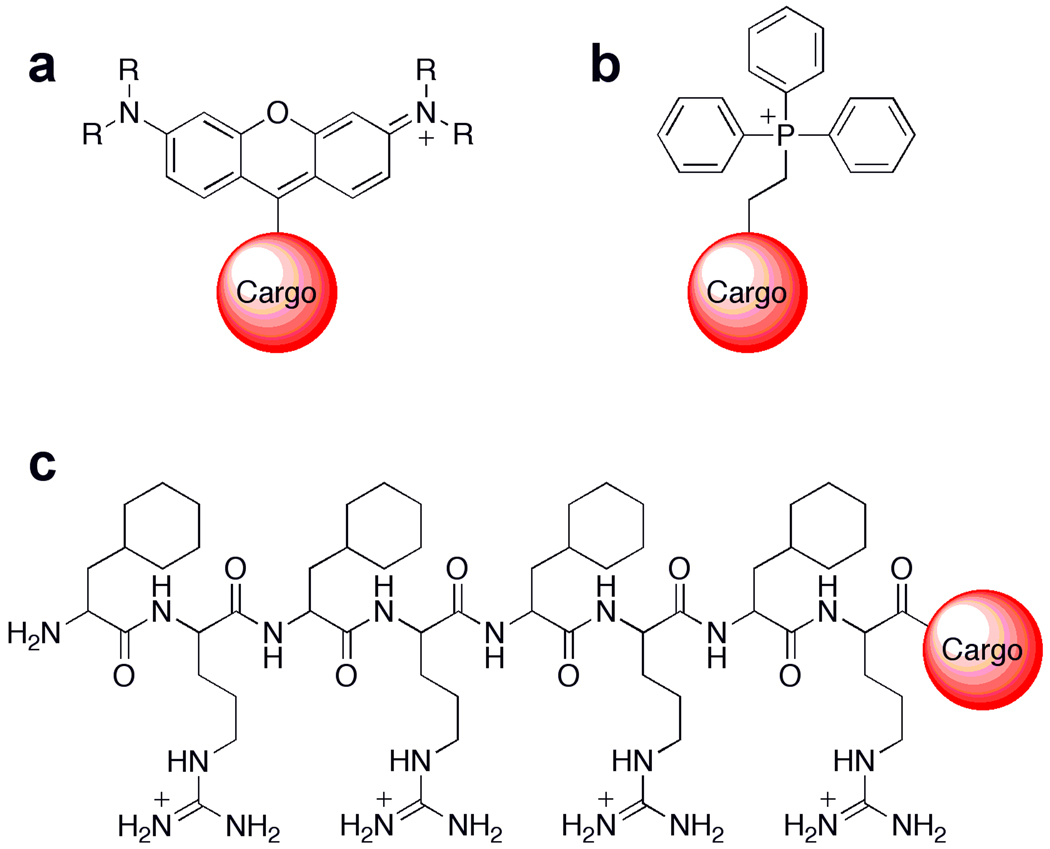
Figure 2 outlines various approaches to deliver small-molecule probes to cellular mitochondria. The most common way to target molecules to the mitochondria of living cells is through the use of lipophilic cations, which are attracted to the negative potential caused by the proton gradient across the inner mitochondrial membrane. For example, rhodamine and the MitoTracker dyes possess an overall positive charge that is delocalized through resonance, allowing for passage through plasma membranes and subsequent accumulation within mitochondria [38]. Through the same general mechanism, Murphy and colleagues have championed the use of triphenylphosphonium (TPP) head groups to deliver a variety of cargoes to the mitochondria, including antioxidants such as vitamin E [39], coenzyme Q [40], S-nitrosothiols [41], as well as SOD and peroxidase mimics [42]. A newer approach developed by Kelley and co-workers utilizes mitochondria penetrating peptides (MMPs). By balancing charge and liophilicity through a combination of natural and synthetic amino acids, various cargoes can be delivered to the mitochondria, including a singlet-oxygen generating fluorophore [43] that allows the localized generation of this ROS. We refer the reader to more comprehensive reviews for TPP [44] and general mitochondrial targeting [45] that have appeared in the recent literature, and the remainder of this review will present a brief survey of probes for various mitochondrial ROS molecules.
Figure 2.
General methods for delivering molecular cargo to the mitochondria. Small molecules can be delivered to the mitochondria through the use of lipophilic cations such as rhodamine dyes (a) and triphenylphosphonium moieties (b), which take advantage of the proton gradient and subsequent electrochemical potential generated within the matrix of mitochondria. Recently, mitochondria-targeted peptides (c) that can contain both natural and unnatural amino acids have been rationally designed and screened as mitochondrial delivery vehicles [45].
A fluorescent probe for mitochondrial superoxide
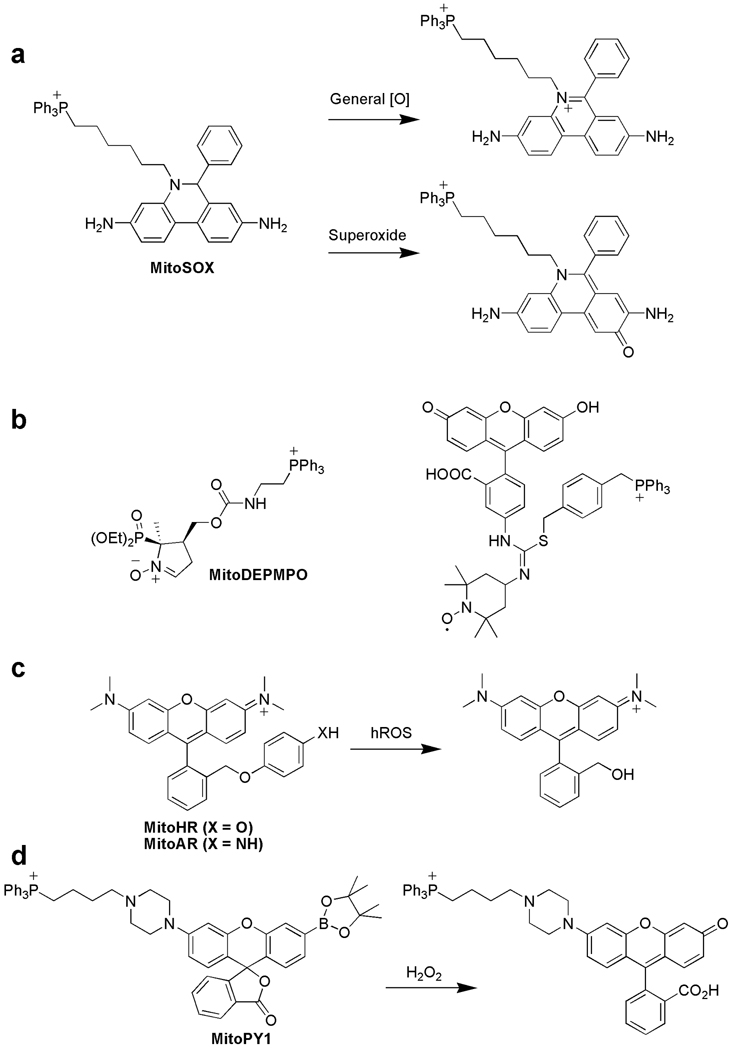
Derived from the one-electron reduction of oxygen, superoxide (O2−) is a marker for early ROS generation in primary or secondary chemical reaction cascades and a major ROS leaking from the respiratory chain. The most widely used fluorescent indicator for O2− is hydroethidium (HE), a two-electron reduced form of the nucleic acid stain ethidium. HE has been employed to detect ROS production during phagocytic respiratory bursts [46] and intracellular oxidative stress [47]. Appending a TPP targeting group to HE yields MitoSOX; the TPP successfully directs the probe away from the nucleus and to the mitochondria of living cells (Figure 3a). A major experimental complication for applying HE probes to monitor O2− specifically is that HE can be oxidized by multiple ROS to yield multiple fluorescent species; however, only O2−-dependent oxidation can generate a hydroxylated product (2-OH-E) that has a unique excitation at 396 nm in addition to the typical ethidium excitation at 510 nm [48]. Results obtained using only ethidium excitation at 510 nm must be interpreted with caution as they are likely to overestimate the contributions of O2− while underestimating the roles of other ROS. Careful control experiments and/or direct excitation of the 2-OH-E product are necessary to distinguish whether O2− is the dominant participating ROS in a given situation [49].
Figure 3.
Selected mitochondrial-targeted probes for detection of reactive oxygen species (ROS). (a) MitoSOX is a dihydroethidium-based probe bearing a triphenylphosphonium (TPP) targeting moiety. This probe reacts with several ROS, but the product from superoxide oxidation can be distinguished from other potentially formed oxidized products by selective excitation of the 2-hydroxylethidium product. (b) Two examples of mitochondrial-targeted nitrone spin-trap probes that utilize TPP localization groups. (c) MitoAR and MitoHR are rhodamine-like probes that react with highly reactive oxygen species (hROS) including hydroxyl radical (•OH), hypochlorous acid (HOCl), and peroxynitrite (ONOO−). (d) MitoPY1 is a boronate-based hybrid rhodamine/fluorescein probe with a TPP targeting group for chemospecific detection of hydrogen peroxide (H2O2).
Mitochondrial-targeted probes for free radicals
Oxygen-derived free radicals are frequently associated with aging and disease, and mitochondria are a major source and target of these damaging molecular oxidants. Electron paramagnetic resonance (EPR) spectroscopy is a useful way to detect free radical ROS and other molecules possessing unpaired electrons [50]. In particular, spin trapping with nitrones like TEMPO can generate, upon reaction with ROS, persistent nitroxide adducts that are detectable by EPR. Several laboratories have described the use of mitochondrial-targeted nitrone traps for EPR detection or antioxidant treatments using TPP (Figure 3b, left) [51] or hemigramicidin fragments of the membrane-active antibiotic Gramicidin S for cargo delivery [52, 53]. A trifunctional reporter combining a TPP for mitochondrial targeting, a TEMPO derivative for spin trapping, and a fluorescein moiety for optical tracking has been reported (Figure 3b, right) [54]. Confocal microscopy experiments show that this probe accumulates within the mitochondria of live RAW264.7 macrophages, but more studies are needed to validate this potential free radical imaging strategy.
Fluorescent mitochondrial probes for highly reactive oxygen species
Highly reactive oxygen species (hROS), including hydroxyl radical (•OH), peroxynitrite (ONOO−), and hypochlorous acid (HOCl), are potent oxidants that are capable of directly oxidizing nucleic acids, proteins, and lipids. hROS are generated in secondary cellular ROS reaction cascades involving O2− and H2O2. For example, H2O2 can trigger uncontrolled Fenton chemistry in the presence of iron or copper centers that are prevalent in the mitochondria to make the exceedingly reactive and damaging •OH species. Likewise, nitric oxide (NO) generated by plasma membrane or mitochondrial nitric oxide synthases can combine with O2− to make the nitrating oxidant ONOO−. In addition, myeloperoxidases (MPOs) catalyze the transformation of H2O2 to HOCl, which can then potentially diffuse into and damage the mitochondria. To monitor mitochondrial hROS, Nagano and co-workers have exploited rhodamine-like fluorophores capped with either a 4-aminophenyl aryl ether (MitoAR) or a 4-hydroxy aryl ether group (MitoHR) (Figure 3c). The ether capping groups quench the fluorophore emission by photoinduced electron transfer (PET). Reaction with hROS cleaves off the PET quenching moiety, resulting in the highly fluorescent rhodamine type reporter [55]. MitoAR responds mainly to •OH and HOCl, whereas MitoHR is most sensitive to •OH. Both probes react with ONOO−, but at a slower rate. Fluorescence imaging experiments establish that MitoAR can accumulate selectively within the mitochondria of live HeLa cells and respond with increased fluorescence to exogenously added HOCl. Importantly, treatment of MitoAR-loaded HeLa cells, which are lacking MPO, with H2O2 does not result in a fluorescence turn-on. However, addition of H2O2 to MitoAR-loaded HL-60 cells, which do possess MPO, causes a turn-on fluorescence increase.
A targetable fluorescent probe for mitochondrial hydrogen peroxide
Major chemical pathways to cellular H2O2 production include the incomplete reduction of O2 to H2O during mitochondrial electron transport, the two-electron, two-proton reduction of O2 by oxidase activity, or the reduction of O2− by spontaneous or SOD-catalyzed processes. Aberrant H2O2 fluxes within mitochondria can trigger apoptosis, but controlled bursts of H2O2 elicited by specific ligand-receptor interactions are beneficial to metabolic function. To help disentangle the diverse contributions of H2O2 to mitochondrial biology, we sought to create new chemical tools for monitoring H2O2 levels within this subcellular locale. Previous work from our laboratory established that the H2O2-mediated conversion of aryl boronates to phenols can provide a chemospecific, biologically compatible reaction method for detecting endogenous H2O2 production [56–60]. Appending a TPP moiety onto a boronate-masked hybrid fluorescein/rhodamine reporter gives MitoPY1 (Figure 3d), which shows good localization to the mitochondria of a variety of mammalian cell types, including HeLa, HEK293, Cos-7, and CHO.K1 [61]. MitoPY1 can respond selectively to rises in H2O2 levels by a turn-on fluorescence increase as determined by both confocal imaging and flow cytometry experiments (Figure 4). Furthermore, MitoPY1 can detect mitochondrially-derived H2O2 in a neurodegenerative oxidative stress model; treatment of HeLa cells with paraquat, an uncoupler of the mitochondrial electron transport chain and small-molecule model for Parkinson’s disease, causes a robust, mitochondrial-localized fluorescence enhancement due to elevated generation of H2O2.
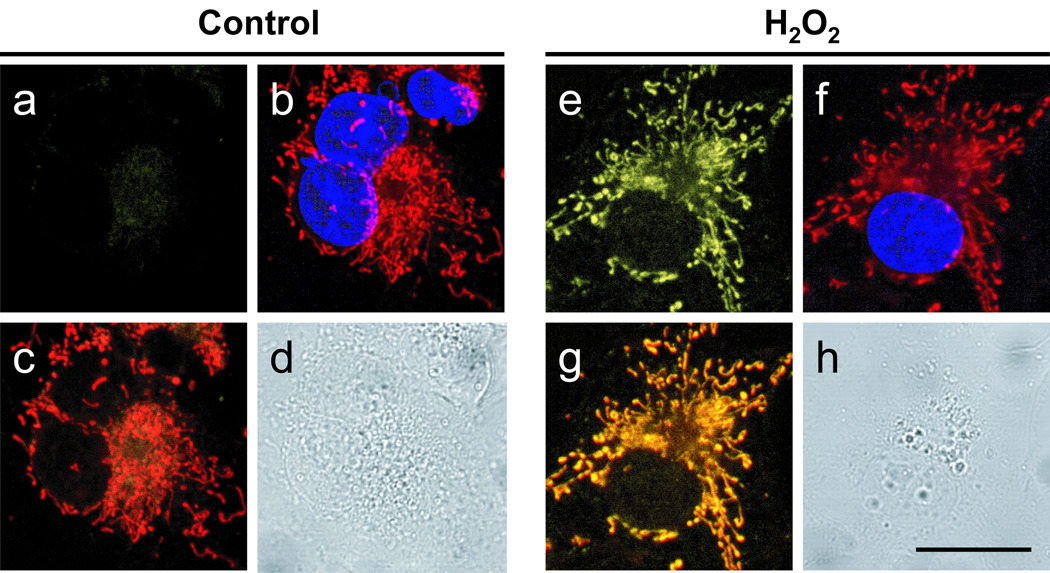
Figure 4.
Confocal fluorescence images of live Cos-7 cells with varying levels of mitochondrial H2O2 as visualized using MitoPY1. Cos-7 cells incubated with 5 µM MitoPY1 for 60 minutes at 37 °C and imaged with either MitoPY1 (a), MitoTracker Deep Red and Hoechst (overlay, b), MitoPY1 with MitoTracker Deep Red (overlay, c), or in brightfield mode (d). Cos-7 cells incubated with 5 µM MitoPY1 with 300 µM H2O2 added for the final 40 minutes and imaged with either MitoPY1 (e), MitoTracker Deep Red and Hoechst (overlay, f), MitoPY1 with MitoTracker Deep Red (overlay, g), or in brightfield mode (h) with a 20 µm scale bar.
Conclusions and outlook
The foregoing examples have highlighted a select but growing number of targeted small-molecule chemical tools for studying the biology of mitochondrial ROS. These indicators, along with emerging approaches toward detecting peroxynitrite [62], superoxide [63], nitric oxide [64, 65], hypochlorous acid [66, 67], ozone [68], and reversible redox changes [69], presages the possibility that one can help elucidate the dynamic cascades involving mitochondrial ROS metabolism using a family of targeted reagents that detect different ROS molecules. In addition, one can also envision using a set of probes with selectivity for a specific ROS to construct a multicolor calibration scale for the mitochondrial redox environment. Combining parallel advances in the delivery of molecular cargoes to the mitochondria with the development of highly specific and selective molecular switches would be of potential broad utility in further deepening our understanding of mitochondria redox biology, as well as delineating their contributions to the more complex oxidation biology of living cells, tissue, and organisms.
Acknowledgements
We thank the National Institutes of Health (GM079465), the University of California at Berkeley (Hellman Faculty Fund), the Beckman, Packard, and Sloan foundations, Amgen, and the Howard Hughes Medical Institute for funding our laboratory’s research in oxidation biology. BCD was supported by a Chemical Biology Training Grant from the NIH (T32 GM066698) and DS was supported by a scholarship from the Ministry of Science, Thailand.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Stone JR, Yang S. Hydrogen peroxide: a signaling messenger. Antioxid Redox Sign. 2006;8:243–270. doi: 10.1089/ars.2006.8.243. [DOI] [PubMed] [Google Scholar]
- 2.Rhee SG. Cell signaling: H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 3.Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 4.D'AutrEaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 5.Giorgio M, Trinei M, Migliaccio E, Pelicci PG. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nat Mol Cell Biol. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- 6.Reddie KG, Carroll KS. Expanding the functional diversity of proteins through cysteine oxidation. Curr Opin Chem Biol. 2008;12:746–754. doi: 10.1016/j.cbpa.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 7.Poole LB, Nelson KJ. Discovering mechanisms of signaling-mediated cysteine oxidation. Curr Opin Chem Biol. 2008;12:18–24. doi: 10.1016/j.cbpa.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 9.Miller EW, Chang CJ. Fluorescent probes for nitric oxide and hydrogen peroxide in cell signaling. Curr Opin Chem Biol. 2007;11:620–625. doi: 10.1016/j.cbpa.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 11.Stadtman ER. Protein oxidation and aging. Free Radic Res. 2006;40:1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 12.Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 14.Barnham KJ, Masters CL, Bush AI. Neurodegenerative diseases and oxidative stress. Nat Drug Rev Discov. 2004;3:205–214. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- 15.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 16.Bae YS, Kang SW, Seo MS, Baines IC, Tekle E, Chock PB, Rhee SG. Epidermal growth factor (EGF)-induced generation of hydrogen peroxide. J Biol Chem. 2005;175:5423–5429. [PubMed] [Google Scholar]
- 17.Woo HA, Chae HZ, Hwang SC, Yang K-S, Kang SW, Kim K, Rhee SG. Reversing the Inactivation of Peroxiredoxins Caused by Cysteine Sulfinic Acid Formation. Science. 2003;300:653–656. doi: 10.1126/science.1080273. [DOI] [PubMed] [Google Scholar]
- 18.Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 19.van Montfort RLM, Congreve M, Tisi D, Carr R, Jhoti H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature. 2003;423:773–777. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- 20.Bao L, Avshalumov MV, Patel JC, Lee CR, Miller EW, Chang CJ, Rice ME. Mitochondria are the source of hydrogen peroxide for dynamic brain-cell signaling. J Neurosci. 2009;29:9002–9010. doi: 10.1523/JNEUROSCI.1706-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T. Requirement for generation of H2O2 for platelet-derived growth-factor signal-transduction. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 22.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 23.Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 24.St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 25.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol (Lond.) 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liochev SI, Fridovich I. The effects of superoxide dismutase on H2O2 formation. Free Radical Biol Med. 2007;42:1465–1469. doi: 10.1016/j.freeradbiomed.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 27. Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. **This review describes the potential production of ROS by mitochondria.
- 28.Addabbo F, Montagnani M, Goligorsky MS. Mitochondria and Reactive Oxygen Species. Hypertension. 2009;53:885–892. doi: 10.1161/HYPERTENSIONAHA.109.130054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Atamna H, Kuratsune H, Ames BN. Delaying brain mitochondrial decay and aging with mitochondrial antioxidants and metabolites. Ann N.Y. Acad Sci. 2002;959:133–166. doi: 10.1111/j.1749-6632.2002.tb02090.x. [DOI] [PubMed] [Google Scholar]
- 30.Sun J, Folk D, Bradley TJ, Tower J. Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics. 2002;161:661–672. doi: 10.1093/genetics/161.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schriner SE, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 32.Ostergaard H, Henriksen A, Hansen FG, Winther JR. Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein. EMBO J. 2001;20:5853. doi: 10.1093/emboj/20.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279:13044. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 34.Cannon MB, Remington SJ. Re-engineering redox-sensitive green fluorescent protein for improved response rate. Protein Sci. 2006;15:45–57. doi: 10.1110/ps.051734306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. *This paper describes the development and application of a Hyper, a genetically-encoded sensor for hydrogen peroxide.
- 36.Gutscher M, Pauleau AL, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, Dick TP. Real-time imaging of the intracellular glutathione redox potential. Nat Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 37. Wang W, et al. Superoxide flashes in single mitochondria. Cell. 2008;134:279–290. doi: 10.1016/j.cell.2008.06.017. *This paper describes the development and application of a genetically-encoded sensor for superoxide.
- 38.Johnson LV, Walsh ML, Chen LB. Localization of mitochondria in living cells with rhodamine 123. P Natl Acad Sci. 1980;77:990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith RAJ, Porteous CM, Coulter CV, Murphy MP. Selective targeting of an antioxidant to mitochondria. Eur J Biochem. 1999;263:709–716. doi: 10.1046/j.1432-1327.1999.00543.x. [DOI] [PubMed] [Google Scholar]
- 40.Kelso GF, Porteous CM, Coulter CV, Hughes G, Porteous WK, Ledgerwood EC, Smith RAJ, Murphy MP. Selective Targeting of a Redox-active Ubiquinone to Mitochondria within Cells antioxidant and antiapoptotic properties. J Biol Chem. 2001;276:4588–4596. doi: 10.1074/jbc.M009093200. [DOI] [PubMed] [Google Scholar]
- 41.Prime TA, et al. A mitochondria-targeted S-nitrosothiol modulates respiration, nitrosates thiols, and protects against ischemia-reperfusion injury. P Natl Acad Sci USA. 2009;106:10764–10769. doi: 10.1073/pnas.0903250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Filipovska A, Kelso GF, Brown SE, Beer SM, Smith RAJ, Murphy MP. Synthesis and characterization of a triphenylphosphonium-conjugated peroxidase mimetic - Insights into the interaction of ebselen with mitochondria. J Biol Chem. 2005;280:24113–24126. doi: 10.1074/jbc.M501148200. [DOI] [PubMed] [Google Scholar]
- 43.Mahon KP, Potocky TB, Blair D, Roy MD, Stewart KM, Chiles TC, Kelley SO. Deconvolution of the cellular oxidative stress response with organelle-specific Peptide conjugates. Chem Biol. 2007;14:923–930. doi: 10.1016/j.chembiol.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 44. Murphy MP, Smith RAJ. Targeting Antioxidants to Mitochondria by Conjugation to Lipophilic Cations. Annu Rev Pharmacol Toxicol. 2007;47:629–656. doi: 10.1146/annurev.pharmtox.47.120505.105110. *This review describes the use of triphenylphosphoium groups to deliver cargoes to the mitochondria.
- 45. Lema FY, Kelly MS, Shana OK. Targeting Mitochondria with Organelle-Specific Compounds: Strategies and Applications. ChemBioChem. 2009;10:1939–1950. doi: 10.1002/cbic.200900185. **This review describes the current approaches to delivering small-molecules to mitochondria.
- 46.Rothe G, Valet G. Flow cytometric analysis of respiratory burst activity in phagocytes with hydroethidine and 2',7'-dichlorofluorescin. J Leukocyte Biol. 1990;47:440–448. [PubMed] [Google Scholar]
- 47.Tarpey MM, Wink DA, Grisham MB. Methods for detection of reactive metabolites of oxygen and nitrogen: in vitro and in vivo considerations. Am J Physiol Regul Integr Comp Physiol. 2004;286:R431–R444. doi: 10.1152/ajpregu.00361.2003. [DOI] [PubMed] [Google Scholar]
- 48. Robinson KM, Janes MS, Pehar M, Monette JS, Ross MF, Hagen TM, Murphy MP, Beckman JS. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. P Natl Acad Sci USA. 2006;103:15038–15043. doi: 10.1073/pnas.0601945103. ** Mitosox is a targetable fluorescent indicator for superoxide and general oxidations.
- 49.Robinson KM, Janes MS, Beckman JS. The selective detection of mitochondrial superoxide by live cell imaging. Nat Protoc. 2008;3:941–947. doi: 10.1038/nprot.2008.56. [DOI] [PubMed] [Google Scholar]
- 50.Villamena FA, Zweier JL. Detection of reactive oxygen and nitrogen species by EPR spin trapping. Antioxid Redox Sign. 2004;6:619–629. doi: 10.1089/152308604773934387. [DOI] [PubMed] [Google Scholar]
- 51.Hardy M, Chalier F, Ouari O, Finet JP, Rockenbauer A, Kalyanaraman B, Tordo P. Mito-DEPMPO synthesized from a novel NH2-reactive DEPMPO spin trap: a new and improved trap for the detection of superoxide. Chem Commun. 2007;2007:1083–1085. doi: 10.1039/b616076j. [DOI] [PubMed] [Google Scholar]
- 52.Wipf P, Xiao J, Jiang J, Belikova NA, Tyurin VA, Fink MP, Kagan VE. Mitochondrial targeting of selective electron scavengers: synthesis and biological analysis of hemigramicidin-TEMPO conjugates. J Am Chem Soc. 2005;127:12460–12461. doi: 10.1021/ja053679l. [DOI] [PubMed] [Google Scholar]
- 53.Fink MP, et al. Hemigramicidin-TEMPO conjugates: novel mitochondria-targeted anti-oxidants. Biochem Pharmacol. 2007;74:801–809. doi: 10.1016/j.bcp.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 54.Ban S, Nakagawa H, Suzuki T, Miyata N. Novel mitochondria-localizing TEMPO derivative for measurement of cellular oxidative stress in mitochondria. Bioorg Med Chem Lett. 2007;17:2055–2058. doi: 10.1016/j.bmcl.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 55. Koide Y, Urano Y, Kenmoku S, Kojima H, Nagano T. Design and synthesis of fluorescent probes for selective detection of highly reactive oxygen species in mitochondria of living cells. J Am Chem Soc. 2007;129:10324–10325. doi: 10.1021/ja073220m. **MitoAR and MitoHR are mitochondrial-localized probes for highly reactive oxygen species (hROS).
- 56.Chang MCY, Pralle A, Isacoff EY, Chang CJ. A selective, cell-permeable optical probe for hydrogen peroxide in living cells. J Am Chem Soc. 2004;126:15392–15393. doi: 10.1021/ja0441716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller EW, Albers AE, Pralle A, Isacoff EY, Chang CJ. Boronate-based fluorescent probes for imaging cellular hydrogen peroxide. J Am Chem Soc. 2005;127:16652–16659. doi: 10.1021/ja054474f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Albers AE, Okreglak VS, Chang CJ. A FRET-based approach to ratiometric fluorescence detection of hydrogen peroxide. J Am Chem Soc. 2006;128:9640–9641. doi: 10.1021/ja063308k. [DOI] [PubMed] [Google Scholar]
- 59.Miller EW, Tulyathan O, Isacoff EY, Chang CJ. Molecular imaging of hydrogen peroxide produced for cell signaling. Nat Chem Biol. 2007;3:263–267. doi: 10.1038/nchembio871. [DOI] [PubMed] [Google Scholar]
- 60.Srikun D, Miller EW, Domaille DW, Chang CJ. An ICT-based approach to ratiometric fluorescence imaging of hydrogen peroxide produced in living cells. J Am Chem Soc. 2008;130:4596–4597. doi: 10.1021/ja711480f. [DOI] [PubMed] [Google Scholar]
- 61. Dickinson BC, Chang CJ. A Targetable Fluorescent Probe for Imaging Hydrogen Peroxide in the Mitochondria of Living Cells. J Am Chem Soc. 2008;130:11561. doi: 10.1021/ja802355u. ** This communication describes the synthesis and application of MitoPY1, a unique small-molecule probe for chemospecific detection of hydrogen peroxide in the mitochondria.
- 62.Yang D, Wang HL, Sun ZN, Chung NW, Shen JG. A highly selective fluorescent probe for the detection and imaging of peroxynitrite in living cells. J Am Chem Soc. 2006;128:6004–6005. doi: 10.1021/ja0603756. [DOI] [PubMed] [Google Scholar]
- 63.Maeda H, et al. A design of fluorescent probes for superoxide based on a nonredox mechanism. J Am Chem Soc. 2005;127:68–69. doi: 10.1021/ja047018k. [DOI] [PubMed] [Google Scholar]
- 64.Sasaki E, Kojima H, Nishimatsu H, Urano Y, Kikuchi K, Hirata Y, Nagano T. Highly sensitive near-infrared fluorescent probes for nitric oxide and their application to isolated organs. J Am Chem Soc. 2005;127:3684–3685. doi: 10.1021/ja042967z. [DOI] [PubMed] [Google Scholar]
- 65.Lim MH, Xu D, Lippard SJ. Visualization of nitric oxide in living cells by a copper-based fluorescent probe. Nat Chem Biol. 2006;2:375–380. doi: 10.1038/nchembio794. [DOI] [PubMed] [Google Scholar]
- 66.Kenmoku S, Urano Y, Kojima H, Nagano T. Development of a highly specific rhodamine-based fluorescence probe for hypochlorous acid and its application to real-time imaging of phagocytosis. J Am Chem Soc. 2007;129:7313–7318. doi: 10.1021/ja068740g. [DOI] [PubMed] [Google Scholar]
- 67.Sun ZN, Liu FQ, Chen Y, Tam PKH, Yang D. A highly specific BODIPY-based fluorescent probe for the detection of hypochlorous acid. Org Lett. 2008;10:2171–2174. doi: 10.1021/ol800507m. [DOI] [PubMed] [Google Scholar]
- 68.Garner AL, St Croix CM, Pitt BR, Leikauf GD, Ando S, Koide K. Specific fluorogenic probes for ozone in biological and atmospheric samples. Nat Chem. 2009;1:316–321. doi: 10.1038/nchem.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller EW, Bian SX, Chang CJ. A fluorescent sensor for imaging reversible redox cycles in living cells. J Am Chem Soc. 2007;129:3458–3459. doi: 10.1021/ja0668973. [DOI] [PMC free article] [PubMed] [Google Scholar]