Abstract
Cytochrome P450 (CYP) 24A1 catalyzes the side-chain oxidation of the hormonal form of vitamin D. Expression of CYP24A1 is up-regulated to attenuate vitamin-D signaling associated with calcium homeostasis and cellular growth processes. The development of therapeutics for disorders linked to vitamin D-insufficiency would be greatly facilitated by structural knowledge of CYP24A1. Here we report the crystal structure of rat CYP24A1 at 2.5 Å resolution. The structure exhibits an open cleft leading to the active site heme prosthetic group on the distal surface that is likely to define the path of substrate access into the active site. The entrance to the cleft is flanked by conserved hydrophobic residues on helices A′ and G′ suggesting a mode of insertion into the inner mitochondrial membrane. A docking model for 1α,25-(OH)2D3 binding in the open form of CYP24A1 is proposed that clarifies the structural determinants of secosteroid recognition and validates the predictive power of existing homology models of CYP24A1. Analysis of CYP24A1's proximal surface identifies the determinants of adrenodoxin recognition as a constellation of conserved residues from helices K, K″ and L that converge with an adjacent lysine-rich loop for binding the redox protein. Overall, the CYP24A1 structure provides the first template for understanding membrane insertion, substrate binding, and redox partner interaction in mitochondrial P450s.
Keywords: cytochrome P450, mitochondria, monotopic membrane protein, vitamin D metabolism, adrenodoxin
Introduction
Mitochondrial P450 enzymes catalyze highly specific reactions in the biosynthesis and degradation of hormones [1,2]. Mitochondrial P450s, i.e. CYP11, 24 and 27 families, cluster into a single evolutionary clan; CYP24A1 itself has less than 40% sequence identity with other P450s [3]. An N-terminal leader sequence targeting these P450s to the mitochondrion is cleaved during import; the mature enzymes bind monotopically to the matrix side of the inner membrane. Mitochondrial P450s exhibit the same monooxygenase activity as the microsomal enzymes (R-H + NADPH + H+ + O2 → R-OH + NADP+ + H2O), but are coupled to a two-component electron transfer system comprised of the soluble iron-sulfur ([2Fe-2S]) protein adrenodoxin (Adx), and adrenodoxin reductase (Adr), a FAD-containing flavoenzyme which binds NADPH [2,4]. In this respect, mitochondrial P450s resemble the soluble bacterial P450 CYP101 from Pseudomonas putida, which utilizes a comparable putidaredoxin (Pdx) and putidaredoxin reductase (Pdr) system [5]. Adrenodoxin is expected to bind to the proximal surface of the enzyme adjacent to the heme prosthetic group [6,7].
Because CYP24A1 degrades vitamin D, it is a target for inhibitor development for treatment of diseases linked to vitamin D insufficiency, such as bone disorders, kidney disease, and cancer [8-11]. Regulation of vitamin D activity is crucial to human health, as the active hormone, 1α,25-dihydroxyvitamin D3 (1α,25-(OH)2D3 or calcitriol), controls gene-expression and signal-transduction processes associated with calcium homeostasis, cellular growth, and the maintenance of heart, muscle, immune, and skin function [12,13]. Evidence suggests that vitamin D insufficiency, due to tissue specific CYP24A1 overexpression, contributes to development of diabetes [14] and cancers of the prostate, breast, colon and lung [15-18]. A number of promising vitamin D analogs have been developed [19], but their administration often induces hormonal resistance. Therefore, a complementary strategy for combating vitamin D insufficiency is to develop specific CYP24A1 inhibitors to retard degradation of the endogenous hormone.
The primary circulating form of vitamin D3 (25-hydroxyvitamin D3, 25-(OH)D3) is converted to calcitriol by mitochondrial CYP27B1 (the 25D3-1α-hydroxylase) under hypocalcemic conditions. Calcitriol elicits pleiotropic effects, in part via the vitamin D nuclear receptor (VDR). The CYP24A1 promoter contains multiple vitamin D response elements (VDREs) that respond to VDR signaling; CYP24A1 expression results in side-chain cleavage and elimination of circulating 25-(OH)D3 and 1α,25-(OH)2D3 by catalyzing hydroxylation at the 23 and 24 positions [12,20]. The mechanism of CYP24A1's six step oxidation pathway for production of calcitriol remains poorly understood [21-22]. Structural knowledge of CYP24A1 would elucidate the basis for secosteroid specificity, and facilitate the rational design of inhibitors, an approach complementary to the synthetic analog design already exploiting structural studies of the VDR [23]. Here we report the crystal structure of the rat CYP24A1 determined to 2.5 Å resolution.
Results
We developed a bacterial over-expression system for mature rat CYP24A1 lacking the mitochondrial import signal (Δ2-32) to enable biochemical characterization of the purified enzyme [24]. Subsequent affinity-labeling studies identified a stabilizing mutation for recombinant CYP24A1 (Ser-57-Asp) that does not alter catalytic function [25], and site-directed mutational analysis using P450 homology modeling established functional roles for conserved residues involved in substrate recognition and catalysis [26]. Recombinant preparations (Δ2-32, S57D) were solubilized from bacterial membranes and purified in the presence of CHAPS (0.8% w/v) by affinity chromatography using adrenodoxin linked to sepharose, yielding monodisperse samples that crystallize readily by vapor diffusion (Supplemental Methods). Detergent exchange from CHAPS into CYMAL®-5 afforded crystals of CYP24A1 which diffracted to 3.0 Ǻ resolution in space group C2 on SSRL beamline 9-2. The structure was solved by molecular replacement (MR) with Phaser [27] using an ensemble of microsomal P450 structures (Supplemental Methods). A model with two molecules in the asymmetric unit was built using COOT [28] and MiFit [29], and refined using the CCP4 suite [30]. Concurrently, an improved “mixed-detergent” method for growing CYP24A1 crystals was developed that eliminated the need for CYMAL®-5 exchange by utilizing a second detergent together with CHAPS. CYP24A1 in mixed-detergent micelles with CHAPS (0.5% w/v) and common detergents (e.g. CYMAL®-5, Foscholine®-12) at 0.5-2-fold their critical micelle concentration, grew readily, but diffracted no better than 4 Å resolution.
However, a novel class of detergents, termed facial amphiphiles (FA) [31], are effective in improving CYP24A1 crystal quality in combination with CHAPS; single, rod-like crystals grown in the presence of CHAPS and the FA, 231-chol (3α-hydroxy-7α,12α-bis[(β-D-maltopyranosyl)ethyloxy]cholane), show diffraction to 2.0 Å resolution. A 7.6-fold redundant 2.5 Å data set was collected on SSRL beamline 12-2 [32], and used to complete and refine the CYP24A1 model including residues 51-514 (Table S1). Residues 33-50 of the S57D construct were not visible in the electron density. The two copies of CYP24A1 in the asymmetric unit are very similar with a root mean square deviation between Cα atoms of 0.62 Ǻ.
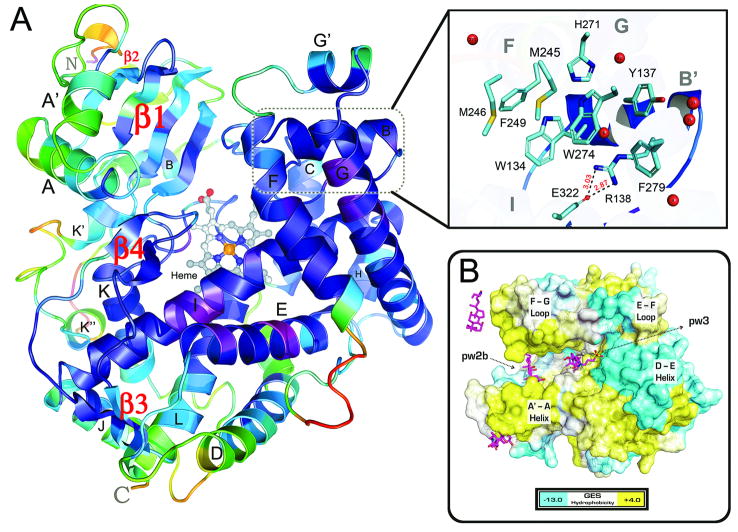
The open form structure of rat CYP24A1 displays the canonical P450 fold, including the 12 α-helices (A-L) and four β-sheet systems (β1-β4), as well as additional helices A′, B′, and G′ on the distal surface, and K′ and K″ between β2 and the conserved heme binding motif (Figs. 1A, S1). The F-helix of CYP24A1 extends for 18 residues and lacks a well defined F′-helix seen for example in structures of microsomal CYP3A4 [34,35]. The structural elements defining the substrate binding cavity, including the β1 and β4 sheets, the B-C loop, and helices E, F, G, I and K surrounding the heme, exhibit lower than average B-factors (Fig. 1A). The B-C loop contains a 3-turn B′ helix that packs closely with helices G and G′. A distinct arrangement of aromatic residues from helices B′ (Trp134, Tyr137), F (Phe249), and G (His271, Trp275, Phe279) cluster together with extensive interactions (Fig. 1A, inset), contributing to the lower B-values in this region of the structure. The interaction between residues of helix G with helix B′ entraps a water molecule hydrogen bonded with the carbonyl oxygen of Trp134 and the side chain of Arg138; the latter residue engages in a salt bridge with Glu322 on the adjacent I helix. The aromatic cluster effectively blocks access to the active site via the solvent-accessible-channel (pw2c) along the I-helix [36]. At the same time, these residues contribute significantly to the hydrophobicity of the active-site interior. The eight residues comprising the aromatic cluster and salt bridge are conserved across all mitochondrial P450s (Fig. S2). Hence, the aromatic cluster is a distinctive feature of CYP24A1, and may be a general feature of mitochondrial P450s, stabilizing the open form.
Fig 1. The Crystal Structure of CYP24A1.
A) The structure of CYP24A1 refined at 2.5 Ǻ resolution (R=0.206, Rfree=0.252) shown colored by B-factor from high (red) to low (violet) temperature. P450 structural elements (α-helices, β-sheets) are labeled. Conserved residues from helices F (M245, F249), G (H271, W275, F279) and B′ (W134, Y137, R138) participate in an “aromatic cluster” centered on a water-molecule bound to R138 that promotes a membrane-directed (pw2a) substrate access channel. B) The distal surface of CYP24A1 is shown colored from negative (cyan) to positive (yellow) hydrophobicity (GES scale [41]) revealing the open channel between the A′-A helix & F-G loop regions. CHAPS molecules (pink sticks) from the crystal structure are overlaid to illustrate a model for substrate diffusion from the membrane-associated channel (pw2a) to a putative exit channel (pw3) between the D-E helix region & E-F loop [36].
The substrate access channel apparent in CYP24A1 is occupied by detergent molecules (Figs. 1B, S4) and lies among the B-B′ segment of the B-C loop, the β1-sheet and the F-G loop (pw2a); this channel appears to partially merge with a secondary channel that also lies between the B-B′ loop and the β1-sheet, and enters from below the F-G loop (pw2b) [36]. A similar substrate access channel, oriented towards the membrane, is present in the structure of the microsomal P-450, CYP46A1, which is a cholesterol 24-hydroxylase [37]. This feature distinguishes CYP24A1 from the non-specific microsomal vitamin D 25-hydroxylase, CYP2R1, which contains a pw2c substrate binding channel typical of enzymes from the CYP2 family [38].
Among currently available structures, the overall topology of structural features that form the substrate binding cavity in CYP24A1 is most similar to that of CYP3A4 [34,35]. A comparison of the two structures suggests that the open cleft observed for CYP24A1 results from the close packing of helix F with helix G and the elevated pitch of both helices that separates helix F from the β4 sheet. This separation is increased by the conformation of the β1-4 sheet region which is similar to that seen in CYP3A4. This conformation exposes a larger portion of the heme surface and a widening of the substrate binding cavity close to the catalytic center, relative to structures of mammalian P450s of families 1 or 2 [34]. In structures of CYP3A4, the helix F does not extend over the substrate binding cavity and the polypeptide chain exhibits a coil structure as it passes across the cavity and connects with helix F′. In contrast, the helix F of CYP24A1 extends across the structure toward helix G′ without a well defined helix F′.
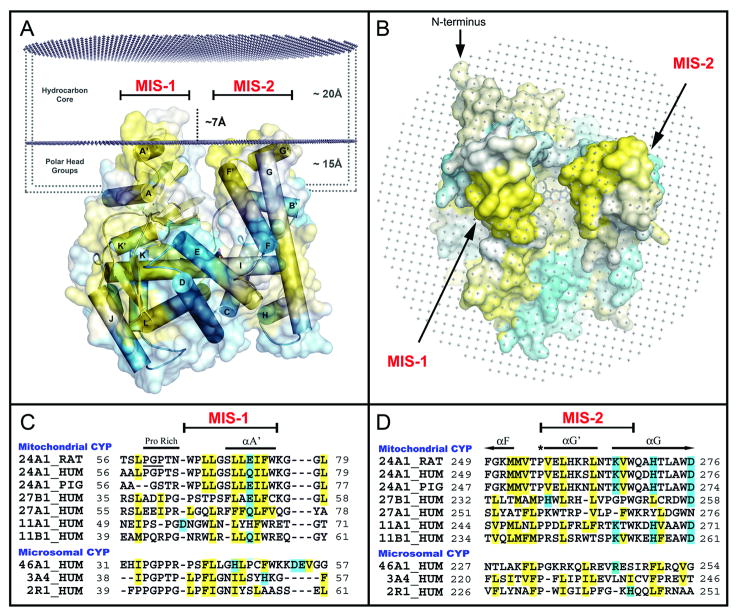
Analysis of the CYP24A1 structure [39-40] identified hydrophobic surfaces of helices A′ and G′ as potential anchors to the mitochondrial inner membrane; together, these short helices flank the substrate access channel (Figs. 1A, 1B). The corresponding A′-A-helix and F-G loop regions of microsomal P450s are also implicated in membrane association [42-45]. Computational prediction of CYP24A1 insertion into a simulated lipid bilayer confirms this analysis (Figs. 2A, 2B). The predicted protein:membrane association transfer free-energy (-9.0 kcal/mol) is within the range of values predicted from other P450 crystal structures (e.g. CYP3A4 (1TQN) [34] = -20.7 kcal/mol; and CYP46A1 (2Q9F) [37] = -4.7 kcal/mol) [46]. This analysis also suggests that residues on helices A′ and G′ can penetrate approximately 7 Å into the alkyl chain region of the bilayer, or up to 22 Å into the outer leaflet (Fig. 2A) [47].
Fig 2. Adaptations for Monotopic Membrane Binding.
A) Interactions between CYP24A1 and the lipid-bilayer were studied computationally with the OPM server [46] and our model for membrane-binding is shown colored by hydrophobicity (GES [41]). Two membrane-insertion-sequences (MIS) are predicted for CYP24A1 that correspond to hydrophobic surface regions (helices A′ and G′) that are modeled to penetrate into the membrane's carbonyl core (∼17-22 Å) with polar lipid head groups reaching deep into the substrate-access channel between the A′-A helix and F-G loop regions. B) An orthogonal view of the OPM model illustrates the width and depth of the hydrophobic substrate access leading to the heme center. C.) Primary sequence alignments of predicted MIS-1 and D) MIS-2 binding regions demonstrate the conservation of membrane binding features across key membrane-bound P-450 forms.
Alignment of the A′ and G′ segments with other mitochondrial P450 sequences identifies each as a membrane insertion sequence (MIS), implying a conserved membrane binding architecture within mitochondrial CYPs 11, 24, and 27 (Fig. 2C, 2D). In MIS-1, the conserved proline-rich (PGP) region is followed by a hydrophobic segment of 9-12 residues bracketed by aromatic amino acids (Trp64 and Trp75, Fig. 2C). MIS-2 encompasses the amphipathic G′-helix, a distinctive feature of the structure (Fig. 1A), in which polar residues are oriented back towards the protein and hydrophobic residues point outward. MIS-2 is delimited by a conserved proline in the F-G loop region (Pro256), and a tryptophan at the N-terminus of the G-helix, present in all mitochondrial P450s (Trp268, Fig. 2D). The alignment in Fig. 2D suggests that the amphipathic nature of the G′-helix is a conserved feature of mitochondrial P450s. A number of conserved, basic residues are also arrayed in the vicinity of the A′ and G′ hydrophobic regions, consistent with interactions with phospholipid head groups (Fig. S3).
Four molecules of CHAPS were observed in copy A of the 2.5 Å structure of CYP24A1. Two molecules are associated with membrane binding regions (MIS-1 & 2) and structural lattice contacts, one is bound in the substrate-access channel and a fourth is positioned above the heme in a non-productive binding orientation (Fig. 1B). Alternatively, two molecules of CYMAL®-5 occupy the pw2a access channel in the independent 3.0 Å structure of CYP24A1 (Fig. S4). The arrangement of either CHAPS or CYMAL®-5 in isomorphous CYP24A1 structures suggest that the open form of CYP24A1 is a biologically-relevant conformation that is not dependent on the specific details of detergent binding. Additionally, the presence of trapped detergent molecules is consistent with the hydrophobic nature of the access channel and binding pocket, and illustrates a potential pathway for lipophillic substrates to transit from the membrane to the active site.
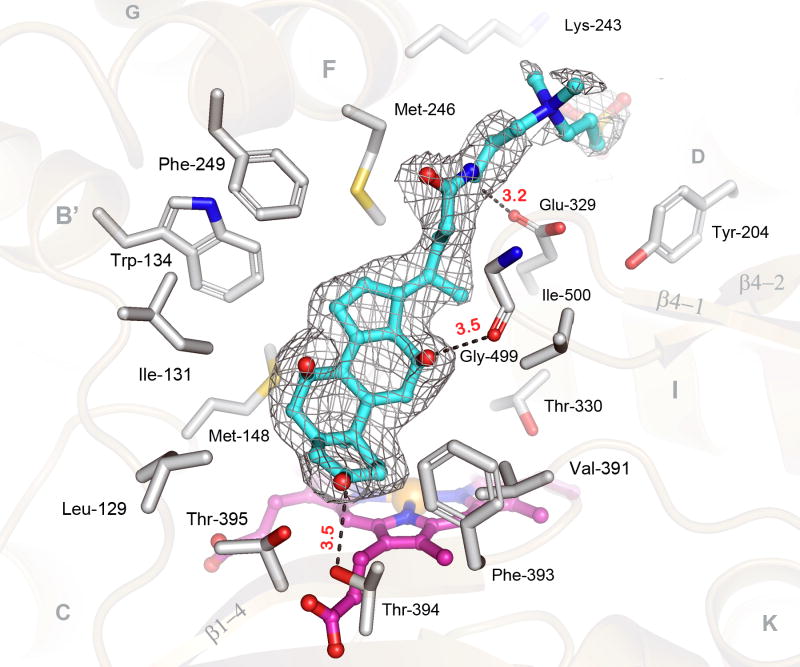
There is well defined electron density for a CHAPS molecule (CPS-600, 3K9V) close to the heme within the substrate-binding pocket of CYP24A1 (Fig. 3), but density for the zwitterionic side-chain extending out from the cavity is weaker. The hydrophobic face of the cholate ring is oriented toward the heme surface and non-polar residues of helices B′ and F, whereas the polar face of CHAPS is oriented toward polar protein groups, allowing three hydrogen bonds to be formed. In solution, CHAPS does not inhibit the catalytic function of CYP24A1 and does not induce a Type-I spectral perturbation of the heme typical of a proper substrate [24-26]. So while detergent association in the active-site appears strong, it likely represents a non-productive interaction that does not promote the enzyme's closed-form. Nevertheless, CHAPS binding is illustrative of the hydrophobic nature of the active site, and its ability to provide specific interactions with amphipathic molecules.
Fig 3. The Active Site of Rat CYP24A1 Bound to CHAPS.
A) The electron density for CHAPS (cyan carbons) (σA−weighted 2|Fo−Fc|-composite-omit map, 1.0σ) is shown in the heme-centered active-site of CYP24A1; important secondary structural elements (tan), CHAPS binding residues (grey carbons), the heme prosthetic group (pink carbons), and notable bond distances (Ǻ, red) are noted. Amino acid residues from the B-C loop (I131, M148) and Helices B′, F, G & I (W134, M246, F249 & T330) the K-helix/β1-4 loop (V391, F393) and the β4-1/β4-2 turn (Ile-500) mediate hydrophobic interactions with CHAPS. Polar/charged residues from the I-helix (E329), β1-4 sheet (T394, T395) and the β4-1/β4-2 turn (G499) mediated bonds to the polar face of CHAPS. The negatively charged tail of CHAPS is bound outside the active site and interacts with residues from the helices D (Y204) and F (K243).
Residues surrounding the active site cavity are contributed from nine regions of the CYP24A1 fold: B-B′ region (Leu129, Ile131), B′-helix (Trp134), B′-C region (Met148), F-helix (Met245, Met246, Phe249), G-helix (His271, Trp275), I-helix (Leu325, Ala326, Glu329, Thr330), K-helix/β1-4 loop (Val391), β1-4 sheet (Phe393, Thr394, Thr395), and the β4-1/β4-2 turn (Gly499, Ile500) (Fig. 3). Mutational analysis of CYP24A1 based on P450 homology modeling studies have attributed roles in substrate-binding and catalysis for 13 of the residues identified in the structure: Ile131, Trp134, Met148, Met245, Met246, Phe249, Ala326, Glu329, Thr330, Val391, Thr394, Gly499, and Ile500 [26,48,49]. Structural analysis also supports predicted roles for Ala326 (I-helix) and Ile-500 (β4-1/β4-2 loop) in modulating regiospecificity of hydroxylation in CYP24A1 [50, 51]. A residue from the β1-3 strand (Thr416) is also linked to this process [50], but is found outside the CHAPS binding site in the open channel. However, this location could be relevant for secosteroid binding by analogy to cholesterol-3-sulfate binding in CYP46A1 [37].
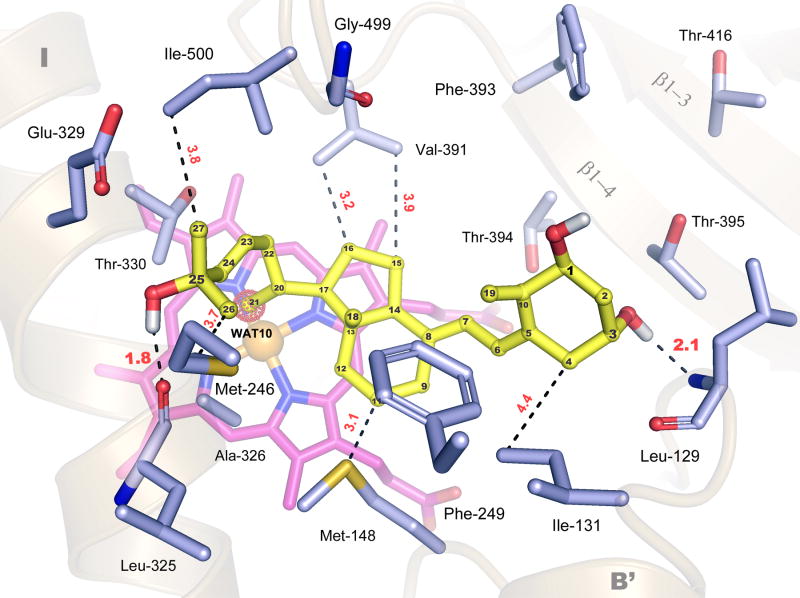
A hybrid homology model for CYP24A1 based on the structures of mammalian CYP2C5 and bacterial CYP102, successfully predicted multiple residues in the CYP24A1 binding pocket, including Met246, Phe249, Val391, Thr394, and Ile500 [26]. However, this model does retain some template bias as it predicts substrate-access and binding similar to CYP2C5, via the solvent-accessible pw2c channel that is occluded in the structure. A P450 structural-motif based method of homology model building utilized by Masuda et al. appears to be highly effective at predicting key elements of the CYP24A1 tertiary structure [48]. This 3D model has been used to make strong predictions concerning the positioning, and putative role, of conserved residues in the CYP24A1 active-site, including: Ile131, Trp134, Met148, Met246, Ala326, and Gly499. To clarify the functional role of putative substrate-binding residues in the open form, flexible ligand docking simulations with CHAPS (as a control) and 1α,25-(OH)2D3 were conducted against the rat CYP24A1 structure (Table S2) using Autodock 4.0 (see Materials and Methods) [70-72]. As illustrated in Fig. 4, 1α,25-(OH)2D3 docks in the open form of CYP24A1 with computed nanomolar affinity (Ki=2.65 nM) in a reproducible conformation stabilized by two hydrogen bonds (between the 3-OH group of the vitamin D A-ring and the B-B′ loop (Leu129), and the 25-OH of the side chain and the I-helix (Leu325)) and multiple hydrophobic interactions among key conserved residues (Ile131, Trp134, Met246, Phe249, Ala326 Val391, Thr394, Thr395, Gly499, and Ile500). The docking calculations confirm that the open form of CYP24A1 is well organized to bind 1α,25-(OH)2D3, in accord with predictions made by homology modeling concerning residues likely to bind the A-C-D ring system (Leu129, Ile131, Trp134, Phe249, Thr394, Val391) or side chain (Leu325, Ala326, Met246 and Ile500) of the vitamin D hormone. In this configuration, the secosteroid is well-positioned to interact with the aromatic cluster of residues among helices B′, F and G via interactions with the B-C loop (Leu129, Ile131, Trp134, Met148) and helix F (Met246, Phe249). However, it is unlikely that this result represents the substrate's terminal binding configuration relevant for the catalytically-active enzyme; the C21 methyl group of 1α,25-(OH)2D3, and not the target C23 or C24 carbons, is positioned over the heme iron, where it is superimposed with an ordered water molecule (WAT10, 3K9V). This result implies that in the open form secosteroid substrates bind over the heme in a fashion that brings the C21-methyl group into position to perturb the water-bound, heme iron (Figs. 4, S5). Further, when WAT-10 is included in the docking simulations a similar solution for 1α,25-(OH)2D3 is obtained (Table S2, Fig. S6), but here the C21-methyl is forced to rotate away from WAT10, forming a hydrophobic interaction with the important catalytic residue Ala326 [51]; the hydrogen bonding network is also altered as the 3-OH group is repositioned to interact with the β1-4 sheet (Thr395), rather than the B-B′ loop (Leu129) (Fig. S6). These derived results suggest that electrostatic interactions between the 25-OH group and the I-helix (Leu325), and hydrophobic interactions among the hormone's C-D ring system and the B′-C loop (Met148) and the K-helix/β1-4 loop (Val391), are essential for high-affinity substrate binding in the open form. This analysis validates published CYP24A1 homology modeling studies concerning the composition of residues in the active-site, and supports our hypothesis that the open form is a biologically relevant conformation, useful for studying substrate binding in mitochondrial P450s.
Fig 4. Secosteroid Docking in the Open Form of Rat CYP24A1.
A model for 1α,25-(OH)2D3 (yellow) binding in the heme-centered (pink) active-site of CYP24A1 was developed with a crystal structure-calibrated docking protocol, using Autodock 4.0 (see methods). Amino acid residues that flank the secosteroid docking site are shown from the B-B′ loop (L129, I131) B′-C loop (M148), helix F (M246, F249), helix I (L325, A326, E329, T330), the K-helix/β1-4 loop (V391, F393), the β1-4 sheet (T394, T395), the β1-3 sheet (T416), and the β4-1/β4-2 turn (G499, I500), and notable hydrogen bond distances (Ǻ, red) are given with respect to the computed H atom positions. Individual carbon atoms on 1α,25-(OH)2D3 are labeled for reference. Multiple hydrophobic interactions and two hydrogen bonds between the 3-OH group and the B-B′ loop (L129) and the 25-OH group and helix I (L325) are predicted to stabilize secosteroid binding in the open form. In this configuration, the C21-methyl group superimposes with a structural water (WAT-10) bound to the heme (shown as red-dotted sphere); this water molecule was excluded from the calibrated docking experiment shown here (Table S2, Fig. S5). An alternative docking model derived using control parameters, which include WAT-10, is presented in Supplemental Fig. S6.
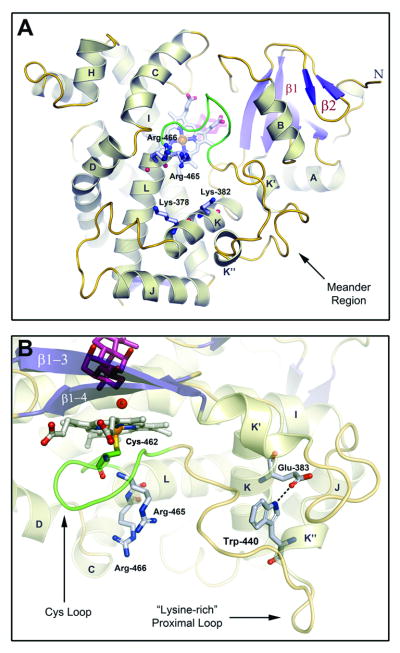
The CYP24A1 structure also reveals structural elements and key residues for interacting with the [2Fe-2S] electron transfer protein, adrenodoxin (Adx) (Fig. 5A). Four basic residues from helices K (Lys378, Lys382) and L (Arg465, Arg466), involved in Adx recognition and electron transfer in CYP27B1 [52], are juxtaposed on the proximal surface. These basic residues are conserved in mitochondrial P450s, but divergent in microsomal forms (Fig. S7). Structural elements on the proximal surface include segments of helices B, C, D, and K, helices K′ and K″ flanking the meander region, and the Cys loop (Fig. 5B), all of which are implicated in redox partner binding [52-55] and circumscribe the four basic residues. Compared to microsomal P450s, all mitochondrial CYPs contain a tryptophan (Trp440 in CYP24A1) in the conserved motif PxRWL in the K″-helix (Fig. S7). Trp440 interacts with the K-helix via another conserved residue, Glu383, within the invariant KExxR motif (Figs. 5B, S7). This conserved tertiary interaction, which anchors the lysine rich loop (residues 442-446, QKEKK), and is next to Lys378 and Lys382, appears to be important for Adx interaction. For example, it links a substrate recognition sequence (SRS5, K-helix/β1-4 sheet [56]) with the lysine rich loop, which mediates redox protein interaction in P450cam [67]. It also displays the conserved residue Leu441 on the protein surface, providing a complement to electrostatic interactions in CYP24A1-Adx recognition.
Fig 5. Adaptations for Adrenodoxin Binding.
A) CYP24A1's proximal surface is shown below the heme prosthetic group with key structural elements implicated in adrenodoxin binding noted. Basic residues from helices B, C, D, J, K and L, the Cys-Loop and the (bacterial) meander region line the positively-charged Adx binding. Fully-conserved residues from helices K (K378,K382) and L (R465,R466) known to mediate adrenodoxin binding and electron transfer in related P450s are labeled [52-55]. B) A conserved tryptophan residue (W440), from the K″ helix of mitochondrial P450s, forms a salt-bridge to the K-helix via a fully-conserved glutamate residue (E383) that may contribute to the display of the meander's lysine-rich, proximal loop, that is associated with Adx binding [57]. Residues from the L-helix (R465,R466) and Cys-loop (M462), implicated in the electron shuttle process are shown below the heme in close proximity to the K-helix. The lower portion of the active-site is also shown with CHAPS (pink) positioned above the water (WAT6) bound heme iron.
Discussion
Mitochondrial CYP24A1 is a monotopic membrane protein that binds tightly to, but does not span, the lipid bilayer [47,58,59]. The CYP24A1 structure indicates that CYP24A1 associates with the membrane using hydrophobic membrane insertion sequences and conserved basic residues surrounding helices A′ and G′ (Figs. 2, S3). Prior studies of both microsomal (CYP2C2, CYP3A4) and mitochondrial (CYP11A1, CYP27A1) P450s have established roles for the N-terminal proline rich motif, A′-A-helix region, and the F-G loop in membrane binding [42-45,60]. The CYP24A1 structure is consistent with these results and clarifies the key role of the conserved membrane-insertion elements (MIS) encompassing helices A′ and G′. However, the MIS-1 and -2 features of CYP24A1 are not shared by bacterial P450-BM3 (CYP102), a fatty acid metabolizing, soluble P450 that otherwise shares common features with mitochondrial P450s [61,62]. MIS elements could also mediate protein:lipid interactions, as postulated for other monotopic membrane proteins [58], such as the C2 domain of cytosolic phospholipase A2 [63]. Penetration of CYP24A1 up to 22 Å into the bilayer could account for the roles of specific lipids, e.g. cardiolipin, in altering mitochondrial P450 function [64,65].
The open conformation of CYP24A1, compatible with substrate diffusion from within the membrane, is uniquely stabilized by a cluster of conserved aromatic amino acids from helices B′, F, and G. The aromatic cluster blocks the solvent accessible channel (pw2c) formed by helices B′, G, and I, and promotes formation of a pw2a channel directed toward the membrane [66]. Stabilization of an open conformation may also contribute to substrate specificity in CYP24A1. Active site collapse to a closed state, as required to exclude solvent during catalysis, may be triggered by solvent displacement and stabilized by the enthalpy of specific interactions with the biological substrate. Hence, the open form would disallow efficient, non-specific oxidation of improper substrates which are not able to stabilize the closed state, defining at least one mechanism by which CYP24A1, and perhaps mitochondrial P450s, may achieve their exquisite specificity. Docking analysis supports this hypothesis, as 1α,25-(OH)2D3 is computed to bind deep within the active-site where it can interact with the heme group and presumably accommodate the cavity collapse required for catalysis. In contrast, the non-substrate CHAPS, which also docks the enzyme with computed nanomolar affinity (Ki=5.79 nM), is bound in a shallow conformation with limited access to the heme that would not be predicted to inhibit secosteroid accessibility. These studies provide new insight into the enzyme's active-site organization before substrate-binding, providing testable hypotheses concerning the determinants of secosteroid recognition. These contributions should prove useful in the evaluation and design of vitamin D analogs and CYP24A1-specific-inhibitors.
The proximal surface of CYP24A1 interacts with Adx and is rich in basic residues, including the invariant arginine pair (Arg465, Arg466) at the N-terminal end of the L-helix (Fig. 5A). These side chains protrude below the heme and are within 8-10 Ǻ of the conserved K-helix residues, Lys378 and Lys382. Mutational analysis of the arginine pair in CYP11A1 and CYP27B1 suggested that the first arginine contributes to protein folding events and/or heme binding, as mutations abolish enzyme function and promote the P-420 form [52,53]; an Arg465 mutant of CYP24A1 (R465F) showed a similar dysfunction [26]. Hence, Arg465 may play a dual role. The second arginine (Arg466) has been more clearly linked to catalytic function and is thought to mediate both electron and oxygen transfer steps [52]. In CYP24A1, Arg466 is positioned to promote electron transfer between Adx and the heme iron via main chain interactions with Cys462, as modeled for the CYP101:Pdx complex [67]. Conserved residues from the K-helix (Lys378, Lys382) and the lysine rich loop are also positioned to interact with the very acidic surface of Adx that surrounds the [2Fe-2S] cluster [5,68]. The link between the K and K″ helices provided by the conserved Glu383-Trp440 tertiary structure interaction suggests that presentation of the PxRWL motif is another key aspect of Adx recognition.
In summary, we have solved the first structure of a mitochondrial P450, CYP24A1, which defines key features for this important class of enzymes. A membrane-directed (pw2a) substrate access channel is stabilized in an open conformation by an aromatic cluster formed among conserved residues on helices B′, F, and G. Two conserved membrane insertion sequences overlap hydrophobic features on the distal surface near helices A′ and G′. Membrane insertion and orientation of the enzyme's hydrophobic channel shows how a lipophillic substrate, in this case vitamin D, can be transferred from the bilayer to the active site. Conserved surface features from helices K, K″ and L, and the lysine-rich proximal loop of the meander region, define components of the basic, adrenodoxin binding site. The composition of residues in the active site is consistent with prior biochemical, mutagenesis, and homology modeling data for CYP24A1, and related mitochondrial P450s, and docking simulations with 1α,25-(OH)2D3 clarify the structural determinants of secosteroid recognition in the enzyme's open conformation.
Materials and Methods
Recombinant rat CYP24A1 enzyme (Δ2-32, S57D mutant) was purified using established protocols [24-26]. Some modifications to our original procedure were required for crystallization (as detailed in Supplemental Methods). In short, a secondary chromatography step was added that allows CYP24A1 samples prepared in CHAPS (Anatrace) to be exchanged into alternative detergents, like CYMAL®-5 (Anatrace), as described by Wester et. al [69]. Detergent-exchange experiments identified conditions for growing a CYMAL®-5-based crystals that diffract to 2.8 Å, and allowed the CYP24A1 structure to be solved by the molecular replacement method using Phaser [27]. Later a mixed-detergent method was established for improving CYP24A1 crystal quality without the need for detergent exchange. Facial amphiphile detergents developed at Scripps [31] were found to produce high-order CYP24A1 crystals that diffracted to 2.0 Å resolution when mixed with CHAPS. Diffraction data for all CYP24A1 crystal forms were collected at the Stanford Synchrotron Radiation Laboratory (SSRL BL9-2 and BL12-2) [32].
Detailed methods for structural determination & refinement, computational (surface and membrane binding) analysis, and flexible-ligand docking studies with Autodock 4.0 [70-72], are provided in the Supplementary materials. All sequence alignments were developed using the program CLUSTALX2 [73]; labeling and format are explained in Supplemental methods. All figures for this manuscript were generated using Pymol [74].
Supplementary Material
Acknowledgments
Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. We are grateful to the Stanford Synchotron Radiation Laboratory (SSRL) staff for their generous support. We express gratitude to the University of New Mexico, Department of Biochemistry & Molecular Biology, for donating materials from the laboratory of the late Dr. John L. Omdahl, that supported this research. We would also like to thank our collaborators Dr. Irina Pikuleva and Dr. Natalia Mast (Case Western Medical Center) for providing valuable guidance in mitochondrial P450 crystallization. This work was supported by the following NIH grants: GM031001 (to E.F.J.), GM41049 (to D.B.G.) and GM073197 (to Q.Z.).
Footnotes
Author Contributions: A.J.A., C.D.S., D.B.G. and E.F.J. designed the research; A.J.A. and C.D.S. performed research; A.J.A., C.D.S., D.B.G. and E.F.J. analyzed data. W.H. and Q.Z. provided new materials; A.J.A., C.D.S., D.B.G. and E.F.J. wrote the paper;
Accession Codes: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3K9V and 3K9Y).
Data Deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3K9V and 3K9Y).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Andrew J. Annalora, Email: annalora@scripps.edu.
C. David Stout, Email: dave@scripps.edu.
References
- 1.Bernhardt R. Cytochromes P450 as versatile biocatalysts. J Biotechnol. 2006;124:128–145. doi: 10.1016/j.jbiotec.2006.01.026. Review. [DOI] [PubMed] [Google Scholar]
- 2.Sigel A, Sigel H, Sigel R. In: Metal Ions in Life Sciences, Vol 3: The Ubiquitous Roles of Cytochrome P450 Proteins. Sigel A, Sigel H, Sigel R, editors. New Jersey: John Wiley and Sons, Ltd.; 2007. [Google Scholar]
- 3.Nelson DR. Metazoan cytochrome P450 evolution. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1998;1-3:15–22. doi: 10.1016/s0742-8413(98)10027-0. Review. [DOI] [PubMed] [Google Scholar]
- 4.Guengerich FP. Mechanisms of cytochrome P450 substrate oxidation: MiniReview. J Biochem Mol Toxicol. 2007;21:163–168. doi: 10.1002/jbt.20174. Review. [DOI] [PubMed] [Google Scholar]
- 5.Grinberg AV, Hannemann F, Schiffler B, Müller J, Heinemann U, Bernhardt R. Adrenodoxin: structure, stability, and electron transfer properties. Proteins. 2000;40:590–612. doi: 10.1002/1097-0134(20000901)40:4<590::aid-prot50>3.0.co;2-p. Review. [DOI] [PubMed] [Google Scholar]
- 6.Pikuleva IA, Tesh K, Waterman MR, Kim Y. The tertiary structure of full-length bovine adrenodoxin suggests functional dimers. Arch Biochem Biophys. 2000;373(1):44–55. doi: 10.1006/abbi.1999.1536. [DOI] [PubMed] [Google Scholar]
- 7.Behlke J, Ristau O, Müller EC, Hannemann F, Bernhardt R. Self-association of adrenodoxin studied by using analytical ultracentrifugation. Biophys Chem. 2007;125(1):159–165. doi: 10.1016/j.bpc.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Prosser DE, Jones G. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem Sci. 2004;12:664–673. doi: 10.1016/j.tibs.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Sakaki T, Kagawa N, Yamamoto K, Inouye K. Metabolism of vitamin D3 by cytochromes P450. Front Biosci. 2005;10:119–134. doi: 10.2741/1514. [DOI] [PubMed] [Google Scholar]
- 10.Masuda S, Jones G. Promise of vitamin D analogues in the treatment of hyperproliferative conditions. Mol Cancer Ther. 2006;4:797–808. doi: 10.1158/1535-7163.MCT-05-0539. [DOI] [PubMed] [Google Scholar]
- 11.Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nature Rev Cancer. 2007;7:684–700. doi: 10.1038/nrc2196. [DOI] [PubMed] [Google Scholar]
- 12.Omdahl JL, Morris HA, May BK. Hydroxylase Enzymes of the Vitamin D Pathway: Expression, Function, and Regulation. Annu Rev Nutr. 2002;22:139–166. doi: 10.1146/annurev.nutr.22.120501.150216. [DOI] [PubMed] [Google Scholar]
- 13.Dusso AS, Brown AJ, Slatopolsky E. Vitamin D. Am J Physiol Renal Physiol. 2005;289:8–28. doi: 10.1152/ajprenal.00336.2004. [DOI] [PubMed] [Google Scholar]
- 14.Holick MF. The vitamin D deficiency pandemic and consequences for nonskeletal health: mechanisms of action. Mol Aspects Med. 2008;29(6):361–368. doi: 10.1016/j.mam.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welsh J. Vitamin D and breast cancer: insights from animal models. Am J Clin Nutr. 2004;80:1721S–4S. doi: 10.1093/ajcn/80.6.1721S. [DOI] [PubMed] [Google Scholar]
- 16.Lou YR, Qiao S, Talonpoika R, Syvala H, Tuohimaa P. The role of Vitamin D3 metabolism in prostate cancer. J Steroid Biochem Mol Biol. 2004;92:317–325. doi: 10.1016/j.jsbmb.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Parise RA, Egorin MJ, Kanterewicz B, Taimi M, Petkovich M, Lew AM, Chuang SS, Nichols M, El-Hefnawy T, Hershberger PA. CYP24, the enzyme that catabolizes the antiproliferative agent vitamin D, is increased in lung cancer. Int J Cancer. 2006;119:1819–1828. doi: 10.1002/ijc.22058. [DOI] [PubMed] [Google Scholar]
- 18.Pendás-Franco N, Aguilera O, Pereira F, González-Sancho JM, Munoz A. Vitamin D and Wnt/beta-catenin pathway in colon cancer: role and regulation of DICKKOPF genes. Anticancer Res. 2008;28(5A):2613–2623. Review. [PubMed] [Google Scholar]
- 19.DeLuca HF. Evolution of our understanding of vitamin D. Nutr Rev. 2008;66:S73–87. doi: 10.1111/j.1753-4887.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 20.Sakaki T, Sawada N, Komai K, Shiozawa S, Yamada Y, Yamamoto K, Ohyama Y, Inouye K. Dual metabolic pathway of 25-hydroxyvitamin D3 catalyzed by human CYP24. Eur J Biochem. 2000;267:6158–6165. doi: 10.1046/j.1432-1327.2000.01680.x. [DOI] [PubMed] [Google Scholar]
- 21.Uskokovic MR, Norman AW, Manchand PS, Studzinski GP, Campbell MJ, Koeffler HP, Takeuchi A, Siu-Caldera ML, Rao DS, Reddy GS. Highly active analogs of 1alpha,25-dihydroxyvitamin D(3) that resist metabolism through C-24 oxidation and C-3 epimerization pathways. Steroids. 2001;66(35):463–471. doi: 10.1016/s0039-128x(00)00226-9. Review. [DOI] [PubMed] [Google Scholar]
- 22.Schuster I, Egger H, Herzig G, Reddy GS, Schmid JA, Schüssler M, Vorisek G. Selective inhibitors of vitamin D metabolism--new concepts and perspectives. Anticancer Res. 2006;26(4A):2653–2668. Review. [PubMed] [Google Scholar]
- 23.Hourai S, Rodrigues LC, Antony P, Reina-San-Martin B, Ciesielski F, Magnier BC, Schoonjans K, Mouriño A, Rochel N, Moras D. Structure-based design of a superagonist ligand for the vitamin D nuclear receptor. Chem Biol. 2008;15(4):383–392. doi: 10.1016/j.chembiol.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Annalora AJ, Bobrovnikova-Marjon E, Serda R, Lansing L, Chiu ML, Pastuszyn A, Iyer S, Marcus CB, Omdahl JL. Rat cytochrome P450C24 (CYP24A1) and the role of F249 in substrate binding and catalytic activity. Arch Biochem Biophys. 2004;425:133–146. doi: 10.1016/j.abb.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 25.Omdahl JL, Swamy N, Serda R, Annalora AJ, Berne M, Ray R. Affinity labeling of rat cytochrome P450C24 (CYP24) and identification of Ser57 as an active site residue. J Steroid Biochem Mol Biol. 2004;89-90:159–162. doi: 10.1016/j.jsbmb.2004.03.107. [DOI] [PubMed] [Google Scholar]
- 26.Annalora AJ, Bobrovnikov-Marjon E, Serda R, Pastuszyn A, Graham SE, Marcus CB, Omdahl JL. Hybrid homology modeling and mutational analysis of cytochrome P450C24A1 (CYP24A1) of the Vitamin D pathway: insights into substrate specificity and membrane bound structure-function. Arch Biochem Biophys. 2007;460:262–273. doi: 10.1016/j.abb.2006.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst (2007) 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.McRee DE. J Struct Biol. Vol. 125. MIFit, Molecular Images, Active Sight; San Diego, CA: 1999. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density; pp. 156–165. [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Q, Ma X, Ward A, Hong W, Jaakola V, Stevens RC. Designing Facial Amphiphiles for the Stabilization of Integral Membrane Proteins. Ange Chemie Internat Ed. 2007;46(37):7023–7025. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]
- 32.Soltis SM, Cohen AE, Deacon A, Eriksson T, González A, McPhillips S, Chui H, Dunten P, Hollenbeck M, Mathews I, Miller M, Moorhead P, Phizackerley RP, Smith C, Song J, van dem Bedem H, Ellis P, Kuhn P, McPhillips T, Sauter N, Sharp K, Tsyba I, Wolf G. New paradigm for macromolecular crystallography experiments at SSRL: automated crystal screening and remote data collection. Acta Crystallogr D Biol Crystallogr. 2008;64(12):1210–1221. doi: 10.1107/S0907444908030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J Biol Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 35.Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 36.Cojocaru V, Winn PJ, Wade RC. The ins and outs of cytochrome P450s. Biochim Biophys Acta. 2007;1770:390–401. doi: 10.1016/j.bbagen.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 37.Mast N, White MA, Bjorkhem I, Johnson EF, Stout CD, Pikuleva IA. Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain. Proc Natl Acad Sci U S A. 2008;105:9546–9551. doi: 10.1073/pnas.0803717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strushkevich N, Usanov SA, Plotnikov AN, Jones G, Park HW. Structural analysis of CYP2R1 in complex with vitamin D3. J Mol Biol. 2008;380(1):95–106. doi: 10.1016/j.jmb.2008.03.065. [DOI] [PubMed] [Google Scholar]
- 39.Claros MG, von Heijne G. TopPred II: An Improved Software For Membrane Protein Structure Predictions. CABIOS. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 40.Néron B, Ménager H, Maufrais C, Joly N, Tufféry P, Letondal C, Maupetit J. Mobyle: a new full web bioinformatics framework. Institue Pasteur (Logiciels et Banques de Données) and RPSB (Ressource Parisienne en Bioinformatique Structurale) 2008 http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py.
- 41.Engelman DM, Steitz TA, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 42.Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol Cell. 2000;5(1):121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- 43.Kemper B. Structural basis for the role in protein folding of conserved proline-rich regions in cytochromes P450. Toxicol Appl Pharmacol. 2004;199(3):305–315. doi: 10.1016/j.taap.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 44.Ozalp C, Szczesna-Skorupa E, Kemper B. Identification of membrane-contacting loops of the catalytic domain of cytochrome P450 2C2 by tryptophan fluorescence scanning. Biochemistry. 2006;45(14):4629–4637. doi: 10.1021/bi051372t. [DOI] [PubMed] [Google Scholar]
- 45.Pikuleva IA, Mast N, Liao WL, Turko IV. Studies of Membrane Topology of Mitochondrial Cholesterol Hydroxylases CYPs 27A1 and 11A1. Lipids. 2008;43(12):1127–1132. doi: 10.1007/s11745-008-3234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of Proteins in Membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. http://opm.phar.umich.edu/ [DOI] [PubMed]
- 47.White SH. Membrane Protein Insertion: The Biology-Physics Nexus. J Gen Physiol. 2007;129:363–369. doi: 10.1085/jgp.200709741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masuda S, Prosser DE, Guo YD, Kaufmann M, Jones G. Generation of a homology model for the human cytochrome P450, CYP24A1, and the testing of putative substrate binding residues by site. Arch Biochem Biophys. 2007;460(2):177–191. doi: 10.1016/j.abb.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 49.Gomaa MS, Simons C, Brancale AJ. Homology model of 1alpha,25-dihydroxyvitamin D3 24-hydroxylase cytochrome P450C24A1 (CYP24A1): active site architecture and ligand binding. Steroid Biochem Mol Biol. 2007;104:53–60. doi: 10.1016/j.jsbmb.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 50.Hamamoto H, Kusudo T, Urushino N, Masuno H, Yamamoto K, Yamada S, Kamakura M, Ohta M, Inouye K, Sakaki T. Structure-Function Analysis of Vitamin D 24-Hydroxylase (CYP24A1) by Site-Directed Mutagenesis: Amino Acid Residues Responsible for Species-Based Difference of CYP24A1 between Humans and Rats. Mol Pharmacol. 2006;70:120–128. doi: 10.1124/mol.106.023275. [DOI] [PubMed] [Google Scholar]
- 51.Prosser DE, Kaufmann M, O'Leary B, Byford V, Jones G. Single A326G mutation converts human CYP24A1 from 25-OH-D3-24-hydroxylase into -23-hydroxylase, generating 1alpha,25-(OH)2D3-26,23-lactone. Proc Natl Acad Sci U S A. 2007;104(31):12673–12678. doi: 10.1073/pnas.0702093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Urushino N, Yamamoto K, Kagawa N, Ikushiro S, Kamakura M, Yamada S, Kato S, Inouye K, Sakaki T. Interaction between mitochondrial CYP27B1 and adrenodoxin: role of arginine 458 of mouse CYP27B1. Biochemistry. 2006;45(14):4405–4412. doi: 10.1021/bi060072o. [DOI] [PubMed] [Google Scholar]
- 53.Usanov SA, Graham SE, Lepesheva GI, Azeva TN, Strushkevich NV, Gilep AA, Estabrook RW, Peterson JA. Probing the interaction of bovine cytochrome P450scc (CYP11A1) with adrenodoxin: evaluating site-directed mutations by molecular modeling. Biochemistry. 2002;41(26):8310–8320. doi: 10.1021/bi0255928. [DOI] [PubMed] [Google Scholar]
- 54.Beilke D, Weiss R, Löhr F, Pristovsek P, Hannemann F, Bernhardt R, Rüterjans H. A new electron transport mechanism in mitochondrial steroid hydroxylase systems based on structural changes upon the reduction of adrenodoxin. Biochemistry. 2002;41(25):7969–7978. doi: 10.1021/bi0160361. [DOI] [PubMed] [Google Scholar]
- 55.Heinz A, Hannemann F, Müller JJ, Heinemann U, Bernhardt R. The interaction domain of the redox protein adrenodoxin is mandatory for binding of the electron acceptor CYP11A1, but is not required for binding of the electron donor adrenodoxin reductase. Biochem Biophys Res Commun. 2005;338(1):491–498. doi: 10.1016/j.bbrc.2005.08.077. [DOI] [PubMed] [Google Scholar]
- 56.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267(1):83–90. [PubMed] [Google Scholar]
- 57.Pikuleva IA, Cao C, Waterman MR. An additional electrostatic interaction between adrenodoxin and P450c27 (CYP27A1) results in tighter binding than between adrenodoxin and p450scc (CYP11A1) J Biol Chem. 1999;274(4):2045–2052. doi: 10.1074/jbc.274.4.2045. [DOI] [PubMed] [Google Scholar]
- 58.Bracey MH, Cravatt BF, Stevens RC. Structural commonalities among integral membrane enzymes. FEBS Lett. 2004;567(23):159–165. doi: 10.1016/j.febslet.2004.04.084. [DOI] [PubMed] [Google Scholar]
- 59.Fowler PW, Balali-Mood K, Deol S, Coveney PV, Sansom MS. Monotopic enzymes and lipid bilayers: a comparative study. Biochemistry. 2007;46(11):3108–3115. doi: 10.1021/bi602455n. [DOI] [PubMed] [Google Scholar]
- 60.Zhao Y, White MA, Muralidhara BK, Sun L, Halpert JR, Stout CD. Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction. J Biol Chem. 2006;281(9):5973–5981. doi: 10.1074/jbc.M511464200. [DOI] [PubMed] [Google Scholar]
- 61.Li H, Poulos TL. The structure of the cytochrome p450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid. Nat Struct Biol. 1997;4(2):140–146. doi: 10.1038/nsb0297-140. [DOI] [PubMed] [Google Scholar]
- 62.Haines DC, Chen B, Tomchick DR, Bondlela M, Hegde A, Machius M, Peterson JA. Crystal structure of inhibitor-bound P450BM-3 reveals open conformation of substrate access channel. Biochemistry. 2008;47(12):3662–3670. doi: 10.1021/bi7023964. [DOI] [PubMed] [Google Scholar]
- 63.Jaud S, Tobias TJ, Falke JJ, White SH. Self-induced docking site of a deeply embedded peripheral membrane protein. Biophys J. 2007;92:517–524. doi: 10.1529/biophysj.106.090704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kisselev P, Wessel R, Pisch S, Bornscheuer U, Schmid RD, Schwarz D. Branched phosphatidylcholines stimulate activity of cytochrome P450SCC (CYP11A1) in phospholipid vesicles by enhancing cholesterol binding, membrane incorporation, and protein exchange. J Biol Chem. 1998;273(3):1380–1386. doi: 10.1074/jbc.273.3.1380. [DOI] [PubMed] [Google Scholar]
- 65.Murtazina DA, Andersson U, Hahn IS, Bjorkhem I, Ansari GA, Pikuleva IA. Phospholipids modify substrate binding and enzyme activity of human cytochrome P450 27A1. J Lipid Res. 2004;45:2345–2353. doi: 10.1194/jlr.M400300-JLR200. [DOI] [PubMed] [Google Scholar]
- 66.Johnson EF. Deciphering substrate recognition by drug-metabolizing Cytochromes P450. Drug Metab Dispos. 2003;31:1532–1540. doi: 10.1124/dmd.31.12.1532. [DOI] [PubMed] [Google Scholar]
- 67.Kuznetsov VY, Poulos TL, Sevrioukova IF. Putidaredoxin-to-cytochrome P450cam electron transfer: differences between the two reductive steps required for catalysis. Biochemistry. 2006;45(39):11934–11944. doi: 10.1021/bi0611154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heinz A, Hannemann F, Müller JJ, Heinemann U, Bernhardt R. The interaction domain of the redox protein adrenodoxin is mandatory for binding of the electron acceptor CYP11A1, but is not required for binding of the electron donor adrenodoxin reductase. Biochem Biophys Res Commun. 2005;338(1):491–498. doi: 10.1016/j.bbrc.2005.08.077. [DOI] [PubMed] [Google Scholar]
- 69.Wester MR, Stout CD, Johnson EF. Purification and crystallization of N-terminally truncated forms of microsomal cytochrome P450 2C5. Methods Enzymol. 2002;357:73–79. doi: 10.1016/s0076-6879(02)57667-9. [DOI] [PubMed] [Google Scholar]
- 70.Goodsell DS, Olson AJ. Automated Docking of Substrates to Proteins by Simulated Annealing. Proteins: Structure, Function and Genetics. 1990;8:195–202. doi: 10.1002/prot.340080302. [DOI] [PubMed] [Google Scholar]
- 71.Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit. 1996;9(1):1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 72.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using Lamarckian genetic algorithm and an empirical binding free energy function. J Comp Chem. 1998;19:1639–62. [Google Scholar]
- 73.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. ClustalW2 and ClustalX version 2. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 74.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC; San Carlos, CA, USA: http://www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.