Summary
Cellular replicases contain a multiprotein ATPase that loads a sliding clamp processivity factor onto DNA. We reveal a new role for a clamp loader: chaperoning of the replicative polymerase onto a clamp newly bound to DNA. We show that chaperoning confers distinct advantages, including marked acceleration of initiation complex formation. We reveal a requirement for the τ form of DnaX complex to relieve inhibition by single-stranded DNA binding protein during initiation complex formation. We propose that, after loading β2, DnaX complex preserves an SSB-free segment of DNA immediately downstream of the primer terminus and chaperones Pol III into that position, preventing competition by SSB. The C-terminal tail of SSB stimulates reactions catalyzed by τ-containing DnaX complexes through contact distinct for the only known contact involving the χ subunit. The chaperoning of Pol III by the DnaX complex provides a molecular explanation for how initiation complexes form when supported by the non-hydrolyzed analog ATPγS.
Introduction
Cellular replicases are tripartite assemblies composed of a replicative polymerase, a sliding clamp processivity factor, and a clamp loader. The clamp loader, RFC in eukaryotes and DnaX complex in bacteria, is a specialized AAA+ ATPase that opens the ring-shaped processivity factor (PCNA in eukaryotes, β2 in bacteria) and closes it around DNA (Bloom, 2009; Hingorani and O'Donnell, 1998; Schmidt et al., 2001; Johnson et al., 2006). Association of replicative polymerases (ε and δ in eukaryotes, DNA polymerase III (Pol III) in bacteria) with the sliding clamp confers the high level of processivity essential for rapid chromosomal replication (LaDuca et al., 1986; Burgers, 1988).
The clamp loading cycle is driven by ATP binding and hydrolysis by the clamp loader. Binding of ATP to the clamp loader is thought to provide the energy for opening the sliding clamp ring, forming an essential intermediate that can be loaded onto DNA (Hingorani and O'Donnell, 1998; Alley et al., 2000). ATP binding also stabilizes a clamp loader conformation with high affinity for both the clamp and primed DNA, facilitating ternary complex formation (Hingorani and O'Donnell, 1998; Bloom, 2009). ATP hydrolysis decreases the affinity of the clamp loader for DNA, leading to dissociation of the loader and assembly of the clamp on DNA (Bloom, 2009). Pol III, a complex of the polymerase catalytic subunit (α), the 3′-5′ proofreader (ε), and θ, associates with β2 on DNA to form a stable initiation complex that is competent for processive elongation in the presence of dNTPs (McHenry and Crow, 1979; Johanson and McHenry, 1982; LaDuca et al., 1986). Although ATP hydrolysis is coupled to efficient replicase initiation, ATPγS can be substituted to drive initiation complex formation for the E. coli system (Johanson and McHenry, 1984; Glover and McHenry, 2001). Probing with ATPγS has revealed functional asymmetry within the dimeric Pol III holoenzyme (Pol III HE). This asymmetry is thought to correlate with unique leading and lagging strand functions (Glover and McHenry, 2001) and has served as a useful mechanistic tool to drive partial reactions with the DnaX complex (Ason et al., 2000).
Clamp loaders contain a core ring of five homologous proteins. In eukaryotes, these subunits arise from five separate genes (Majka and Burgers, 2004). In E. coli, the five subunits are encoded by three genes, with unique copies of δ and δ′, encoded respectively by holA and holB, and three copies of the dnaX product (τ and/or γ). δ and δ′ are similar to the DnaX ATPase subunits but lack sites competent for ATP binding and hydrolysis (Jeruzalmi et al., 2001; Bullard et al., 2002). τ is the full length dnaX translation product, and γ arises by translational frameshifting and contains about two-thirds of the sequence found in τ. γ and τ share three domains that bind ATP, β2, and primed DNA and are involved in β2 loading (Williams et al., 2003). γ-complex (DnaX complex lacking τ) has been used as a model system to study clamp loading and was once thought to be the physiologically-relevant clamp loader. However, γ-complex has severe deficiencies if asked to support full replicative function. Cells expressing only the γ form of DnaX are not viable (Blinkova et al., 1993). τ contains two domains absent in γ. Domain IV binds DnaB, the replicative helicase that reversibly binds primase during lagging strand synthesis (Tougu and Marians, 1996), and domain V binds to the α subunit of Pol III (Gao and McHenry, 2001b; Gao and McHenry, 2001a). Since at least two copies of τ are found in cellular DnaX complex, the τ subunits dimerize Pol III within the replicase (Kim and McHenry, 1996; McHenry, 1982; Studwell-Vaughan and O'Donnell, 1991). Thus, the τ subunits function as the central replisome organizers, linking the leading- and lagging-strand polymerases with the helicase and priming activities necessary for replication of double-stranded DNA (Kim et al., 1996a; Kim et al., 1996b). The E. coli clamp loader complex contains one copy each of two peripheral subunits, χ and ψ, with ψ binding three DnaX subunits asymmetrically in a unique orientation relative to one DnaX subunit in the core pentameric ring and with χ binding to ψ (Glover and McHenry, 2000; Simonetta et al., 2009).
In addition to its role in organizing the replication fork, τ protects elongating complexes from premature removal of β2 by exogenous DnaX complex, helping to ensure high processivity for the leading strand polymerase (Kim et al., 1996c). τ stabilizes δ and δ′ in a complex with the elongating replicase; dissociation of δ and δ′ leads to elongation defects (Song et al., 2001). τ also serves as a link between Pol III and the χ subunit, which binds to single-strand binding protein (SSB) and aids in polymerase progression on SSB-coated templates (Glover and McHenry, 1998). Since τ-containing DnaX complexes tightly bind Pol III, we explored another potential role of τ: preferentially introducing its bound polymerase into initiation complexes, conferring efficiencies to the overall initiation process.
Results
The DnaX complex of E. coli has a well-established role in loading the β2 sliding clamp processivity factor onto DNA (Davey et al., 2002). The tight association between Pol III and τ-containing forms of the DnaX complex (Kim and McHenry, 1996) suggested that the DnaX complex might preferentially attach the associated Pol III to a newly-loaded β2 clamp. To test this possibility, we exploited a version of the α subunit of Pol III containing an inhibitory mutation in the polymerase active site (α-D403E). This α subunit retains the domains that interact with τ and β, but cannot extend a primer if assembled into an initiation complex (Pritchard and McHenry, 1999). We tested whether α-D403E could successfully compete with wild-type Pol III preassembled into a complex with the DnaX complex.
Primer Extension Assay for Active Pol III Initiation Complexes
To distinguish active from inactive initiation complexes, we probed the complexes functionally by their ability to extend a primer. We annealed a synthetic 30 nt, 5′-32P end-labeled primer to a single-stranded circular template in front of a 22 nucleotide T-less stretch of the template. An RNA primer was used to prevent digestion by the proofreading exonuclease activity of the Pol III ε subunit (Figure S1). To separate the effects of τ, SSB, and other subunits on initiation complex formation from their effects on extensive DNA elongation, we limited the primer extension to a short distance by omitting dATP from the extension mixture. This yielded the expected 52 nt product but also longer products, presumably due to misincorporation of other nucleotides in place of the missing dATP, complicating quantification (Figure S1). Inclusion of ddATP in the extension step solved the problem and yielded a single 53 nt product (Figure S2). We formed initiation complexes in the absence of dNTPs, blocked further initiation complex formation by adding excess unlabeled primer-template, extended initiation complexes containing the labeled primer-template by adding three dNTPs and ddATP, and separated and analyzed the reaction products by polyacrylamide gel electrophoresis (Figure S3). Both β2 and ATP were required to observe primer extension, demonstrating that the assay detected only fully assembled initiation complexes (Figure S4).
τ-Containing DnaX Complex Chaperones Pol III into Initiation Complexes
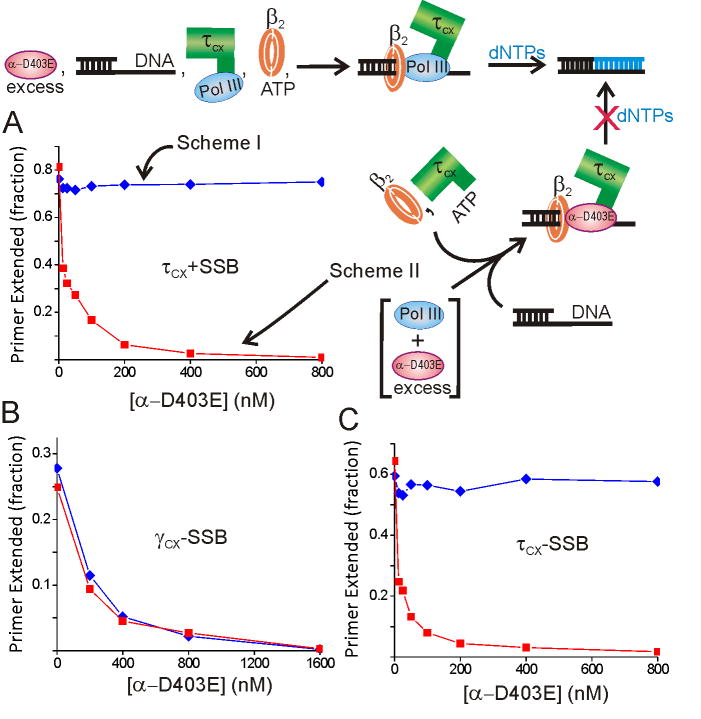
To investigate how the interaction between Pol III and the τ complex affects the initiation mechanism, we performed an experiment monitoring initiation complex formation in the presence of varying levels of the inhibitory polymerase subunit α-D403E. If we added a mixture of wild-type Pol III and α-D403E to the τ-complex (Figure1, Scheme II), the level of initiation complexes formed that were competent for extension was reduced proportionally to the ratio of α-D403E to wild-type Pol III (Figure 1A). An approximately two-fold molar excess of α-D403E over the τ subunit was required for 50% inhibition. A strikingly different result was obtained if an assembly of τ-complex and wild-type Pol III was formed before addition of α-D403E (Figure 1, Scheme I): active initiation complex formation was completely resistant to the α-D403E challenge up the highest level tested (400-fold excess). A similar result was obtained if the Pol III subunits ε and θ were added with α-D403E to reconstitute an inhibitory Pol III complex, confirming that the mutant α competes similarly to full Pol III for τ binding (Figure S5). These results show that the Pol III bound to the DnaX complex is selectively attached to the β2 clamp loaded by the same DnaX complex, preventing competition from a large exogenous pool of inhibitory polymerase.
Figure 1. Pol III bound to τ-complex is resistant to competition from inhibitory α-D403E.
A) Initiation complex formation with the τ-complex pre-incubated with wild-type Pol III (blue, scheme I) or exposed to α-D403E and wild-type Pol III simultaneously (red, scheme II). These reactions contained 0.25 μM SSB4. B) Initiation complex formation with the γ-complex pre-incubated with wild-type Pol III (blue, scheme I) or exposed to α-D403E and wild-type Pol III simultaneously (red, scheme II). C) Same procedure as (A) conducted in the absence of SSB.
τ forms a very tight complex with the α subunit of Pol III (KD ∼70 pM) in an interaction that is slow to dissociate (t½∼3 h) (Kim and McHenry, 1996). Consistent with this slow dissociation, we found that preformed complexes of Pol III and τ-complex retained full activity up to an hour after addition of α-D403E (data not shown). If the resistance to competition from exogenous α-D403E is due to this tight association, we would not expect the γ-complex to afford the same protection, since γ lacks the domain required bind Pol III (Kim and McHenry, 1996). Initially, we repeated the experiment described above with γ-complex substituted for τ-complex, but we discovered that the reaction was inefficient in the presence of SSB, even in the absence of α-D403E (pursued further in next section). Thus, we performed the γ-complex reaction in the absence of SSB. In contrast to the τ-complex, we observed that preincubation of γ-complex with wild-type Pol III afforded no protection from the α-D403E challenge (Fig 1B). Performing the τ-complex experiment in the absence of SSB gave the same qualitative result as in its presence (Fig 1C). These results show that the τ subunit is required for the DnaX complex to selectively attach Pol III to the newly loaded β2. Pol III must reach the loaded β2 by a different mechanism with the γ-complex.
As an important technical note, we observed that γ-complexes overexpressed and purified from E. coli by standard methods in some cases contained trace levels of τ, which contributed a troublesome background to certain experiments (Figure S6). The source was apparently the low level of endogenous τ expression from chromosomal dnaX. To ensure our γ-complex sample was completely free of τ, we further purified the γ-complex by treating it with beads carrying immobilized α. These beads bound τ tightly, yielding γ-complexes free of τ contaminants (Figure S6). This α-affinity purification procedure could be valuable for future studies with γ-complex, since the results may be complicated by a background arising from the unique properties of the τ subunit, especially at high DnaX complex concentrations.
Initiation Complex Formation is Enhanced by SSB with τ-Complex and Inhibited by SSB with the γ-Complex
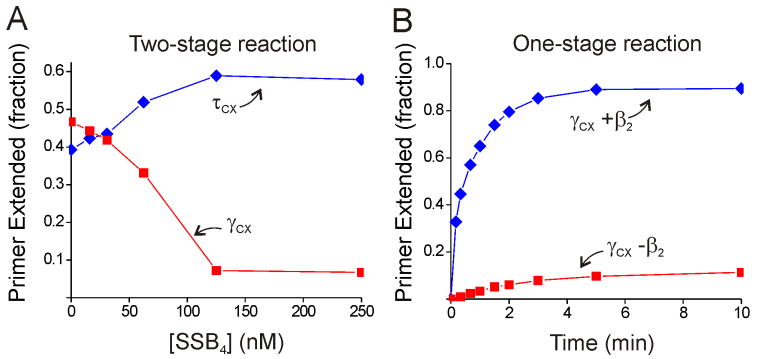
The result that γ-complex-catalyzed initiation complex formation is inhibited by SSB was surprising, since SSB is routinely included in reactions where Pol III HE-like activity is reconstituted using γ-complex. Indeed, the γ-complex used in this study was fully active with SSB in standard Pol III HE reconstitution assays (see Supplemental Data). Thus, we further investigated how SSB affects initiation complex formation. A comparison of reactions with τ- or γ-complex, assayed by our procedure of conducting initiation complex formation and primer extension as separate steps, showed drastically different behavior with increasing concentrations of SSB (Figure 2A). Initiation complex formation supported by τ-complex was stimulated as SSB concentration increased, whereas the reaction supported by γ-complex was markedly inhibited. At 0.13 μM SSB4 (SSB binds DNA as a tetramer), the template DNA will be ∼95% coated with SSB4 (assuming a binding site of 65 nt for SSB4 (Lohman and Ferrari, 1994)), and under these conditions we observed nearly complete inhibition of the γ-complex-catalyzed reaction and maximal stimulation of the τ-complex-catalyzed reaction.
Figure 2. SSB has different effects on the τ- and γ-complex initiation complex formation reactions.
A) Influence of SSB concentration on initiation complex formation for the τ- (blue) and γ-(red) complexes (labeled γcx and τcx in all figures). Initiation complex formation and primer extension were conducted as separate reaction steps. B) Time course for the γ-complex reaction with (blue) and without (red) β2 conducted with initiation complex formation and primer extension in a single reaction (i.e., with dNTPs present during initiation complex formation). The experiments in (B) were conducted with 0.25 μM SSB4.
A major difference between our primer extension assay and many assays reconstituting Pol III HE activity with γ-complex is that our assay separates initiation and primer extension into two separate steps. We re-examined the γ-complex in our primer extension assay conducted in one reaction step, with initiation complexes formed in the presence of dNTPs to immediately extend the complexes. In this one-step reaction, which more closely resembles other Pol III HE reconstitution assays, the γ-complex is active in the presence of 0.25 μM SSB4 (Figure 2B). This γ-complex activity required β2, confirming that the one-step assay still detects full initiation complexes.
Our interpretation of these γ-complex results is that, in the absence of dNTPs, SSB significantly reduces the steady-state population of initiation complexes that is detected in a separate primer elongation reaction. This reduced population could arise from SSB slowing the initiation rate, increasing the rate of the initiation complex dissociation, or both. The remaining experiments with the γ-complex described herein were conducted in the absence of SSB.
ATPγS Supports Initiation Complex Formation Chaperoned by the τ Complex
To further test the conclusion that τ chaperones Pol III into initiation complexes, we reinvestigated initiation complex formation in the presence of the non-hydrolyzed ATP analogue ATPγS. It has been shown previously that ATPγS supports initiation complex formation with the τ-complex at 50% of ATP-supported levels and does not support initiation with the γ-complex (Glover and McHenry, 2001; Johanson and McHenry, 1984). One explanation for this effect is that ATPγS mimics the positive allosteric effect of ATP in increasing the affinity of the DnaX complex for β2 and primed DNA (Bertram et al., 1998). In the absence of the ATP hydrolysis proposed to drive closing of the β2 ring, the quaternary ATPγS–τ-complex–β2–DNA complexes may exist in an internal equilibrium where the majority of β2 is in an open conformation but a small population is closed (reversibly) around the DNA. Our first prediction from this model is that if τ chaperones Pol III onto DNA, the Pol III could trap these spontaneously loaded β2-DNA complexes and form initiation complexes. Thus, we tested whether the ATPγS reaction can withstand an α-D403E challenge. Second, if the reaction depends on Pol III trapping transiently closed β2, the unchaperoned γ-complex-catalyzed reaction should be even less efficient for the ATPγS-aided reaction than for the ATP reaction.
Using our two-step primer extension assay, we observed ATPγS effects similar to those described above for previous studies, with the τ-complex forming active initiation complexes at approximately half the ATP-driven levels and with initiation complex formation greatly reduced for the γ-complex (Figure 3A). The ATPγS-supported γ-complex reaction is greatly diminished compared to the ATP-supported reaction, with primer extension reduced from nearly 50% with ATP to only ∼7% with ATPγS in the absence of SSB and with no product detected at all in the presence of saturating SSB (Figure 3A). These findings are consistent with the above predictions.
Figure 3. ATPγS supports SSB-dependent chaperoning of Pol III by the τ-complex.
A) Dependence of initiation on SSB4 concentration for the τ- (blue) and γ- (red) complexes in the presence of ATPγS. B) ATPγS-supported initiation complex formation with the τ-complex pre-incubated with wild-type Pol III (blue; scheme I in Figure 1, with ATPγS substituted for ATP) or exposed to α-D403E and wild-type Pol III simultaneously (red; scheme II in Figure 1). The experiments in (B) were conducted with 0.25 μM SSB4.
Challenging the ATPγS-supported τ-complex initiation complex formation with α-D403E showed the same effects as the ATP-supported reaction. Formation of active initiation complexes is completely inhibited by α-D403E if τ complex is simultaneously exposed to α-D403E and wild-type Pol III, whereas the system is unaffected by large excesses of α-D403E if τ complex is pre-bound to wild-type Pol III (Figure 3B). As with ATP, only the polymerases bound to the τ-complex form initiation complexes in the ATPγS reaction. These findings are consistent with the first prediction above. We conclude that the reason τ-complex supports relatively efficient initiation complex formation with ATPγS is that by chaperoning Pol III to β2 and DNA, the τ-complex enables Pol III to trap any β2 transiently loaded onto the DNA independently from ATP hydrolysis. This model provides a molecular explanation of why τ is required for ATPγS-supported initiation complex formation.
SSB Enhancement of the τ-Complex Does Not Require χ-ψ
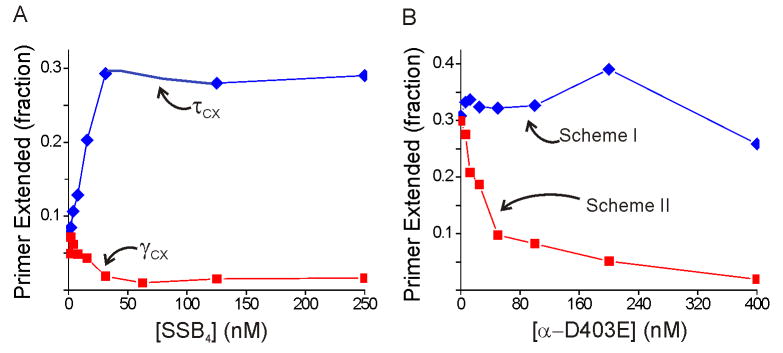
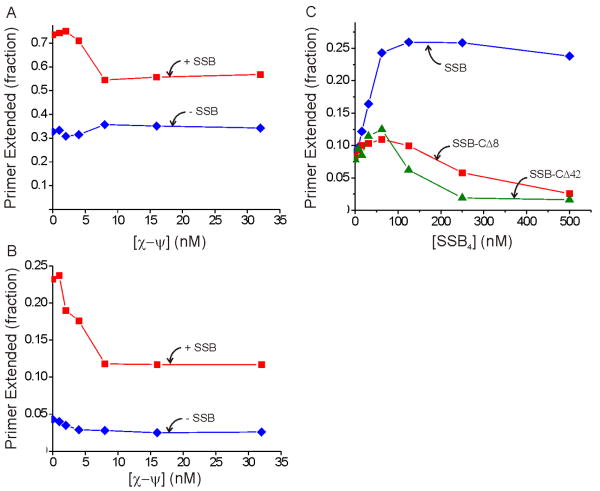
Notably, the ATPγS-supported process with the τ-complex depends strongly on SSB, with a 3-fold greater yield at saturating SSB levels compared to the yield without SSB (Figure 3A). The effect of SSB is more pronounced for this reaction than for ATP-driven initiation complex formation shown in Figure 2A. Since it has been shown previously that the χ subunit of the DnaX complex binds to SSB and increases the affinity of the DnaX complex for DNA (Kelman et al., 1998; Glover and McHenry, 1998), we hypothesized that the stimulation of the τ-complex by SSB was mediated through the χ and ψ subunits (ψ couples χ to the DnaX complex). To test this hypothesis, we reconstituted τ-complex from fixed concentrations of τ, δ, and δ′ and varying concentrations of χ-ψ and probed these complexes for initiation complex formation in our primer extension assay. The results showed that SSB enhanced both ATP- and ATPγS-driven initiation complex formation even in the absence of χ-ψ (Figures 4A and B, respectively). Thus, the SSB stimulation is independent of the interaction between SSB and χ.
Figure 4. SSB enhancement for the τ-complex does not require χ-ψ.
A) ATP-driven initiation complex formation with the τ-complex components reconstituted with varying concentrations of χ-ψ without SSB (blue) and with 0.25 μM SSB4 (red). B) ATPγS-driven initiation complex formation with the τ-complex components reconstituted with varying concentrations of χ-ψ without SSB (blue) and with 0.25 μM SSB4 (red). The concentrations in (A) and (B) refer to the χ–ψ heterodimer, the form in which these subunits are purified. C) ATPγS-supported initiation complex formation for purified full τ-complex with varying concentrations of SSB (blue), SSB-CΔ8 (red), and SSB-CΔ42 (green).
The χ–ψ-independent stimulation by SSB might reflect a heretofore uncharacterized interaction between SSB and other subunits of the Pol III HE or may arise from a less direct effect such as SSB rearranging the DNA template into a configuration more accessible to the Pol III HE components. The C-terminus of SSB is responsible for the interaction between SSB and various proteins, including χ (Kelman et al., 1998; Witte et al., 2003). Mutations of the C-terminal region do not disrupt DNA binding but obviate SSB-protein interactions (Shereda et al., 2008). To test whether this region was important for the SSB enhancement of the τ-complex, we measured ATPγS-driven initiation complex formation with two previously characterized SSB proteins with 8 (SSB-CΔ8) or 42 (SSB-CΔ42) residues deleted from their C-termini (Roy et al., 2007; Hobbs et al., 2007). Neither SSB-CΔ8 nor SSB-CΔ42 stimulated initiation complex formation as effectively as full-length SSB, and both of these proteins inhibited initiation complex formation at higher concentrations (Figure 4C). Thus, the C-terminal region of SSB is required to stimulate initiation complex formation catalyzed by the τ-complex, suggesting that the effect likely arises from an unknown interaction between SSB and a Pol III HE component other than χ.
τ Reduces the Pol III Concentration Required for Initiation Complex Formation
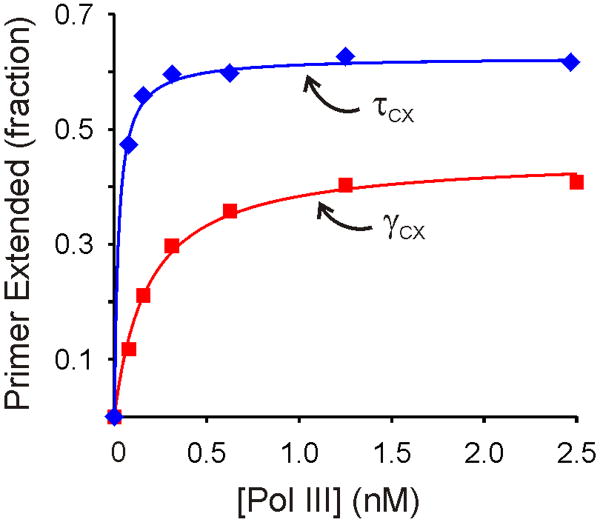
If τ-complex loads a β2 clamp and the chaperoned Pol III binds to that β2 in a tightly coupled reaction, then the concentration of Pol III required for initiation complex formation might be lower than for an unchaperoned reaction where free Pol III must reach the loaded β2 without assistance. We tested this hypothesis by comparing the concentrations of Pol III required for efficient initiation with the τ- and γ-complexes at 1.0 nM DnaX complex and 0.10 nM primer/template concentrations, with excess β2 and ATP. We observed stoichiometric association of Pol III and primer-template into initiation complexes with the τ-complex (Figure 5). Fitting the data to a binding isotherm yielded an apparent dissociation constant, K1/2, of ∼20 pM for Pol III associating with the initiation complex, but the true K1/2 value could be lower since the binding appears stoichiometric. By contrast, Pol III associated into initiation complexes with a K1/2 of 180 pM for γ-complex catalyzed initiation (Figure 5). These data show that the ability of τ-complex to chaperone Pol III to the initiation complex decreases the required concentration of Pol III at least 10-fold.
Figure 5. Chaperoning by the τ-complex lowers the concentration requirement for Pol III.
The Pol III concentration dependences for initiation with the τ-complex (blue) and γ-complex (red). The solid lines represent fits to a standard binding isotherm, yielding K1/2 values of ∼20 and 180 pM for the τ- and γ-complexes, respectively. Experimental details are provided in Supplemental Data.
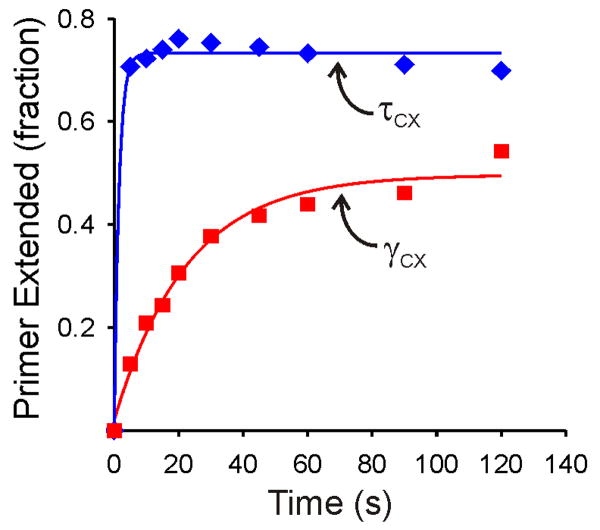
τ Enhances the Initiation Rate
If τ-complexes chaperone Pol III into initiation complexes, one would expect a kinetic advantage over an unchaperoned reaction. Thus, we measured the rates of initiation complex formation with the τ- and γ- complexes. The kinetics experiments were initiated with the primer/template as the last component added to ensure that association of any of the holoenzyme proteins was not rate-limiting, and the reactions were conducted under single-turnover conditions with limiting primer/template. The results for the γ-complex showed initiation complex formation proceeding on a 100 s timescale (Figure 6). The data were well fit by a single exponential function, yielding kobs = 0.045 s-1. By contrast, initiation complex formation with the τ-complex was so rapid that it had proceeded to completion before the first manually sampled time point of 5 s, indicating a rate constant >0.5 s-1 (Figure 6). These results show that the ability of the τ-complex to chaperone Pol III to the initiation complex enhances the rate of initiation complex formation by at least an order of magnitude, which could have important functional consequences in vivo.
Figure 6. Chaperoning by the τ-complex accelerates initiation complex formation.
The data for the τ- (blue) and γ- (red) complexes were each fit to a single exponential (solid lines). The τ-complex reaction reached completion before the first manually sampled time point (5 s) and could not be fit accurately. The kobs for this reaction was estimated as >0.5 s-1. The fit for the γ-complex yielded kobs = 0.045 s-1. Both reactions were performed under single turnover conditions, with 10-fold excess DnaX complex over DNA substrate and 5-fold excess Pol III over DnaX complex. Experimental details are provided in Supplemental Data.
Discussion
The AAA+ ATPases that load bracelet-like sliding clamp processivity factors around DNA are well characterized (Davey et al., 2002). The change in processivity conferred by the association of a replicative DNA polymerase with a sliding clamp has also been well established (LaDuca et al., 1986; Fay et al., 1981; Burgers, 1988). In this work, we reveal an important intermediate step linking these two processes. The τ-containing form of the DnaX complex chaperones the replicative polymerase onto a newly DNA-bound sliding clamp in an efficient coupled process. This conclusion is most dramatically supported by the ability of τ-complex bound to wild-type Pol III to withstand inhibition by large excesses of α-D403E, which can form initiation complexes but cannot elongate. The chaperoning effect is also observed for initiation complex formation supported by the non-hydrolyzed ATP analog, ATPγS, with which initiation complex formation is less efficient and presumably slower and thus should be more susceptible to competition by exogenous αD403E.
For Pol III bound to DnaX complex to preferentially reach the initiation complex over an exogenous challenge, the rate of association of this bound Pol III would have to be much greater than for free polymerase. Direct measurements of the rate of initiation complex formation catalyzed by τ-complex and γ-complex support this concept. The rate of the γ-complex-catalyzed reaction is slow (kobs = 0.045 s-1), requiring one minute for completion at concentrations of Pol III that likely approximate free cellular levels. Under equivalent conditions, the τ-complex mediated reaction was complete before the first time point could be taken (ca. 5 seconds). Thus, the τ-complex-catalyzed reaction is at least 10-fold faster. During lagging strand synthesis, a 1-2 kb Okazaki fragment is synthesized every 1-2 seconds. Since most of this time would be required for the elongation reaction, the release from a completed fragment and formation of an initiation complex for the next fragment would have to occur in a fraction of this cycle time. Thus, it would appear that γ-complex could not function with acceptable kinetics, making the function of τ at the replication fork essential for this reason alone, in addition to its other important contributions.
Association with τ would greatly increase the effective local concentration of Pol III during initiation complex formation, allowing Pol III-dependent reactions to occur at lower overall concentrations of Pol III. Our data support this prediction. Initiation complex formation catalyzed by τ-complex proceeded with stoichiometric association between Pol III and primer-template, indicating a maximum K1/2 of 20 pM. This value could be much lower, since stoichiometric binding precludes accurate determination of the apparent dissociation constant. Under identical conditions, the K1/2 was at least 10-fold higher with the γ-complex.
It is interesting to note that a role for a clamp loader as a polymerase chaperone has been considered before. For the bacteriophage T4 replication system, a chaperoning role was proposed to explain the observations that all of the polymerase enters a productive complex if the T4 clamp loader (p44/p62) is in excess whereas simulations suggested that 50% of the polymerase is trapped in a nonproductive complex with the clamp if p44/p62 was limiting (Kaboord and Benkovic, 1996). Thus, it was suggested that p44/p62, if present at stoichiometric levels, steers the polymerase away from a nonproductive state. Most of our experiments were performed with DnaX complex in excess over Pol III, so we would not have observed this benefit of chaperoning, if extant, in the E. coli system.
An important finding arising from our studies was that SSB, if probed independently from its known positive effects on priming and extensive elongation, has a significant role in initiation complex formation. When initiation complex formation and primer extension are conducted as separate reactions, SSB stimulates initiation complex formation catalyzed by τ-complex but markedly inhibits the same reaction with the γ-complex. However, if initiation complex formation and primer extension are conducted in a single reaction (i.e., with dNTPs present during initiation complex formation), an efficient conversion of primers to product is observed for the γ-complex in the presence of SSB. Our interpretation of these results is that both τ- and γ-complexes can form initiation complexes in the presence of SSB, but that the γ-complex does so much less efficiently. Without dNTPs present, the γ-complex-catalyzed reaction reaches a steady state in which only a small proportion of the primer/template is associated with initiation complexes. In the one-step reaction with dNTPs present throughout, the initiation complexes are elongated before they can dissociate, so most of the primer/template is eventually converted to elongated product.
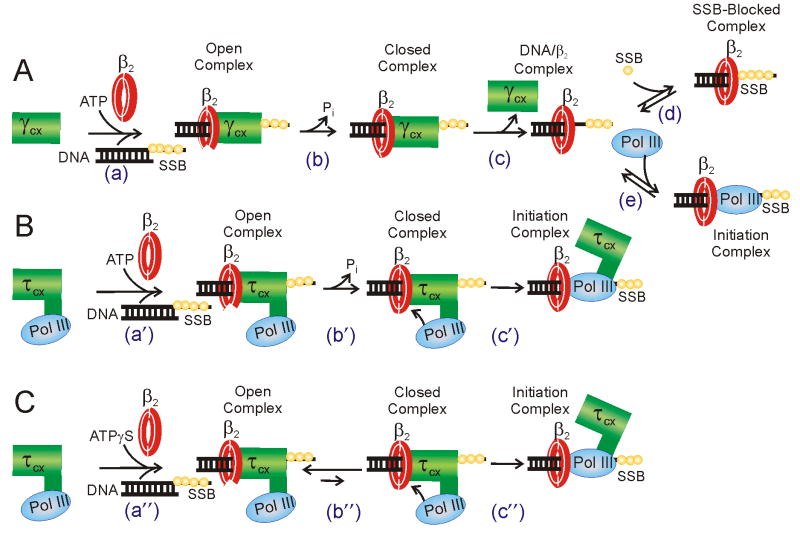
The reduced efficiency of initiation for the γ-complex could arise from SSB slowing initiation complex formation, from SSB increasing the dissociation of the complexes once formed, or from both effects. A reduced rate of initiation is consistent with our previous result showing that SSB inhibits DNA synthesis by Pol III in the absence of τ, ψ, and χ, suggesting that SSB prevents free Pol III from binding to the single-stranded template just beyond the primer terminus (Glover and McHenry, 1998). If the γ-complex dissociates from the DNA prior to Pol III binding to β2 (Figure 7A, step c), then SSB could fill the vacated template and compete with Pol III (Figure 7A, step d). The τ-complex could resist this inhibition since it is likely still bound to the template when Pol III binds to the newly loaded β2 (Figure 7B, step c′). The presence of the τ-complex during this step could serve to keep the primer terminus cleared of SSB and enable access by the Pol III.
Figure 7. Models for unchaperoned and chaperoned initiation complex formation.
A) Model for unchaperoned initiation complex formation catalyzed by the γ-complex. Most features of this reaction have been established by previous work and are discussed and cited in the text. To provide a plausible explanation for why initiation is inhibited by SSB, step d includes a process where SSB binds the template DNA in place of the dissociated γ-complex. In this mechanism, free Pol III diffuses to a β2/DNA complex after γ-complex dissociates. B) Model for chaperoned initiation complex formation catalyzed by the τ-complex. Steps a′ and b′ of this reaction are the same as the analogous steps for γ-complex in (A) except that Pol III is associated with the DnaX complex. In step c′ Pol III binds to the newly loaded β2, permitting concerted Pol III loading. In contrast to the γ-complex mechanism, the contact of the τ-complex around the primer terminus is preserved in the Pol III/β2 binding step, providing an explanation for why this reaction is not inhibited by SSB. C) Model for chaperoned initiation complex formation in the absence of ATP hydrolysis. The mechanism is the same as (B) except that in a″ ATPγS substitutes for the allosteric effects of ATP and in b″ the closing of β2 is energetically unfavorable in the absence of ATP hydrolysis. The coupled equilibria of steps b″ and c″ drive the reaction to form an initiation complex, competent for extension upon the addition of dNTPs.
The initiation complexes formed between Pol III and β2, assembled on DNA, by γ-complex are also likely to dissociate faster due to the absence of an associated DnaX complex. Previously, we have demonstrated that elongating complexes are unstable if the δ and δ′ subunits, normally bound to Pol III by a τ tether, are removed from an associated DnaX complex (Song et al., 2001). The presence of τ in the complex has also been shown to prevent premature removal of β2 by exogenous DnaX complex (Kim et al., 1996c). Thus, initiation complexes formed by γ-complex are likely less stable, since they lack the protective association of the DnaX complex enabled by τ.
τ-complex-catalyzed initiation complex formation not only resists inhibition by SSB, it is enhanced by SSB. Since we have shown previously that the χ subunit of the DnaX complex binds to SSB and increases the affinity of the DnaX complex for DNA (Glover and McHenry, 1998), we were surprised to find here that the SSB enhancement of the τ-complex does not require χ-ψ. Further exploration of this phenomenon with SSB-CΔ8 and SSB-CΔ42 revealed that the C-terminal region of SSB, which is responsible for interactions between SSB and other proteins, is necessary for the SSB enhancement of τ-complex. This result raises the intriguing possibility that SSB forms interactions with a Pol III HE component(s) other than χ that are important for initiation complex formation. Since the rest of the Pol III HE components are required to observe any DNA synthesis readout in our assay, the relevant subunit(s) interacting with SSB could not readily be deconvoluted. A strong SSB effect is also observed with τ-complex for initiation complex formation in a pathway driven by ATPγS binding. This pathway is likely slower than that driven by ATP binding and hydrolysis and is therefore likely even more sensitive to the positive contributions conferred by SSB. These results for SSB underscore the significant differences between the initiation mechanisms with the τ and γ complexes and further suggest that physically coupling Pol III to the DNA substrate via the τ-complex enhances initiation complex formation.
We propose a model to explain the mechanistic differences between initiation complex formation driven by the γ- and τ-complexes (Figure 7). ATP binding to the DnaX complex has the allosteric affect of increasing the affinity of the DnaX complex for primed DNA and β2 (Davey et al., 2002). The energy of ATP binding is thought to be coupled to opening the β2 ring (Figure 7A, step a). ATP hydrolysis is thought to be coupled to closing of the clamp around the DNA (step b) and the loss of affinity of the γ-complex for β2-loaded DNA (step c). Pol III then associates with the loaded clamp in a separate reaction (step e). If SSB is present, it may bind to the template after γ-complex dissociation and occlude Pol III from binding the DNA (step d), inhibiting synthesis (Glover and McHenry, 1998). Step e may be reversible in the absence of dNTPs and active elongation, leading to instability of the initiation complex.
For the τ-complex-catalyzed reaction (Figure 7B), the first two steps (designated a′ and b′) are analogous to the γ-complex steps a and b except Pol III is bound to the DnaX complex by the extra C-terminal domains present in τ. Having Pol III in the same complex with the DnaX clamp loader significantly changes the downstream steps. Direct attack of Pol III upon the loaded clamp could be responsible for part of the chaperoning reaction, allowing a direct swap of the DnaX complex for Pol III on the primer terminus (step c′). Since the γ-complex would be dissociated from the DNA in the analogous reaction step (Figure 7A, step e), the β2-loaded template could be subject to additional SSB binding, leading to SSB inhibition (Figure 7A, step d). Thus, the chaperoning activity of τ-containing DnaX complexes may arise not only from the high local concentration of bound Pol III near the newly loaded β2, but also from the presence of DnaX complex during Pol III/β2 binding to preserve a clearance of SSB from the DNA immediately downstream of the primer terminus. The recent structure of DnaX complex bound to a primer-template shows a contact between δ and the template immediately distal to the 3′-primer terminus that may be responsible for the clearance of SSB (Simonetta et al., 2009).
We also present a model for ATPγS-assisted initiation complex formation without ATP hydrolysis formulated in light of our findings (Figure 7C). Step a″ is equivalent to steps a and a′ in Figure 7A and B, with ATPγS binding substituting for ATP binding in stabilizing a DnaX–β2–DNA ternary complex. Step b″ is similar to steps b and b′ except that it does not involve ATP hydrolysis and is presumably energetically uphill. However, the closed state of this unfavorable internal equilibrium could still have a mechanistically relevant population. If Pol III is chaperoned to the closed β2 clamp, it could bind and trap the closed state and form an initiation complex (Figure 7C, step c″). This step would be analogous to step c′ in Figure 7B. The coupling of the energy of Pol III binding to the closed β2 would make the overall initiation complex formation reaction more energetically favorable. An alternative model would be that Pol III attacks the open β2 complex and drives the complex closed.
In the ATPγS reaction, SSB interactions appear to be even more important than for the natural, ATP-driven reaction. This could be explained by the reaction intermediates forming more slowly and/or being less stable in the absence of ATP hydrolysis. If these steps are less efficient, then factors that enhance lifetimes of these intermediates will become more important. Both the open and closed complexes could be stabilized by a Pol III HE-SSB interaction, making the intermediates longer lived and increasing the time available for closed complex formation and/or Pol III attack. Detailed kinetic and structural studies will be required to establish the reaction pathway followed for this complex system.
A form of the DnaX complex containing only γ (γ complex, γ3δδ′χψ) has often used as a model for DnaX complex action with the assumption that τ-containing DnaX complex and γ-complex are interchangeable. However, important functions of τ, not shared with γ, have been previously revealed, including (i) formation of a dimeric replicative complex containing the leading and lagging strand polymerase (McHenry, 1982; Kim et al., 1996a), (ii) association with the replicative helicase accelerating its rate of progression and serving as the central replisome organizer (Kim et al., 1996a), (iii) protecting β2 associated with the elongating replicase from removal by exogenous protein factors (Kim et al., 1996c),(iv) holding χψ in the elongating complex allowing stabilization by enabling association with SSB (Kelman et al., 1998; Glover and McHenry, 1998), and (v) a proposed role for τ as a sensor of the conversion of a gap to a nick, facilitating cycling upon completion of Okazaki fragment formation (Leu et al., 2003). The results reported in this paper add (vi) chaperoning of the associated polymerase to the newly loaded β2, (vii) accelerating the rate of initiation complex formation in the presence of physiological protein concentrations to a rate that is required to support the in vivo rate of DNA replication, and (viii) overcoming inhibition by SSB during initiation complex formation, to the list of critical functions contributed uniquely by τ. Together, these functions suggest that γ-complex, even if it exists in the cell, does not participate directly in DNA replication.
Both a γ and τ form of DnaX are produced by translational frameshifting in E. coli and the presence of both forms is of sufficient importance that a different mechanism (site-specific transcriptional slippage) evolved in another organism to accomplish the same goal [(Larsen et al., 2000) and references therein]. Pol III holoenzyme purified from wild-type cells contains both γ and τ (McHenry, 1982). A question arose about a potential artifactual source of γ arising from τ, because of the observation that τ can be cleaved to a protein nearly the same size as γ by the E. coli OmpT protease (Pritchard et al., 1996). However, more recent preparations of Pol III holoenzyme isolated from ompT cells have eliminated this as a possibility (Chen and McHenry, unpublished observation). It has been observed that intact τ3δδ′χψ, when expressed from an artificial operon containing a non-frameshifting mutant of dnaX, can be isolated from ompT cells, eliminating the possibility that a γ-like protein is generated by an alternative protease (Pritchard et al., 1996). Given that at least two τ protomers are required for a physiologically relevant association of Pol III holoenzyme and the replicative helicase (Gao and McHenry, 2001a), the most common DnaX complex stoichiometry within Pol III holoenzyme is likely τ2γδδ′χψ. This could serve to limit most replisomes to two polymerases while preserving a complex that benefits from the essential contributions of τ. A mutant that only encodes the τ form of DnaX is viable (Blinkova et al., 1993), but competition experiments with the wild-type counterpart have not been conducted to determine the relative fitness of the τ-only mutant. Thus, it is certain that the E. coli replicase must contain τ, but determining the stoichiometry of γ and its function will require further investigation.
The experimental demonstration of chaperoning of the E. coli polymerase to the loaded clamp was facilitated by the tight association of τ and Pol III. Many other clamp loaders, such as RFC in eukaryotes or even the τ-complex ortholog in Gram-positive bacteria (Bruck and O'Donnell, 2000), do not bind their cognate polymerases tightly. However, the advantages conferred by chaperoning at a replication fork could prove essential in all organisms. A weaker interaction between a clamp loader and its cognate polymerase does not preclude a chaperoning function, it only makes this function more difficult to detect. A transient interaction between a clamp loader and a polymerase during initiation complex formation could be detected by establishing the detailed mechanism using kinetics experiments or other techniques that permit detection of transient protein-protein interactions. The polymerase chaperoning performed by the E. coli clamp loader could serve as a prototype for a broader test for the conservation of this mechanism in the replication of chromosomes of other life forms.
Experimental Procedures
Primer Extension Assay for Initiation Complex Formation
Initiation complex formation was instigated by combining Pol III, DnaX complex, β2, and ATP with 32P-labeled primer/template (SSB coated where applicable). All reactions were conducted under single turnover conditions. After the reaction times described, initiation complex formation was stopped by addition of activated calf thymus DNA. Simultaneously, primer extension was initiated by addition of 3 dNTPs and ddATP. After 10 s, the reaction was stopped by adding formamide and EDTA. Details are provided in Supplemental Data.
α-D403E Challenge Experiments
Polymerase challenge reactions were conducted with 0.5 nM 32P-labeled primer/template, 0.25 μM SSB4 (where applicable), 50 nM β2, 2.0 nM DnaX complex, and 2.0 nM Pol III. The DnaX complex was either pre-incubated with Pol III for 10 min prior to adding α-D403E (Figure 1, Scheme I), or Pol III and α-D403E were mixed together before addition to the DnaX complex (Scheme II). These DnaX/Pol III/α-D403E mixtures were then combined with the other reagents for the primer extension assay. Initiation complex formation was conducted for 10 s with the τ-complex and 60 s with the γ-complex.
ATPγS-Driven Reactions
Initiation complex formation reactions with adenosine-5′-O-(3-thiotriphosphate) (ATPγS, Roche) were conducted under conditions identical to the analogous reactions with ATP, but with 0.20 mM ATPγS substituted for ATP. For all τ-complex catalyzed reactions with ATPγS, initiation complex formation times were increased to 30 s.
SSB Dependence of Initiation Complex Formation
SSB dependence was probed under conditions of 1 nM 32P-labeled primer/template, 50 nM β2, 4.0 nM DnaX complex, and 4.0 nM Pol III. Varying concentrations of SSB4 were added to the primer/template prior to conducting initiation complex formation. Initiation complex formation was run for 10 s with the τ-complex and 5 min with the γ-complex. Full activity for initiation complex formation was demonstrated for the γ-complex in the presence of 0.25 μM SSB4 by modifying our primer extension assay to combine initiation and extension into a single reaction step, which was done by including 40 μM each of dTTP, dGTP and dCTP and 2 μM ddATP during initiation complex formation. A single large-scale initiation complex formation reaction was conducted with 25 μL aliquots withdrawn at various time points and stopped with 25 μL 96% formamide/25 mM EDTA solution. This case is the only time the primer extension assay was conducted as a one-step reaction.
χ–ψ Concentration Dependence of Initiation Complex Formation
The χ–ψ concentration dependence was probed under conditions of 1 nM 32P-labeled primer/template, 50 nM β2, and 4.0 nM Pol III. τ-complex lacking χ–ψ was reconstituted by combining equal concentrations of τ monomer, δ, and δ′ and incubating at room temperature for 15 min prior to combining with varying concentrations of χ–ψ. The final concentration of τ, δ, and δ′ in the reactions was 12 nM (4 nM τ3-complex). The initiation complex formation times were 10 s and 30s for the ATP and ATPγS-driven reactions, respectively.
Supplementary Material
Acknowledgments
We thank Anna Wiktor-Becker for experimental support and Drs. Garry Dallmann and Paul Dohrmann for valuable discussions. We are grateful to Drs. Tim Lohman and Mike Cox, who provided the SSB-CΔ42and SSB-CΔ8 proteins, respectively. This work was supported by NIH grants R01 GM035695 and F32GM084697 (postdoctoral fellowship for C.D.D.). Melissa Stauffer, PhD, of Scientific Editing Solutions, provided editorial assistance with the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alley SC, Abel-Santos E, Benkovic SJ. Tracking Sliding Clamp Opening and Closing During Bacteriophage T4 DNA Polymerase Holoenzyme Assembly. Biochemistry. 2000;39:3076–3090. doi: 10.1021/bi992377r. [DOI] [PubMed] [Google Scholar]
- Ason B, Bertram JG, Hingorani MM, Beechem JM, O'Donnell ME, Goodman MF, Bloom LB. A Model for Escherichia coli DNA Polymerase III Holoenzyme Assembly at Primer/Template Ends. DNA triggers a change in binding specificity of the γ complex clamp loader. J Biol Chem. 2000;275:3006–3015. doi: 10.1074/jbc.275.4.3006. [DOI] [PubMed] [Google Scholar]
- Bertram JG, Bloom LB, Turner J, O'Donnell ME, Beechem JM, Goodman MF. Pre-Steady State Analysis of the Assembly of Wild Type and Mutant Circular Clamps of Escherichia coli DNA Polymerase III onto DNA. J Biol Chem. 1998;273:24564–24574. doi: 10.1074/jbc.273.38.24564. [DOI] [PubMed] [Google Scholar]
- Blinkova A, Hervas C, Stukenberg PT, Onrust R, O'Donnell ME, Walker JR. The Escherichia coli DNA Polymerase III Holoenzyme Contains Both Products of the dnaX Gene, τ and γ, but Only τ Is Essential. J Bacteriol. 1993;175:6018–6027. doi: 10.1128/jb.175.18.6018-6027.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom LB. Loading clamps for DNA replication and repair. DNA Repair (Amst) 2009;8:570–578. doi: 10.1016/j.dnarep.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck I, O'Donnell ME. The DNA Replication Machine of a Gram-positive Organism. J Biol Chem. 2000;275:28971–28983. doi: 10.1074/jbc.M003565200. [DOI] [PubMed] [Google Scholar]
- Bullard JM, Pritchard AE, Song MS, Glover BP, Wieczorek A, Chen J, Janjic N, McHenry CS. A Three-domain Structure for the δ Subunit of the DNA Polymerase III Holoenzyme δ Domain III Binds δ′ and Assembles into the DnaX Complex. J Biol Chem. 2002;277:13246–13256. doi: 10.1074/jbc.M108708200. [DOI] [PubMed] [Google Scholar]
- Burgers PMJ. Mammalian Cyclin/Pcna (DNA Polymerase δ Auxilliary Protein) Stimulates Processive DNA Synthesis by Yeast DNA Polymerase III. Nucleic Acids Res. 1988;16:6297–6307. doi: 10.1093/nar/16.14.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MJ, Jeruzalmi D, Kuriyan J, O'Donnell ME. Motors and Switches: AAA+ Machines within the Replisome. Nat Rev Mol Cell Biol. 2002;3:826–835. doi: 10.1038/nrm949. [DOI] [PubMed] [Google Scholar]
- Fay PJ, Johanson KO, McHenry CS, Bambara RA. Size Classes of Products Synthesized Processively by DNA Polymerase III and DNA Polymerase III Holoenzyme of Escherichia coli. J Biol Chem. 1981;256:976–983. [PubMed] [Google Scholar]
- Gao D, McHenry CS. τ Binds and Organizes Escherichia coli Replication Proteins through Distinct Domains. Domain IV, Located within the Unique C Terminus of τ, Binds the Replication Fork Helicase, DnaB. J Biol Chem. 2001a;276:4441–4446. doi: 10.1074/jbc.M009830200. [DOI] [PubMed] [Google Scholar]
- Gao D, McHenry CS. τ Binds and Organizes Escherichia coli Replication Proteins through Distinct Domains: Partial Proteolysis of Terminally Tagged τ to Determine Candidate Domains and to Assign Domain V as the α Binding Domain. J Biol Chem. 2001b;276:4433–4440. doi: 10.1074/jbc.M009828200. [DOI] [PubMed] [Google Scholar]
- Glover BP, McHenry CS. The χψ Subunits of DNA Polymerase III Holoenzyme Bind to Single-stranded DNA-binding Protein (SSB) and Facilitate Replication of a SSB-coated Template. J Biol Chem. 1998;273:23476–23484. doi: 10.1074/jbc.273.36.23476. [DOI] [PubMed] [Google Scholar]
- Glover BP, McHenry CS. The DnaX-binding Subunits δ′ and ψ are bound to γ and not τ in the DNA Polymerase III Holoenzyme. J Biol Chem. 2000;275:3017–3020. doi: 10.1074/jbc.275.5.3017. [DOI] [PubMed] [Google Scholar]
- Glover BP, McHenry CS. The DNA Polymerase III Holoenzyme: An Asymmetric Dimeric Replicative Complex with Leading and Lagging Strand Polymerases. Cell. 2001;105:925–934. doi: 10.1016/s0092-8674(01)00400-7. [DOI] [PubMed] [Google Scholar]
- Hingorani MM, O'Donnell ME. ATP Binding to the Escherichia coli Clamp Loader Powers Opening of the Ring-Shaped Clamp of DNA Polymerase III Holoenzyme. J Biol Chem. 1998;273:24550–24563. doi: 10.1074/jbc.273.38.24550. [DOI] [PubMed] [Google Scholar]
- Hobbs MD, Sakai A, Cox MM. SSB protein limits RecOR binding onto single-stranded DNA. J Biol Chem. 2007;282:11058–11067. doi: 10.1074/jbc.M611007200. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D, O'Donnell ME, Kuriyan J. Crystal Structure of the Processivity Clamp Loader Gamma Complex of E. coli DNA Polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- Johanson KO, McHenry CS. The β Subunit of the DNA Polymerase III Holoenzyme Becomes Inaccessible to Antibody after Formation of an Initiation Complex with Primed DNA. J Biol Chem. 1982;257:12310–12315. [PubMed] [Google Scholar]
- Johanson KO, McHenry CS. Adenosine 5′-O-(3-Thiotriphosphate) Can Support the Formation of an Initiation Complex between the DNA Polymerase III Holoenzyme and Primed DNA. J Biol Chem. 1984;259:4589–4595. [PubMed] [Google Scholar]
- Johnson A, Yao NY, Bowman GD, Kuriyan J, O'Donnell M. The Replication Factor C Clamp Loader Requires Arginine Finger Sensors to Drive DNA Binding and Proliferating Cell Nuclear Antigen Loading. J Biol Chem. 2006;281:35531–35543. doi: 10.1074/jbc.M606090200. [DOI] [PubMed] [Google Scholar]
- Kaboord BF, Benkovic SJ. Dual Role of the 44/62 Protein as a Matchmaker Protein and DNA Polymerase Chaperone during Assembly of the Bacteriophage T4 Holoenzyme Complex. Biochemistry. 1996;35:1084–1092. doi: 10.1021/bi9520747. [DOI] [PubMed] [Google Scholar]
- Kelman Z, Yuzhakov A, Andjelkovic J, O'Donnell ME. Devoted to the Lagging Strand-The χ Subunit of DNA Polymerase III Holoenzyme Contacts SSB to Promote Processive Elongation and Sliding Clamp Assembly. EMBO J. 1998;17:2436–2449. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DR, McHenry CS. Biotin Tagging Deletion Analysis of Domain Limits Involved in Protein-Macromolecular Interactions: Mapping the τ Binding Domain of the DNA Polymerase III α Subunit. J Biol Chem. 1996;271:20690–20698. doi: 10.1074/jbc.271.34.20690. [DOI] [PubMed] [Google Scholar]
- Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a Replicative Polymerase and Helicase: a τ-DnaB Interaction Mediates Rapid Replication Fork Movement. Cell. 1996a;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- Kim S, Dallmann HG, McHenry CS, Marians KJ. τ Couples the Leading- and Lagging-strand Polymerases at the Escherichia coli DNA Replication Fork. J Biol Chem. 1996b;271:21406–21412. doi: 10.1074/jbc.271.35.21406. [DOI] [PubMed] [Google Scholar]
- Kim S, Dallmann HG, McHenry CS, Marians KJ. τ Protects β in the Leading-strand Polymerase Complex at the Replication Fork. J Biol Chem. 1996c;271:4315–4318. doi: 10.1074/jbc.271.8.4315. [DOI] [PubMed] [Google Scholar]
- LaDuca RJ, Crute JJ, McHenry CS, Bambara RA. The β Subunit of the Escherichia coli DNA Polymerase III Holoenzyme Interacts Functionally with the Catalytic Core in the Absence of Other Subunits. J Biol Chem. 1986;261:7550–7557. [PubMed] [Google Scholar]
- Larsen B, Wills NM, Nelson C, Atkins JF, Gesteland RF. Nonlinearity in Genetic Decoding: Homologous DNA Replicase Genes Use Alternatives of Transcriptional Slippage or Translational Frameshifting. Proc Natl Acad Sci U S A. 2000;97:1683–1688. doi: 10.1073/pnas.97.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu FP, Georgescu R, O'Donnell ME. Mechanism of the E. coli τ Processivity Switch during Lagging-Strand Synthesis. Mol Cell. 2003;11:315–327. doi: 10.1016/s1097-2765(03)00042-x. [DOI] [PubMed] [Google Scholar]
- Lohman TM, Ferrari M. Escherichia coli Single-Stranded DNA-Binding Protein: Multiple DNA-Binding Modes and Cooperativities. Annu Rev Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- Majka J, Burgers PM. The PCNA-RFC Families of DNA Clamps and Clamp Loaders. Prog Nucleic Acid Res Mol Biol. 2004;78:227–260. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- McHenry CS. Purfication and Characterization of DNA Polymerase III′: Identification of τ as a Subunit of the DNA Polymerase III Holoenzyme. J Biol Chem. 1982;257:2657–2663. [PubMed] [Google Scholar]
- McHenry CS, Crow W. DNA Polymerase III of Escherichia coli: Purification and Identification of Subunits. J Biol Chem. 1979;254:1748–1753. [PubMed] [Google Scholar]
- Pritchard AE, Dallmann HG, McHenry CS. In Vivo Assembly of the τ-Complex of the DNA Polymerase III Holoenzyme Expressed from a Five-Gene Artificial Operon: Cleavage of the τ-Complex to Form a Mixed γ–τ-Complex by the OmpT Protease. J Biol Chem. 1996;271:10291–10298. doi: 10.1074/jbc.271.17.10291. [DOI] [PubMed] [Google Scholar]
- Pritchard AE, McHenry CS. Identification of the Acidic Residues in the Active Site of DNA Polymerase III. J Mol Biol. 1999;285:1067–1080. doi: 10.1006/jmbi.1998.2352. [DOI] [PubMed] [Google Scholar]
- Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic Structural Rearrangements Between DNA Binding Modes of E. coli SSB Protein. J Mol Biol. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt SL, Gomes XV, Burgers PMJ. ATP Utilization by Yeast Replication Factor C. III. The ATP-binding domains of Rfc2, Rfc3, and Rfc4 are essential for DNA recognition and clamp loading. J Biol Chem. 2001;276:34784–34791. doi: 10.1074/jbc.M011633200. [DOI] [PubMed] [Google Scholar]
- Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an Organizer/Mobilizer of Genome Maintenance Complexes. Crit Rev Biochem Mol Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, Seyedin SN, Makino DL, O'Donnell M, Kuriyan J. The Mechanism of ATP-dependent Primer-template Recognition by a Clamp Loader Complex. Cell. 2009;137:659–671. doi: 10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Pham PT, Olson M, Carter JR, Franden MA, Schaaper RM, McHenry CS. The δ and δ′ Subunits of the DNA Polymerase III Holoenzyme Are Essential for Initiation Complex Formation and Processive Elongation. J Biol Chem. 2001;276:35165–35175. doi: 10.1074/jbc.M100389200. [DOI] [PubMed] [Google Scholar]
- Studwell-Vaughan PS, O'Donnell ME. Constitution of the Twin Polymerase of DNA Polymerase III Holoenzyme. J Biol Chem. 1991;266:19833–19841. [PubMed] [Google Scholar]
- Tougu K, Marians KJ. The Interaction between Helicase and Primase Sets the Replication Fork Clock. J Biol Chem. 1996;271:21398–21405. doi: 10.1074/jbc.271.35.21398. [DOI] [PubMed] [Google Scholar]
- Williams CR, Snyder AK, Kuzmic P, O'Donnell ME, Bloom LB. Mechanism of Loading the Escherichia coli DNA Polymerase III Sliding Clamp I: Two Distinct Activities for Individual ATP Sites in the γ Complex. J Biol Chem. 2003;279:4376–4385. doi: 10.1074/jbc.M310429200. [DOI] [PubMed] [Google Scholar]
- Witte G, Urbanke C, Curth U. DNA polymerase III χ subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 2003;31:4434–4440. doi: 10.1093/nar/gkg498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.