Abstract
Background
Specific populations of highly tumorigenic cells are thought to exist in many human tumors, including pancreatic adenocarcinoma. However, the clinical significance of these tumor-initiating (ie, cancer stem) cells remains unclear. Aldehyde dehydrogenase (ALDH) activity can identify tumor-initiating cells and normal stem cells from several human tissues. We examined the prognostic significance and functional features of ALDH expression in pancreatic adenocarcinoma.
Methods
ALDH expression was analyzed by immunohistochemistry in 269 primary surgical specimens of pancreatic adenocarcinoma and examined for association with clinical outcomes and in paired primary tumors and metastatic lesions from eight pancreatic cancer patients who had participated in a rapid autopsy program. The clonogenic growth potential of ALDH-positive pancreatic adenocarcinoma cells was assessed in vitro by a colony formation assay and by tumor growth in immunodeficient mice (10–14 mice per group). Mesenchymal features of ALDH-positive pancreatic tumor cells were examined by using quantitative reverse transcription–polymerase chain reaction and an in vitro cell invasion assay. Gene expression levels and the invasive potential of ADLH-positive pancreatic cancer cells relative to the bulk cell population were examined by reverse transcription–polymerase chain reaction and an in vitro invasion assays, respectively. All statistical tests were two-sided.
Results
ALDH-positive tumor cells were detected in 90 of the 269 primary surgical specimens, and their presence was associated with worse survival (median survival for patients with ALDH-positive vs ALDH-negative tumors: 14 vs 18 months, hazard ratio of death = 1.28, 95% confidence interval = 1.02 to 1.68, P = .05). Six (75%) of the eight patients with matched primary and metastatic tumor samples had ALDH-negative primary tumors, and in four (67%) of these six patients, the matched metastatic lesions (located in liver and lung) contained ALDH-positive cells. ALDH-positive cells were approximately five- to 11-fold more clonogenic in vitro and in vivo compared with unsorted or ALHD-negative cells, expressed genes consistent with a mesenchymal state, and had in vitro migratory and invasive potentials that were threefold greater than those of unsorted cells.
Conclusions
ALDH expression marks pancreatic cancer cells that have stem cell and mesenchymal features. The enhanced clonogenic growth and migratory properties of ALDH-positive pancreatic cancer cells suggest that they play a key role in the development of metastatic disease that negatively affects the overall survival of patients with pancreatic adenocarcinoma.
CONTEXT AND CAVEATS
Prior knowledge
Aldehyde dehydrogenase (ALDH) activity identifies normal stem cells and tumor-initiating (ie, cancer stem) cells in several human malignancies, including pancreatic adenocarcinoma. However, the clinical significance of ALDH-expressing cancer stem cells is unclear.
Study design
The clonogenic growth potential of ALDH-positive pancreatic adenocarcinoma cells was assessed in vitro and by tumor growth in immunodeficient mice. Immunohistochemistry was used to analyze ALDH expression in primary pancreatic adenocarcinoma specimens and metastases from patients and its association with survival. Reverse transcription–polymerase chain reaction and in vitro cell invasion assays were used to examine mesenchymal features and the invasive potential of ADLH-positive pancreatic cancer cells relative to the bulk cell population.
Contribution
ALDH expression in pancreatic adenocarcinoma was associated with worse overall survival in patients undergoing resection for early-stage disease. ALDH-positive pancreatic cancer cells were more tumorigenic than the bulk cell population both in vitro and in vivo, expressed genes consistent with a mesenchymal state, and were more frequently detectable in metastatic lesions than in the primary tumor from the same patient.
Implications
ALDH expression marks pancreatic cancer cells that have stem cell and mesenchymal features. The enhanced clonogenic growth of ALDH-positive pancreatic cancer cells suggest that they may play a role in the long-term outcomes of patients with pancreatic adenocarcinoma by mediating the development of metastatic disease.
Limitations
The clinical conclusions about ALDH expression as a prognostic marker in pancreatic adenocarcinoma were based on retrospective analyses and were not independently validated. Differences in tumorigenic capacity between distinct cell populations in immunodeficient mice may be limited by the inherent limitations of a xenotransplantation assay.
From the Editors
Pancreatic adenocarcinoma is highly lethal, and the median survival for newly diagnosed patients is 9–12 months (1). Long-term survival for these patients is best achieved through surgery, but even after complete resection, the median survival is only 18 months (2,3). A major factor that contributes to the high mortality rates among these patients is the propensity of pancreatic cancer cells to invade local tissues and disseminate widely throughout the body; indeed, most pancreatic cancer patients have metastatic lesions at the time of diagnosis or soon thereafter. Gross pathological factors that are predictive of overall survival include tumor size, involvement of the surgical margins, lymph node status, and histological grade (3). Tumor-initiating cells (also known as cancer stem cells [CSCs]) have been identified in an ever-increasing number of human malignancies, including pancreatic adenocarcinoma, and their increased growth potential has suggested that they have a central role in dictating clinical outcomes (4,5). However, the contribution of CSCs to cancer progression is not clearly understood, and it is possible that biological features of these cells may serve as novel prognostic factors in pancreatic adenocarcinoma.
CSCs are functionally defined by two properties: their ability to produce differentiated progeny that are capable of recapitulating the original tumor and their potential for self-renewal, which maintains the long-term proliferative capacity of the malignant clone (6). The cellular processes that regulate the self-renewal of human cancers including pancreatic adenocarcinoma are beginning to be defined and include signaling pathways that are active during normal embryonic development, such as the Hedgehog, Notch, and Wnt pathways (5,7–11). In addition to these pathways, normal pancreatic development requires specific isoforms of aldehyde dehydrogenase (ALDH), specifically ALDH1A1, to synthesize retinoic acid (12,13). Moreover, ALDH is capable of detoxifying xenobiotics and conferring drug resistance (13–15), which suggests that it may play a role in the pathogenesis of pancreatic cancer. Although these properties suggest that CSCs have a central role in disease initiation, relapse, and progression, their existence and clinical relevance have been the subject of continued debate (6,16–19). Moreover, although CSCs may be relatively resistant to standard anticancer therapies (13,20–22), their exact role in disease progression has not been definitively demonstrated.
We have studied the prognostic role of ALDH in pancreatic adenocarcinoma and its association with survival. ALDH activity can also identify tumor cells that have enhanced clonogenic growth potential in many human malignancies, including breast cancer, multiple myeloma, and Hodgkin lymphoma (13,22–27). We also examined the tumorigenic capacity of ALDH-positive cells in vitro and in vivo. Moreover, we studied the expression of genes associated with the epithelial-to-mesenchymal transition and the migratory and invasive capacity of ALDH-positive tumor cells in vitro.
Materials and Methods
Immunohistochemical Analysis
Tissue microarrays were constructed as previously described (28) by using 0.6-mm-diameter samples from formalin-fixed paraffin-embedded blocks of primary pancreatic cancer specimens from 269 patients who had undergone surgery at the Johns Hopkins Hospital between April 1998 and June 2003. In addition, a separate tissue microarray was generated from matched samples of the primary tumor and metastases (from lymph nodes, liver, omentum, and lung) collected from eight patients with metastatic pancreatic cancer who had participated in a rapid autopsy program at the Johns Hopkins Hospital (29). Slides containing the tissue microarrays with three distinct sampling sites from each individual tumor were deparaffinized in xylene and rehydrated in graded alcohol washes. Antigen retrieval was performed by incubating the slides in boiling sodium citrate buffer (10 mM, pH 6.0) for 30 minutes, and endogenous peroxidases were quenched by incubating the slides in 3% hydrogen peroxide in methanol for 10 minutes at room temperature. The slides were then incubated with a mouse monoclonal antibody against human ALDH1 (clone 44; BD Biosciences, San Jose, CA) diluted 1:50 at room temperature for 1 hour followed by antibody detection with the use of a Vectastain Elite developer kit (Vector Labs, Burlingame, CA) and 3,3′-diamniobenzidine, as recommended by the manufacturer. Positive staining was defined as intense 3,3′-diamniobenzidine signal in malignant pancreatic epithelial cells at least twofold greater than normal pancreatic acinar cells that were weakly to moderately positive. Slides were independently scored for anti-ALDH1 staining by two trained pancreatic cancer pathologists (S.-M. Hong and A. Maitra) who were blinded to patient outcomes. Samples with any evidence of positive staining according to the criteria described above were considered positive for the survival analysis.
Cell Culture
We used the following human pancreatic cancer cell lines: CAPAN-1 and Panc1 (American Type Culture Collection, Manassas, VA), DAN-G (DSMZ, Braunschweig, Germany), and L3.6pl (from Dr I. Fidler, Department of Cancer Biology, MD Anderson Cancer Center, Houston, TX). All cell lines were cultured in Dulbecco’s modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (Harlan, Indianapolis, IN), penicillin–streptomycin (Invitrogen), and L-glutamine (Invitrogen). Routine mycoplasma testing of all cell lines was performed by using a MycoProbe Mycoplasma Detection Kit (R&D Systems, Minneapolis, MN) as per the manufacturer's protocol. The authenticity of the cell lines was verified by the American Type Culture Company and the DSMZ.
Pancreatic Cancer Xenografts
All experiments using mice were approved by the Johns Hopkins University Animal Care and Use Committee, and the mice were maintained in accordance with the American Association of Laboratory Animal Care guidelines. We used 10 different human low-passage xenografts from surgical specimens obtained from 10 patients who underwent surgery for pancreatic adenocarcinoma at the Johns Hopkins Hospital. The freshly collected tumors were minced into 2- to 3-mm pieces, and the pieces were implanted subcutaneously into the flanks of 6-week-old female athymic (nu+/nu+) mice (Harlan); when the tumor pieces had grown to 1.5 cm3, they were excised and transplanted to secondary recipient mice (F2; n = 5–6 mice per group) and allowed to grow to 1.5 cm3 as previously described (30).
For immunohistochemical analysis, the tumors were harvested from F2 mice, fixed in neutral buffered 10% formalin, and embedded in paraffin. Tumor sections (10 μm thick) were stained with hematoxylin and eosin and subjected to histopathologic evaluation. For fluorescence-activated cell sorting, the tumors were minced with the use of sterile razor blades to generate single-cell suspensions, which were incubated in 0.6 U/mL dispase and 200 U/mL collagenase type IV (both from Sigma, St Louis, MO) at 37°C for 2 hours with agitation. Debris and necrotic cells were removed by passing the mixture through a 70-μm filter (BD Biosciences) followed by density centrifugation using Ficoll-Paque Plus (GE Healthcare, Uppsala, Sweden). The purified tumor cells were washed twice in cold Dulbecco's modified Eagle medium and counted with the use of a hemocytometer.
Flow Cytometry and Cell Sorting
Cells from the four pancreatic cancer cell lines (CAPAN-1, Panc1, Dan-G, and L3.6pl) and from low-passage xenografts were stained with the ALDEFLUOR reagent system (Stem Cell Technologies, Vancouver, BC, Canada), in which the cell-permeable fluorescent enzyme substrate BODIPY aminoacetaldehyde is intracellularly retained following conversion by ALDH. Briefly, 10 million cells were resuspended in 1 mL Aldefluor buffer and 1 μL Aldefluor reagent in the presence or absence of the ALDH1 inhibitor, diethylamino-benzaldehyde, for 30 minutes in a 37°C water bath according to the manufacturer's protocol. The cells were washed, then incubated at 4°C for 15 minutes with monoclonal anti-CD44-allophycocyanin (APC) (1:20 dilution; clone G44-26; BD Biosciences), anti-CD24-phycoerythrin (PE) (1:20 dilution; clone ML5; BD Biosciences), anti-mouse CD31-biotin (1:100 dilution; BD Biosciences), anti-mouse lineage-biotin (1:100 dilution; Miltenyi Biotec, Auburn, CA), anti-mouse H-2Kd-biotin (1:100 dilution; BD Biosciences), and mouse-specific IgG2b κ-APC (1:100 dilution; BD Biosciences) and IgG2a κ-PE (1:100 dilution; BD Biosciences) antibodies. The cells were subsequently washed and incubated with streptavidin–peridinin chlorophyll protein (0.1 μg/100 μL; BD Biosciences) for 15 minutes at 4°C. The cells were washed once again, resuspended in ALDEFLUOR buffer containing 2 μg/mL propidium iodide, and passed through a 30-μm filter. A FACSAria flow cytometer (BD Biosciences) was used for all cell sorting. The cells were first gated based on side scatter and forward scatter properties, followed by exclusion of mouse-derived (PerCP-positive) and nonviable (PI-positive) cells. The ALDH-positive gate was created based on diethylamino-benzaldehyde–treated cells stained with ALDEFLUOR, anti-CD24-PE, and anti-CD44-APC. The CD44- and CD24-positive gate was created based on cells that stained with ALDEFLUOR and the mouse-specific antibodies IgG2b κ-APC (the isotypic control for anti-CD44-APC) and IgG2a κ-PE (the isotypic control for anti-CD24-PE). Sorted cells were subsequently used for in vitro colony formation, in vivo tumor formation, real-time reverse transcription–polymerase chain reaction (RT-PCR), and cell migration assays.
Colony Formation Assay
Unsorted cells and ALDH-negative, ALDH-positive, CD44- and CD24-positive, and ALDH-, CD44-, and CD24-positive cells that were isolated by fluorescence-activated cell sorting were suspended in 1 mL of methylcellulose medium (1.2% methylcellulose, 30% fetal calf serum, 1% bovine serum albumin, 10−4 M 2-mercaptoethanol, and 2 mM l-glutamine) and plated in quadruplicate onto 35-mm tissue culture dishes with an ultra-low attachment surface (1000 cells per dish) (Corning, Lowell, MA) and incubated at 37°C in 5% CO2. Colonies were scored after 10 days of incubation. Serial replating was performed by washing the dishes three times with Dulbecco's modified Eagle medium and resuspending the pooled cells in 1 mL of methylcellulose medium and replating as described above. Cells were replated for up to four generations, and each experiment was performed three times.
Tumor Formation Assay
Varying numbers of unsorted, ALDH-positive, CD44- and CD24-positive, and ALDH-, CD44-, and CD24-positive cells were isolated from six human pancreatic cancer xenografts (JH015, JH102, Panc496, Panc219, Panc253, and Panc140) by cell sorting. The sorted and unsorted cells from each xenograft were serially diluted and resuspended in 100 μL of a 1:1 mixture of serum-free Dulbecco's modified Eagle medium and Matrigel (BD Biosciences) and then injected subcutaneously into the right and left flanks of 6-week-old sex-matched nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (bred and maintained in the Johns Hopkins Medical Institutes animal care facility). Each unique xenograft was treated as an individual experiment, and 10–14 mice (one to two injections per mouse, depending on the availability of sufficient cell numbers) were used to evaluate the clonogenic growth potential of each starting xenograft. If sufficient cells were available to carry out two injections per mouse, each of the two sites was injected with the same number of cells from the same starting xenograft. The tumor-initiating capacity of the different cell populations was compared by monitoring tumor growth in mice daily for up to 20 weeks. Tumor formation was considered positive if a mass greater than 1 cm in diameter was detected by palpation. After 20 weeks after injection, the mice were killed by carbon dioxide asphyxiation and cervical dislocation and the tumors were harvested for serial retransplantation in fresh mice (up to four times) or for immunohistochemical analysis. Each unique human xenograft was examined once.
Real-Time Reverse Transcription–Polymerase Chain Reaction
One thousand unsorted, ALDH-positive, CD44- and CD24-positive, and ALDH-, CD44-, and CD24-positive cells were isolated by fluorescence-activated cell sorting from four different human pancreatic cancer xenografts (Panc253, JH015, Panc185, and Panc410). The cells were lysed and subjected to reverse transcription without purification of RNA by using a Cells-to-Ct kit (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. Quantitative PCR was performed in triplicate on a MyiQ real-time PCR thermocycler (BioRad, Hercules, CA) with the use of the TaqMan Gene Expression Assays (Applied Biosystems) for ACTB (β-actin), CDH1 (e-cadherin) (Hs00170423_m1), SNAI2 (slug) (Hs00161904), ALDH1A1 (aldehyde dehydrogenase 1A1) (Hs00167445_m1), CD24 (Hs02379687_s1), CD44 (Hs00174139_m1), CDH2 (n-cadherin) (Hs00983062_m1), SNAI1 (snail) (Hs00195591_m1), and TWIST1 (twist homolog 1) (Hs00361186_m1) according to the manufacturer's instructions. Comparative gene expression analysis was performed by using the 2(−ΔΔCt) method with normalization to ACTB. One experiment was performed for each unique starting human xenograft.
Cell Migration Assay
Twenty thousand unsorted, ALDH-positive, CD44- and CD24-positive, and ALDH-, CD44- and CD24-positive cells were isolated by fluorescence-activated cell sorting, suspended in Dulbecco's modified Eagle medium supplemented with 1% fetal bovine serum (Harlan), applied to 24-well cell culture inserts containing 8-μm pores (BD Biosciences) in duplicate, and incubated for 72 hours. Cells that remained above the filter (ie, nonmigrating cells) were removed, and the number of cells below the filter (ie, migrating cells) was counted with the use of an inverted light microscope. Migration assays were repeated three times for each group of cells.
Cell Invasion Assay
Single-cell suspensions of pancreatic cancer cells derived from low-passage xenografts (Panc410 and Panc496) were incubated for 10 minutes on ice with the three biotin-labeled, anti-mouse antibodies (anti-CD31, anti-mouse lineage, and anti-H-2Kd; each diluted 1:100), followed by incubation with streptavidin-coupled magnetic microbeads (Miltenyi Biotec). The cell suspensions were magnetically depleted of mouse cells using a MACS Cell Separation LD Column (Miltenyi Biotec), and the remaining cells were applied to 24-well Matrigel Invasion Chambers in duplicate wells (5 × 105 cells per well; BD Biosciences) and incubated for 72 hours. Invading and noninvading cells from below and above the invasion chamber membrane, respectively, were harvested by using a cell scraper and cell lysis buffer. ACTB, ALDH1A1, CD24, CD44, and CDH1 mRNA levels in the invading and noninvading cells were analyzed by using the Cells-to-Ct kit (Applied Biosystems) and quantitative real-time PCR, as described above. The experiment was performed once for each unique human xenograft.
Statistical Analysis
The Kaplan–Meier method and the log-rank test were used to compare survival, defined as the time from surgery until death (patients alive were censored at the time of their last follow-up), of patients with ALDH-positive and patients with ALDH-negative primary tumors. A χ2 test was used to examine associations between tumor ALDH activity and tumor size greater than 3 cm, surgical margin involvement, tumor differentiation, and lymph node metastasis, which are known prognostic factors for adverse outcome following surgical resection for pancreatic carcinoma (3,31). A multivariable Cox proportional hazards model was used to study associations of all variables with patient survival as well as independent associations between ALDH activity or lymph node metastasis and patient survival. A test of the proportional hazards assumption was based on scaled Schoenfeld residuals. We found no strong evidence of nonproportional hazards for either ALDH expression (P = .663) or lymph node metastasis (P = .687).
To account for correlation between the two injection sites per mouse, a logistic regression model was implemented with generalized estimating equations. The hypotheses of equal probability of tumor formation comparing bulk unsorted cells with ALDH-positive cells, with CD44- and CD24-positive cells, and with ALDH-, CD44-, and CD24-positive cells were tested based on a χ2 test. All statistical tests were two-sided, and statistical significance was defined as a P value less than or equal to .05.
Results
Prognostic Role of ALDH Expression in Pancreatic Adenocarcinoma
Several pathological features are associated with the long-term outcomes of patients following surgical resection for pancreatic adenocarcinoma, including tumor size, lymph node involvement, surgical margin status, and histological grade (2,3). We hypothesized that cellular processes that are involved in the normal development of the pancreas might serve as prognostic factors in pancreatic cancer. We therefore examined the expression of ALDH because it is required for the development of the pancreas during mouse embryogenesis and is associated with drug resistance in multiple myeloma and colorectal adenocarcinoma (13,22,32).
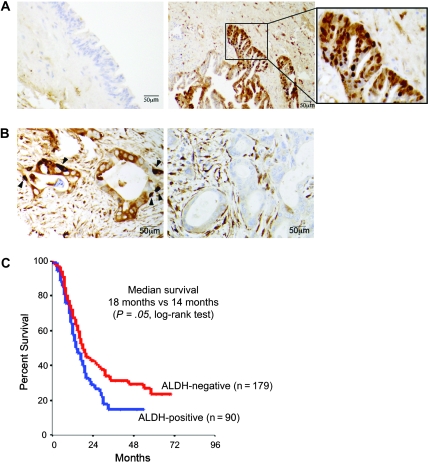
We examined ALDH expression by immunohistochemistry in surgical specimens that represented all stages of pancreatic cancer development. We detected no neoplastic cells that stained positive with an antibody against ALDH in low-grade pancreatic intraepithelial neoplastic lesions (ie, grade 1 or 2) (33). However, we found occasional ALDH-positive cells in high-grade pancreatic intraepithelial neoplastic lesions (ie, grade 3, also known as carcinoma in situ) (Figure 1, A). We also examined ALDH expression in pancreatic adenocarcinomas by immunohistochemistry in resected primary tumors from 269 pancreatic cancer patients. Although all tumor specimens displayed ALDH staining within the tumor stroma, we detected two general patterns of staining among the glandular cancer cells. In 90 tumors (34%), we observed cells located primarily at the base of the cancerous pancreatic glands that stained intensely with the ALDH antibody. Tumors with these intensely stained cells were considered ALDH positive (arrow, Figure 1, B). The remaining 179 specimens (66%) lacked these intensely stained cells within the three distinct sections studied from each tumor and were considered ALDH negative. In a Kaplan–Meier survival analysis, the patients with ALDH-positive primary tumors had a median survival of 14 months compared with 18 months for the patients with ALDH-negative primary tumors (hazard ratio [HR] of death = 1.28, 95% confidence interval [CI] = 1.02 to 1.68, P = .05 [log-rank test]) (Figure 1, C). In addition, ALDH expression was associated with long-term survival: The 25% survival rate was 28.4 months in patients with ALDH-positive primary tumors vs 46 months in patients with ALDH-negative specimens.
Figure 1.
Association between aldehyde dehydrogenase (ALDH) expression in primary pancreatic tumors and patient survival. A) Immunohistochemical localization of ALDH protein expression in pancreatic intraepithelial neoplastic lesions. Representative images show staining for ALDH (brown) in a high-grade pancreatic intraepithelial neoplastic lesion (ie, PanIN-3 or carcinoma in situ; right panel and inset) and in a low-grade pancreatic intraepithelial neoplastic lesion (ie, PanIN-2; left panel). B) Immunohistochemical staining of ALDH in primary human pancreatic adenocarcinoma. Left panel shows an ALDH-positive tumor in which intensely staining cells are found mostly at the base of cancerous glands (arrowheads). Right panel shows an ALDH-negative tumor. C) Kaplan–Meier analysis of overall survival for patients who underwent pancreatectomy based on tumor ALDH expression by immunohistochemistry. At 24 months, the number of patients at risk with ALDH-positive and ALDH-negative tumors was 27 (proportion surviving = 0.30, 95% confidence interval [CI] = 0.22 to 0.42) and 67 (proportion surviving = 0.39, 95% CI = 0.32 to 0.47), respectively. At 48 months, the number of patients at risk with ALDH-positive and ALDH-negative tumors was 9 (proportion surviving = 0.15, 95% CI = 0.09 to 0.25) and 33 (proportion surviving = 0.23, 95% CI = 0.18 to 0.31), respectively. At 72 months, the number of patients at risk with ALDH-positive and ALDH-negative tumors was 2 (proportion surviving = 0.11, 95% CI = 0.05 to 0.21) and 16 (proportion surviving = 0.17, 95% CI = 0.12 to 0.24).
We next examined the combined and independent associations between tumor ALDH positivity by immunohistochemistry and other predictors of poor prognosis in pancreatic cancer on overall survival in a multivariable model. Initially, we examined the association between ALDH positivity and tumor size, the extent of tumor differentiation, the presence of lymph node metastasis, and surgical margin involvement. We found that tumor ALDH positivity was statistically significantly associated with tumor size greater than 3 cm and with poorly differentiated tumors (for both, P = .03) but not with surgical margin involvement (P = .11). Furthermore, statistically significant associations were found between tumor size greater than 3 cm and surgical margin involvement (P = .02) and between tumor size greater than 3 cm and extent of tumor differentiation (P = .03). On the basis of these strong associations, we developed a multivariable Cox proportional hazards model that included ALDH expression and lymph node metastasis as covariates to obtain an estimate of the independent association between ALDH expression and overall survival. Overall, patients with ALDH-positive primary tumors had worse survival compared with patients with ALDH-negative primary tumors (HR of death = 1.32, 95% CI = 1.00 to 1.74, P = .054) (Table 1).
Table 1.
Multivariable Cox proportional hazards analysis of overall survival*
| Characteristic | HR of death (95% CI) | P |
| No. of involved lymph nodes | ||
| 0 | 1.00 (referent) | |
| ≥1 | 1.66 (1.14 to 2.41) | .009 |
| Tumor ALDH expression status | ||
| Negative | 1.00 (referent) | |
| Positive | 1.32 (1.00 to 1.74) | .054 |
ALDH = aldehyde dehydrogenase; CI = confidence interval; HR = hazard ratio.
Tumorigenic Potential of ALDH-Positive Pancreatic Cancer Cells
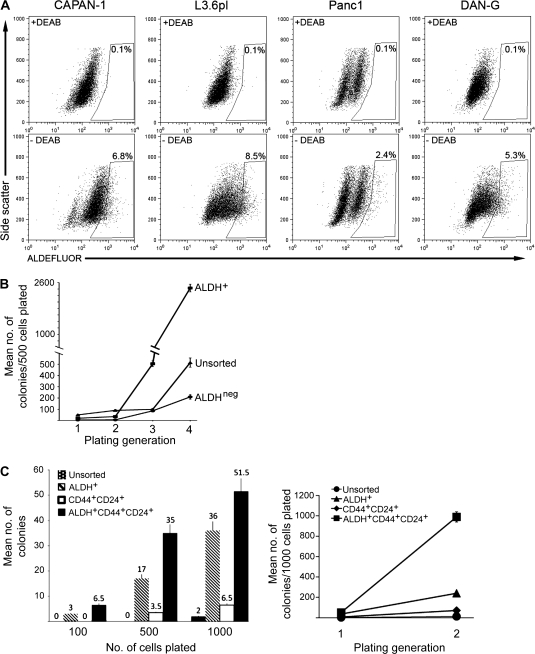
Cells with enhanced clonogenic growth potential can be identified in a variety of normal and malignant tissues based on relative ALDH activity (22–24,27). We examined the relationship between ALDH expression and tumorigenic potential in pancreatic cancer cells. We initially studied four human pancreatic cancer cell lines (CAPAN-1, DAN-G, Panc1, and L3.6pl) by using the ALDEFLUOR flow cytometric assay, which consists of a freely permeable fluorescent enzyme substrate that is retained by cells following conversion by ALDH, and found that each of these cell lines contained small populations of ALDH-positive cells (2.4%–8.5% of the total cell population; Figure 2, A). We next assessed the in vitro clonogenic growth potential of the well-studied CAPAN-1 cell line as a function of ALDH expression detected using the ALDEFLUOR assay. We found that ALDH-positive cells (defined as those with greater ALDEFLUOR fluorescence compared with a control staining reaction using the specific ALDH enzyme inhibitor diethylamino-benzaldehyde) formed statistically significantly greater numbers of colonies during serial rounds of replating (a surrogate measure of self-renewal potential) compared with ALDH-negative or unsorted cells. For example, compared with ALDH-negative cells, ALDH-positive cells formed 5.9-fold more colonies (95% CI = 4.4- to 7.9-fold more, P < .001) during the third generation of replating and 11.6-fold more colonies during the fourth round of replating (95% CI = 9.7- to 13.8-fold more, P < .001) (Figure 2, B). Several studies have reported that tumorigenic pancreatic cancer cells can be identified based on their expression of specific cell surface antigens, including CD44 and CD24 (4,5). We therefore compared the in vitro clonogenic capacity of CAPAN-1 cell populations that were isolated based on their expression of ALDH only, of CD44 and CD24, or of ALDH, CD44, and CD24. Each of these cell populations formed more colonies than the bulk unsorted tumor cells, and the cells that coexpressed ALDH, CD44, and CD24 had the highest clonogenic potential during both initial and secondary platings (Figure 2, C).
Figure 2.
Cancer stem cell properties of aldehyde dehydrogenase (ALDH)–positive pancreatic cancer cells. A) Flow cytometry. ALDH activity in human pancreatic cancer cell lines CAPAN-1, DAN-G, Panc1, and L3.6pl was measured by flow cytometry by using the ALDEFLUOR reagent in the presence (top row) and absence (bottom row) of the ALDH1 inhibitor diethylamino-benzaldehyde (DEAB). The frames represent gates that depict ALDH-positive (ALDH+) cells that were created based on the flow cytometry plot from cells treated with DEAB and then applied to the plots of untreated cells. The percentages of ALDH+ cells are shown in or above the gate. B) Colony formation assay. ALDH+, ALDH-negative (ALDHneg), and unsorted CAPAN-1 cells were isolated by fluorescence-activated cell sorting and evaluated for clonogenic growth in vitro. Five hundred cells were plated in methylcellulose, and colonies were counted 10 days later. Serial replating was carried out for four generations. Error bars correspond to 95% confidence intervals. C) Colony formation assay with serially diluted cells. Unsorted, ALDH+, CD44- and CD24-positive (CD44+CD24+), and ALDH-, CD24-, and CD44-positive (ALDH+CD44+CD24+) CAPAN-1 cells were isolated by fluorescence-activated cell sorting and evaluated for clonogenic growth as described above. The left panel depicts the number of colonies that were counted after varying numbers of cells (100, 500, or 1000) were plated in methylcellulose. The right panel depicts the number of colonies that grew after 1000 cells were plated in methylcellulose and serially replated for a second generation. Mean values (n = 3) with 95% confidence intervals (error bars) are depicted.
To further study whether the ALDH-positive and CD44- and CD24-positive cells represented overlapping or distinct cell populations, we next examined four low-passage xenografts derived from resected primary tumor specimens (30). Single-cell suspensions were stained with the ALDEFLUOR reagent and anti-CD44 and anti-CD24 monoclonal antibodies and then analyzed by flow cytometry. Although relatively small populations of ALDH-positive cells and CD44- and CD24-positive cells could be identified in all xenografts examined, the two cell populations were largely nonoverlapping with cells that were positive for ALDH, CD44, and CD24, representing less than 0.1% of the total cell population (Supplementary Figure 1, A, and Supplementary Table 1, available online). We compared the in vivo growth potential of unsorted, ALDH-positive, CD44- and CD24-positive, and ALDH-, CD44-, and CD24-positive cells by examining their ability to form tumors following the subcutaneous injection of decreasing numbers of cells into immunodeficient NOD/SCID mice (n = 2–4 mice per cell group). As was reported in other studies that examined clonogenic pancreatic cancer growth (4,5), we found that unsorted tumor cells from each of the six low-passage xenografts studied formed tumors only when at least 10 000 cells were injected, whereas injection of as few as 50 cells (range = 50–5000 cells) from each fluorescence-activated cell sorter-isolated cell population (eg, ALDH-positive, CD44- and CD24-positive, and ALDH-, CD44-, and CD24-positive cells) produced tumors that were histologically identical to the original xenograft (Table 2; Supplementary Figure 1, B, available online). Both ALDH-positive cells and CD44- and CD24-positive cells were statistically significantly more tumorigenic than unsorted tumor cells. Tumor incidence in mice injected with ALDH-positive vs unsorted cells was 33.3% vs 6.9% (P = .005) and 33.3% vs 6.9% for mice injected with CD44- and CD24-positive vs unsorted cells (P = .007); however, neither sorted population had a greater tumor-initiating capacity than the other (P = 1.0). Cells that were positive for ALDH, CD44, and CD24 were more tumorigenic than both the ALDH-positive and the CD44- and CD24-positive cell populations, but the differences were not statistically significant (P = .30 and P = .35, respectively). The tumorigenic potential of all three cell populations was maintained after multiple rounds of harvesting, resorting, and injection (data not shown), indicating their self-renewal potential.
Table 2.
Tumor-initiating capacity of sorted pancreatic cancer cells*
| No. of cells injected | Tumor incidence† |
|||
| Unsorted cells | ALDH+ cells | CD44+CD24+ cells | ALDH+CD44+CD24+ cells | |
| 10 000 | 3/17 | ND | ND | ND |
| 1250–5000 | 0/14 | 6/14 | 4/8 | 8/15 |
| 250–1000 | 0/12 | 2/6 | 2/6 | 6/14 |
| 50–200 | ND | 2/10 | 2/10 | 5/14 |
| Total | 3/43 | 10/30 | 8/24 | 19/43 |
| OR (95% CI) | 1.00 (referent) | 6.64 (1.76 to 24.97) | 6.64 (1.67 to 26.35) | 10.38 (3.02 to 35.63) |
| P‡ | .005 | .007 | <.001 | |
Unsorted and sorted human pancreatic cancer cells derived from low-passage xenografts (n = 6) were subcutaneously injected into nonobese diabetic/severe combined immunodeficient mice (one to two injections per mouse). Each xenograft was treated as an independent experiment (or group). For each xenograft, 10–14 mice were used. This table shows the data for all of the xenografts combined. The mice were monitored daily for 20 weeks for tumor formation (arbitrarily scored as a tumor >1 cm in the greatest dimension), at which time they were killed. CI = confidence interval; ND = not done; OR = odds ratio.
Tumor incidence = the number of tumors formed/the total number of injections.
Two-sided χ2 test from a generalized estimation equation approach to modeling tumor formation that accounts for the correlation between injections per mouse.
ALDH-Positive Tumor Cells in Metastatic Lesions
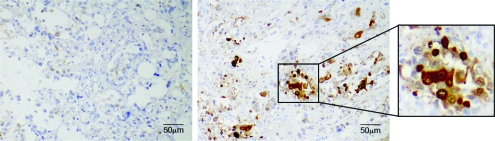
For most solid tumors including pancreatic adenocarcinoma, the development of distant metastases marks disease progression and portends poor prognosis. The association we observed between tumor ALDH expression and worse median survival suggested that ALDH-positive cells may be involved in this process. We therefore analyzed ALDH expression by immunohistochemistry in paired primary tumors and metastatic lesions from eight pancreatic cancer patients who had participated in a rapid autopsy program. Primary tumors from six (75%) of the eight patients were ALDH negative, but in four (67%) of these six cases, the matched metastatic lesions (located in liver and lung) contained ALDH-positive cells (Figure 3 and Table 3). In the two ALDH-positive primary tumors, all of the matched metastatic lesions (located in periaortic lymph nodes, liver, and kidney) contained ALDH-positive cells. These data suggest that ALDH-positive cells may play a role in disease progression.
Figure 3.
Aldehyde dehydrogenase (ALDH) expression in a primary tumor and metastatic lesion from a pancreatic cancer patient who participated in the rapid autopsy program. Immunohistochemical staining for ALDH (brown) in the primary tumor (left panel) and in a lung metastasis (right panel).
Table 3.
Aldehyde dehydrogenase (ALDH) expression in paired primary and metastatic tumors from pancreatic adenocarcinoma patients*
| Patient | ALDH expression |
|
| Primary tumor | Metastatic lesion (location) | |
| A10 | Positive | Positive (periaortic lymph nodes) |
| A19 | Positive | Positive (kidney and liver) |
| A2 | Negative | Positive (liver) |
| A6 | Negative | Positive (liver) |
| A14 | Negative | Positive (liver) |
| A18 | Negative | Positive (lung) |
| A7 | Negative | Negative |
| A17 | Negative | Negative |
ALDH expression was examined by immunohistochemistry in paired primary and metastatic tumors from eight patients with pancreatic adenocarcinoma who participated in a rapid autopsy program. The lymph nodes, liver, kidney, omentum, and lungs were examined for metastases.
Phenotype and Invasiveness of ALDH-Positive Pancreatic Tumor Cells
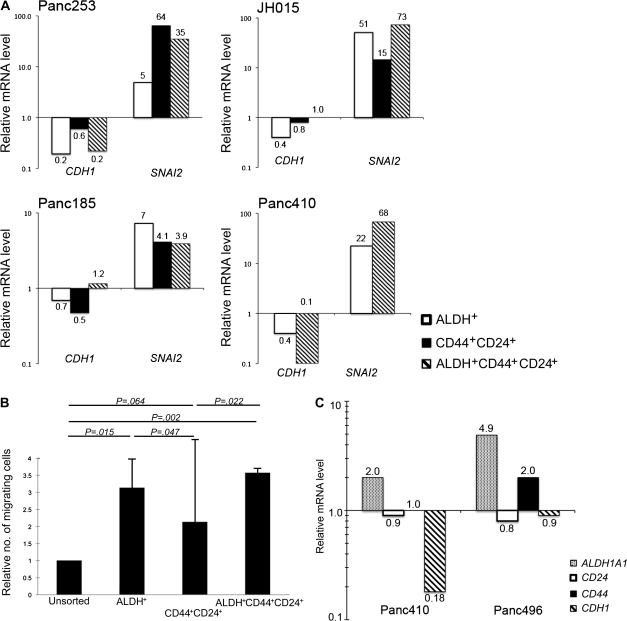
The development of metastatic disease requires tumor cell dissemination from the primary tumor and clonogenic growth at the ectopic site. The epithelial-to-mesenchymal transition is marked by the loss of cell adhesion and increased migratory potential, which regulate cellular movement during normal embryonic development. Because of this increased cellular motility, the epithelial-to-mesenchymal transition has also been implicated in cancer metastasis and is associated with advanced stage for many solid tumors (34,35). During the epithelial-to-mesenchymal transition, mesenchymal cells are characterized by decreased expression of E-cadherin (encoded by CDH1) and increased expression of genes that encode members of the Snail family of transcriptional repressors, such as TWIST (TWIST1), SNAIL (SNAI1), and SLUG (SNAI2). We used quantitative real-time RT-PCR to examine the expression of genes associated with the epithelial-to-mesenchymal transition among the sorted cell populations isolated from four low-passage xenografts. Although expression of CDH2, TWIST1, and SNAI1 could not be consistently detected in these xenografts, CDH1 expression was decreased up to twofold in CD44- and CD24-positive cells, up to fivefold in ALDH-positive cells, and up to 10-fold in cells positive for ALDH, CD44, and CD24 compared with unsorted xenograft tumor cells (Figure 4, A). Conversely, SNAI2 expression was increased up to 64-fold in CD44- and CD24-positive cells, up to 51-fold in ALDH-positive cells, and up to 73-fold in cells positive for ALDH, CD44, and CD24 compared with unsorted xenograft tumor cells. The decreased expression of CDH1 and the increased expression of SNAI2 suggest that these highly clonogenic cell populations have mesenchymal features.
Figure 4.
Properties of pancreatic cancer stem cells (CSCs). A) E-cadherin and SLUG mRNA expression in pancreatic CSC. Unsorted, ALDH-positive (ALDH+), CD44- and CD24-positive (CD44+CD24+), and ALDH-, CD24-, and CD44-positive (ALDH+CD44+CD24+) cells from four human xenografts (Panc253, Panc185, JH015, and Panc410) were isolated by fluorescence-activated cell sorting and analyzed for ACTB (β-actin), CDH1, and SNAI2 mRNA levels by real-time quantitative reverse transcription–polymerase chain reaction (RT-PCR) in triplicate. Results are depicted as normalized gene expression in ALDH+, CD44+CD24+, and ALDH+CD44+CD24+ cells relative to unsorted cells. One experiment was performed for each xenograft. B) Cell migration assay. Unsorted, ALDH+, CD44+CD24+, and ALDH+CD44+CD24+ CAPAN-1 cells were isolated by fluorescence-activated cell sorting and applied to a transwell chambers containing filters with 8-μm pores. Cells that had migrated through the filter were counted 72 hours later. The relative number of migrating cells from each cell population compared with the number of migrating cells from the unsorted population is plotted. The results plotted are from three separate experiments, each containing duplicate wells. Error bars correspond to 95% confidence intervals. P values (two-sided) are from a Student t test. C) Cell invasion assay. Low-passage human pancreatic cancer xenografts from mice (Panc410 and Panc496) were made into single-cell suspensions, depleted of mouse cells by using an LD cell separation column, applied to a Matrigel-coated invasion chamber, and incubated for 72 hours. ALDH1A1, CD44, and CD24 mRNA levels in the invasive and noninvasive cells were measured by quantitative RT-PCR in triplicate. Results are depicted as normalized gene expression in invasive cells relative to noninvasive cells. One experiment was performed for each xenograft.
Tumor cells undergoing the epithelial-to-mesenchymal transition are thought to have increased migratory and invasive potential (35). We next compared the migratory and invasive properties of unsorted, ALDH-positive, CD44- and CD24-positive, and ALDH-, CD44-, and CD24-positive cells isolated from CAPAN-1 cells (Figure 4, B) and Panc1 and DAN-G cells (data not shown). Compared with unsorted cells, cells positive for ALDH, CD44, and CD24 and ALDH-positive cells displayed more migratory capacity in a transwell assay system (unsorted vs ALDH-, CD44-, and CD24-positive cells: 3.6-fold increase, 95% CI = 3.4- to 3.7-fold increase, P = .002; unsorted vs ALDH-positive cells: 3.1-fold increase, 95% CI = 2.3- to 4.0-fold increase, P = .015) (Figure 4, B). It is interesting that CD44- and CD24-positive cells also displayed only slightly increased migration compared with unsorted cells (2.1-fold increase, 95% CI = −0.6- to 4.8-fold increase, P = .064) but displayed statistically significantly less migration than cells positive for ALDH, CD44, and CD24 (1.7-fold decrease, 95% CI = 0.6- to 2.8-fold decrease, P = .022) or ALDH-positive cells (1.5-fold decrease, 95% CI = 0.4- to 2.6-fold decrease, P = .047). These results suggest that the equally clonogenic ALDH-positive and CD44- and CD24-positive cells are biologically distinct because they are not functionally equivalent with respect to migratory capacity.
We also examined ALDH1A1, CD44, and CD24 mRNA expression in invasive tumor cells derived from two low-passage xenografts from which we could obtain sufficient cell numbers for subsequent in vitro analysis. Single-cell suspensions were made from each xenograft, depleted of mouse-derived cells, and subjected to an in vitro invasion assay, and the invading and noninvading cells were harvested and analyzed for gene expression by quantitative RT-PCR. Compared with noninvading tumor cells, cells capable of invading through a Matrigel-coated membrane in vitro displayed approximately two- to fivefold higher levels of ALDH1A1 mRNA but less than a twofold higher level in CD44 mRNA expression and no change in CD24 mRNA expression (Figure 4, C). These data suggest that a substantial population of tumor-initiating pancreatic cells, especially those that express ALDH, has functional features consistent with epithelial-to-mesenchymal transition.
Discussion
In this study, we report that ALDH expression in pancreatic adenocarcinoma is associated with worse overall survival in patients who are undergoing resection for early-stage disease. Furthermore, we found that ALDH-positive pancreatic cancer cells are statistically significantly more tumorigenic than ALDH-negative cells both in vitro and in vivo and are more frequently detectable in metastatic lesions than in the primary tumor from the same patient. These results suggest that CSCs may play a role in the long-term outcomes of patients with pancreatic adenocarcinoma by mediating the development of metastatic disease.
The identification of CSCs in most experimental systems has relied on their expression of specific cell surface antigens. However, although this strategy can enrich for tumorigenic cells, it may not be able to identify all of the clonogenic cells in an individual tumor. Both normal stem cells and CSCs may also be distinguished from non–stem cells by using assays based on physiological properties, such as homing properties and increased drug efflux and ALDH activity (27,36,37). We found little overlap between the ALDH-positive and the CD44- and CD24-positive cell populations despite the fact that they had a similar tumor formation capacity in vivo, which is reminiscent of recent findings in breast cancer (24). We found that sorting cells on the basis of both ALDH activity and cell surface antigen expression slightly enhanced the isolation of clonogenic pancreatic cancer cells compared with sorting on the basis of either ALDH activity or cell surface antigen expression alone. Therefore, it is possible that multiple phenotypically distinct cell populations are clonogenic in an individual tumor. Alternatively, it is possible that the phenotype of CSCs changes in response to cellular activation status, interactions with the external microenvironment, or disease stage. Thus, additional studies that examine whether these factors influence the relationship between expression of specific CSC markers and functional capacities, such as clonogenic growth or cellular migration, are needed.
The tumorigenic and self-renewal capabilities of CSCs have suggested that these rare cell populations are determinants of clinical outcomes, such as the time to relapse and overall survival; however, evidence supporting their clinical importance is limited. It is possible that the CSC-specific features, such as ALDH activity, or their relative frequency within tumors may predict clinical outcomes, which would suggest that these cells are relevant. For example, in breast cancer, the expression of a specific gene signature in CSCs that express CD44 but have low or undetectable levels of CD24 was correlated with metastasis-free and overall survival (38). Furthermore, recent studies have demonstrated that increased frequency of CSCs in breast cancer and in brain tumors is associated with overall survival (24,39). We found that the presence of ALDH-positive cells in resected primary tumors was associated with a 22% decrease in median survival compared with ALDH-negative tumors (14 vs 18 months). We also found that ALDH expression was strongly associated with other predictors of poor prognosis, including tumor size greater than 3 cm, poor differentiation, and positive surgical margins. Thus, ALDH expression may be a marker for pancreatic adenocarcinomas that have a greater chance of locally recurring or metastasizing. Lymph node involvement was also associated with poor outcomes, and in a multivariable model, ALDH expression was independently associated with decreased overall survival. These findings suggest that ALDH may serve as a biomarker that predicts long-term outcomes in pancreatic cancer; however, large prospective analyses are needed to confirm this possibility.
The majority of the primary tumor specimens in this study lacked expression of ALDH-positive cells. The lack of ALDH-positive pancreatic cancer cells in some of the primary specimens suggests that these tumors lack CSCs. It is possible that the ALDH-negative tumors we studied contained rare ALDH-positive cells that were not detected because of variability in tumor sampling in tissue microarrays and that both the relative frequency and distribution of ALDH-positive cells may ultimately be associated with clinical outcomes. Alternatively, ALDH-negative tumors may contain CSCs that are phenotypically and functionally distinct from ALDH-positive tumor cells and are associated with relatively less aggressive disease. We also detected ALDH expression using immunohistochemistry in the stromal cells of all pancreatic adenocarcinoma specimens examined. Given the uniform presence of ALDH-positive stromal cells, this pattern of staining is not likely to be associated with clinical outcomes, and the biological significance of this finding is unclear.
The possibility that ALDH-positive tumor cells represent a subset of CSCs with increased metastatic potential may explain their association with worse prognosis. We found that metastases were commonly ALDH positive, even in paired tissue specimens in which the primary tumors were ALDH negative. These observations support a role for ALDH-positive cells in disease progression. Our findings do not provide a mechanism for how ALDH-negative primary tumors might give rise to ALDH-positive metastases. However, it is possible that ALDH activity is induced during the evolution of clones that mark the acquisition of metastatic potential.
To initiate metastases, tumor cells must be able to disseminate from the primary tumor, invade the surrounding environment, migrate to distant sites, and form macroscopic lesions. Mesenchymal features have been ascribed to invasive cells, and expression of epithelial-to-mesenchymal transition–related proteins is associated with metastases in many cancers, including pancreatic cancer (34,40). We found that ALDH-positive and ALDH-, CD44-, and CD24-positive pancreatic cancer cells isolated from low-passage xenografts had a mesenchymal gene expression profile and were more migratory compared with unsorted tumor cells. Similar findings were reported in a study that demonstrated that induction of the epithelial-to-mesenchymal transition in immortalized human mammary epithelial cells by ectopic expression of TWIST or SNAI1 or exposure to TGF-β can generate breast CSCs (41). We also found that pancreatic cancer cells capable of migration and invasion in vitro also expressed more ALDH1A1 mRNA than cells without these functional abilities but not more mRNA for the cell surface antigens CD44 and CD24. Further evidence that ALDH-positive pancreatic CSCs play a role in the development of metastatic disease is provided by our previous findings that the pharmacological inhibition of the Hedgehog signaling pathway selectively eliminates ALDH-positive pancreatic adenocarcinoma cells and results in reduced pancreatic cancer invasion and metastasis in vivo (42).
In several human tumors, including breast cancer, colorectal carcinoma, and acute pre-B cell lymphoblastic leukemia, distinct sets of markers may identify CSCs (13,24,43–46); however, the relationship between each of these phenotypically distinct CSC subpopulations is unclear. Similarly, in pancreatic adenocarcinoma, reports from other groups (4,5) and the data presented here demonstrate that ALDH-positive, CD44- and CD24-positive, and CD133-positive pancreatic cancer cells all have enhanced tumorigenic and self-renewal capabilities compared with unsorted tumor cells. Despite representing largely nonoverlapping populations, ALDH-positive and CD44- and CD24-positive pancreatic cancer cells were both highly tumorigenic. However, our data also show that ALDH-positive cells are more invasive than CD44- and CD24-positive pancreatic cancer cells in vitro. Our data, as well as those from a study that examined CD133-positive pancreatic tumor cells (4), suggest that phenotypically different CSCs have specific functional capacities in addition to their clonogenic growth potential. It is possible that these different CSC populations are interrelated by a retained hierarchical arrangement in which the expression of each specific marker is restricted to a specific cellular compartment, which is reminiscent of the structured relationship between long- and short-term stem cells and progenitors in normal hematopoiesis (47). ALDH-positive pancreatic cancer cells may represent more primitive cells that give rise to CD44- and CD24-positive cells (or vice versa), and the expression of all of these markers is lost in differentiated epithelial cells. Alternatively, a stochastic model of CSCs may exist in which acquisition of defined functions may generate phenotypically distinct cell types, such as invasive ALDH-positive cells.
These results provide evidence for the clinical relevance of CSCs in pancreatic adenocarcinoma and may explain how the detection of these cells in primary tumors may result in shortened overall survival. However, these findings may be limited by the retrospective nature of our survival analysis and by our use of immunodeficient mice for the functional studies because it is possible that the mouse microenvironment may not fully support the growth of pancreatic tumor cells in the same manner found in humans. Prospective studies that examine the correlation between ALDH expression and survival, and improved in vivo models that alleviate potential bias due to xenografting barriers, may ultimately confirm these findings.
Funding
National Institutes of Health (CA127574, CA107040, CA09071 to W.M., and CA113669 to A.M.); Sidney Kimmel Foundation for Cancer Research to W.M.
Supplementary Material
Footnotes
The study sponsors had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Sohn TA, Yeo CJ, Cameron JL, et al. Resected adenocarcinoma of the pancreas-616 patients: results, outcomes, and prognostic indicators. J Gastrointest Surg. 2000;4(6):567–579. doi: 10.1016/s1091-255x(00)80105-5. [DOI] [PubMed] [Google Scholar]
- 3.Winter JM, Cameron JL, Campbell KA, et al. 1423 pancreaticoduodenectomies for pancreatic cancer: a single-institution experience. J Gastrointest Surg. 2006;10(9):1199–1210. doi: 10.1016/j.gassur.2006.08.018. discussion 1210–1211. [DOI] [PubMed] [Google Scholar]
- 4.Hermann PC, Huber SL, Herrler T, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 6.Jordan CT. Cancer stem cells: controversial or just misunderstood? Cell Stem Cell. 2009;4(3):203–205. doi: 10.1016/j.stem.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman DM, Karhadkar SS, Maitra A, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425(6960):846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 8.Heiser PW, Cano DA, Landsman L, et al. Stabilization of beta-catenin induces pancreas tumor formation. Gastroenterology. 2008;135(4):1288–1300. doi: 10.1053/j.gastro.2008.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peacock CD, Wang Q, Gesell GS, et al. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc Natl Acad Sci U S A. 2007;104(10):4048–4053. doi: 10.1073/pnas.0611682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pignatelli M, Ansari TW, Gunter P, et al. Loss of membranous E-cadherin expression in pancreatic cancer: correlation with lymph node metastasis, high grade, and advanced stage. J Pathol. 1994;174(4):243–248. doi: 10.1002/path.1711740403. [DOI] [PubMed] [Google Scholar]
- 11.Zeng G, Germinaro M, Micsenyi A, et al. Aberrant Wnt/beta-catenin signaling in pancreatic adenocarcinoma. Neoplasia. 2006;8(4):279–289. doi: 10.1593/neo.05607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134(6):921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dylla SJ, Beviglia L, Park IK, et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS One. 2008;3(6):e2428. doi: 10.1371/journal.pone.0002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeda S, Shinchi H, Kurahara H, et al. Clinical significance of midkine expression in pancreatic head carcinoma. Br J Cancer. 2007;97(3):405–411. doi: 10.1038/sj.bjc.6603879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh B, Murphy RF, Ding XZ, Roginsky AB, Bell RH, Jr, Adrian TE. On the role of transforming growth factor-beta in the growth inhibitory effects of retinoic acid in human pancreatic cancer cells. Mol Cancer. 2007;6:82. doi: 10.1186/1476-4598-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell LL, Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle. 2007;6(19):2332–2338. doi: 10.4161/cc.6.19.4914. [DOI] [PubMed] [Google Scholar]
- 17.Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 18.Hill RP. Identifying cancer stem cells in solid tumors: case not proven. Cancer Res. 2006;66(4):1891–1895. doi: 10.1158/0008-5472.CAN-05-3450. discussion 1890. [DOI] [PubMed] [Google Scholar]
- 19.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea—a paradigm shift. Cancer Res. 2006;66(4):1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. discussion 1895–1896. [DOI] [PubMed] [Google Scholar]
- 20.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 21.Jimeno A, Feldmann G, Suarez-Gauthier A, et al. A direct pancreatic cancer xenograft model as a platform for cancer stem cell therapeutic development. Mol Cancer Ther. 2009;8(2):310–314. doi: 10.1158/1535-7163.MCT-08-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsui W, Wang Q, Barber JP, et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008;68(1):190–197. doi: 10.1158/0008-5472.CAN-07-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corti S, Locatelli F, Papadimitriou D, et al. Identification of a primitive brain-derived neural stem cell population based on aldehyde dehydrogenase activity. Stem Cells. 2006;24(4):975–985. doi: 10.1634/stemcells.2005-0217. [DOI] [PubMed] [Google Scholar]
- 24.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones RJ, Gocke CD, Kasamon YL, et al. Circulating clonotypic B cells in classical Hodgkin's lymphoma. Blood. 2009;113(23):5920–5926. doi: 10.1182/blood-2008-11-189688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma S, Chan KW, Lee TK-W, et al. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol Cancer Res. 2008;6(7):1146–1153. doi: 10.1158/1541-7786.MCR-08-0035. [DOI] [PubMed] [Google Scholar]
- 27.Storms RW, Trujillo AP, Springer JB, et al. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci U S A. 1999;96(16):9118–9123. doi: 10.1073/pnas.96.16.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manley S, Mucci NR, De Marzo AM, Rubin MA. Relational database structure to manage high-density tissue microarray data and images for pathology studies focusing on clinical outcome: the prostate specialized program of research excellence model. Am J Pathol. 2001;159(3):837–843. doi: 10.1016/S0002-9440(10)61759-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Embuscado EE, Laheru D, Ricci F, et al. Immortalizing the complexity of cancer metastasis: genetic features of lethal metastatic pancreatic cancer obtained from rapid autopsy. Cancer Biol Ther. 2005;4(5):548–554. doi: 10.4161/cbt.4.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubio-Viqueira B, Jimeno A, Cusatis G, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res. 2006;12(15):4652–4661. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 31.Yeo CJ, Cameron JL, Lillemoe KD, et al. Pancreaticoduodenectomy for cancer of the head of the pancreas. 201 patients. Ann Surg. 1995;221(6):721–731. doi: 10.1097/00000658-199506000-00011. discussion 731–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molotkov A, Molotkova N, Duester G. Retinoic acid generated by Raldh2 in mesoderm is required for mouse dorsal endodermal pancreas development. Dev Dyn. 2005;232(4):950–957. doi: 10.1002/dvdy.20256. [DOI] [PubMed] [Google Scholar]
- 33.Hruban RH, Maitra A, Goggins M. Update on pancreatic intraepithelial neoplasia. Int J Clin Exp Pathol. 2008;1(4):306–316. [PMC free article] [PubMed] [Google Scholar]
- 34.Natalwala A, Spychal R, Tselepis C. Epithelial-mesenchymal transition mediated tumourigenesis in the gastrointestinal tract. World J Gastroenterol. 2008;14(24):3792–3797. doi: 10.3748/wjg.14.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 36.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183(4):1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanzkron SM, Collector MI, Sharkis SJ. Homing of long-term and short-term engrafting cells in vivo. Ann N Y Acad Sci. 1999;872:48–54. doi: 10.1111/j.1749-6632.1999.tb08452.x. discussion 54–56. [DOI] [PubMed] [Google Scholar]
- 38.Liu R, Wang X, Chen GY, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356(3):217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 39.Zeppernick F, Ahmadi R, Campos B, et al. Stem cell marker CD133 affects clinical outcome in glioma patients. Clin Cancer Res. 2008;14(1):123–129. doi: 10.1158/1078-0432.CCR-07-0932. [DOI] [PubMed] [Google Scholar]
- 40.Nakajima S, Doi R, Toyoda E, et al. N-cadherin expression and epithelial-mesenchymal transition in pancreatic carcinoma. Clin Cancer Res. 2004;10(12, pt 1):4125–4133. doi: 10.1158/1078-0432.CCR-0578-03. [DOI] [PubMed] [Google Scholar]
- 41.Mani SA, Guo W, Liao M-J, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feldmann G, Dhara S, Fendrich V, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67(5):2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hotfilder M, Rottgers S, Rosemann A, Jurgens H, Harbott J, Vormoor J. Immature CD34+CD19- progenitor/stem cells in TEL/AML1-positive acute lymphoblastic leukemia are genetically and functionally normal. Blood. 2002;100(2):640–646. doi: 10.1182/blood.v100.2.640. [DOI] [PubMed] [Google Scholar]
- 45.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 46.Wang L, O'Leary H, Fortney J, Gibson LF. Ph+/VE-cadherin+ identifies a stem cell like population of acute lymphoblastic leukemia sustained by bone marrow niche cells. Blood. 2007;110(9):3334–3344. doi: 10.1182/blood-2007-01-068122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241(4861):58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.