Summary
Background:
Factor VIIa (FVIIa) binding to tissue factor (TF) induces cell signaling via the protease activity of FVIIa and protease-activated receptor 2 (PAR2).
Objective:
We examined how the gene-expression profile induced by FVIIa corresponds to the profiles induced by protease-activated receptor 1 (PAR1) or PAR2 agonists using MDA-MB-231 breast carcinoma cells that constitutively express TF, PAR1 and PAR2.
Results and conclusions:
Out of 8500 genes, FVIIa stimulation induced differential regulation of 39 genes most of which were not previously recognized as FVIIa regulated. All genes regulated by FVIIa were similarly regulated by a PAR2 agonist peptide confirming FVIIa signaling via PAR2. An appreciable fraction of the PAR2-regulated genes was also regulated by a PAR1 agonist peptide suggesting extensive redundancy between FVIIa/PAR2 signaling and thrombin/PAR1 signaling. The FVIIa regulated genes encode cytokines, chemokines and growth factors, and the gene repertoire induced by FVIIa in MDA-MB-231 cells is consistent with a role for TF–FVIIa signaling in regulation of a wound healing type of response. Interestingly, a number of genes regulated exclusively by FVIIa/PAR2-mediated cell signaling in MDA-MB-231 cells were regulated by thrombin and a PAR1 agonist, but not by FVIIa, in the TF-expressing glioblastoma U373 cell line.
Keywords: factor VIIa, gene transcription, protease-activated receptor 1, protease-activated receptor 2, signaling
Introduction
Tissue factor (TF) is an integral membrane glycoprotein that acts as the cellular receptor for clotting factor VII (FVII). TF is present in the subendothelial layers of the vessel wall where it is constitutively expressed by many cell types, including fibroblasts and pericytes (see [1] for review). TF is expressed on several cell types in human atherosclerotic plaques and expression is also induced in monocytes under certain inflammatory conditions [2]. TF is thought to contribute to the pathogenesis of a variety of diseases by its participation in both coagulopathic [3,4] and non-coagulopathic processes [5,6]. TF is also expressed on tumor cells in many solid tumors [7]. Accumulating data indicate that the underlying TF-FVIIa-induced effect on cell physiology involves a variety of intracellular signaling events, including phosphorylation of mitogen-activated kinases (MAPK) [8] leading to alterations of gene expression profiles (see [6] for review). TF–FVIIa-induced signaling is known to require the FVIIa catalytic activity, and several lines of evidence suggest that TF–FVIIa transmits cell signaling via activation of protease-activated receptors (PARs), primarily PAR2 [9,10]. At present, conclusions drawn about the mechanism of FVIIa-mediated intracellular signaling and gene transcription are based mainly on measurements of single parameters such as Ca2+ mobilization, MAPK phosphorylation or expression levels of a single or a few selected genes leaving an incomplete picture of intracellular events. Gene-expression profiling permits simultaneous measurement of an abundance of transcripts and makes it possible to study more systematically the down-stream results of complex regulatory processes within a cell. To delineate the involvement of specific PARs in FVIIa-induced cell signaling, we compared the FVIIa-induced transcriptional repertoire with transcription induced by selective PAR agonist peptides. As a cell model system we utilized the breast carcinoma cell line, MDA-MB-231, as it constitutively expresses TF, protease-activated receptor 1 (PAR1) and PAR2 and is highly responsive to PAR1 and PAR2 agonists.
Materials and methods
Reagents
FVIIa [11] and active site-inactivated FVIIa [12] (FFR-FVIIa) were prepared as previously described. FX, FXa, hirudin, and thrombin were from Enzyme Research Laboratories (South Bend, IN, USA). Culture media, fetal calf serum (FCS) were from Gibco (Invitrogen, Carlsbad, CA, USA). PAR1 agonist (TFLLRNPNDK-NH2) and PAR2 agonist (SLIGKV-NH2) were synthesized on an Applied Biosystems 431A peptide synthesizer (Applied Biosystems, Perkin Elmer, Foster City, CA, USA). Tick anticoagulant protein (TAP) was a gift from Dr George Vlasuk (Corvas, La Jolla, CA, USA).
Cell cultures
Cells were grown at 37 C in a humidified environment with 5% CO2. MDA-MB-231 cells (ATCC: HTB-26) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS, 100 IU mL−1 penicillin and 100 μg mL−1 streptomycin. U373 cells (ATCC: HTB-17) were grown in Eagle's medium (MEM) with Earl's salts substituted with 1% non-essential acids and 1% sodium pyruvate (Gibco, Invitrogen). Cells were grown to near confluence (~80%), washed twice with serum-free medium and maintained in serum-free medium for 2 h before treatment.
cDNA microarray analysis
MDA-MB-231 cells were exposed to serum-free medium or serum-free medium supplemented with FFR-FVIIa (100 nm), FVIIa (100 nm), TFLLRNPNDK (10 μm) or SLIGKV (50 μm) for 1 or 6 h. Total RNA was isolated using TriZol (Invitrogen), followed by purification using RNeasy (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's instruction. Equal amounts of RNA from two independent experiments were pooled and Cy3- and Cy5-labeled cDNAs were prepared from 15 μg total RNA using a CyScribe cDNA postlabeling kit (Amersham Bioscience, Bucks, UK) according to the manufacturer's instructions. Gaps II slides (Corning B.V, Schiphol-Rijk, The Netherlands) were spotted with polymerase chain reaction (PCR) products of approximately 8500 human genes (Unigene Human Cloneset Version 2.0) (Incyte Genomics, CA, USA) and prehybridized for 1.5 h in 2 × SSPE buffer (300 mm NaCl, 20 mm NaH2PO4, 2 mm EDTA, pH 7.4) with 0.1% sodium dodecylsulfate (SDS). Next, 1 μg polyA75 and 15 pmol Cy3- and Cy5-labeled cDNAs were mixed in Version 2 microarray hybridization buffer (Amersham Pharmacia Biotech, Piscataway, NJ, USA) and formamide (1:1, vol/vol) in a total volume of 40 μL, denatured at 95°C for 3 min and hybridized in a ISO20 microarray hybridization incubator (Grant Instruments, Cambridge, UK) at 42°C for 16 h. Slides were subsequently washed at 55°C in 1 × sodium chloride/sodium citrate buffer (SSC) (150 mm NaCl, 15 mm sodium citrate, pH 7.0) containing 0.2% SDS for 10 min, then twice in 0.1 × SSC, 0.2% SDS for 10 min, and in 0.1 × SSC for 1 min. Finally, slides were dried and scanned in an Axon4000B (Axon Instruments, Union City, CA, USA).
Data analysis of cDNA microarrays
After DNA microarray hybridization, spots were automatically identified and signals as well as background signals surrounding the spots were measured by the use of ARRAYVISION Software (Amersham Bioscience). To correct for spots with negative net intensities a smoothing function was used as described previously [13]. The threshold values (δ) were chosen in a slide/dye-specific fashion so that the proportion of spots with net intensities between 0 and δ was 10% of the proportion with negative net intensities. For each drug/time-point combination, quantile normalization [14] was performed on the log-intensities from the Cy3 and Cy5 channels. To determine if differences between compared treatments were statistically significant, estimates of the log-expression ratio for each probe, together with the associated SE and t-tests for the hypothesis that the log expression ratio is zero, were calculated.
Real-time quantitative PCR analysis
Total RNA samples were reverse transcribed to cDNA using Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Two (U373 cells) and three (MDA-MB-231 cells) individual reverse transcription reactions were made for each sample. Expression of selected genes was determined using an ABI PRISM 7000 sequence detection system (Applied Biosystems) as previously described [15,16]. Briefly, a five-point serial standard curve (each point performed in triplicate) was made using cDNA from MDA-MB-231 cells treated with FVIIa for 1 or 6 h, with the final assay concentration ranging from 76 pg to 120 ng total RNA (in 25 μL reaction volume). This curve was used to calculate the amount of target gene mRNA in all samples based on qPCR performed with 48 ng total RNA (in 25 μL) reaction volume. Universal PCR master mix and the primer/probe assays listed below were used. mRNA levels of all genes were normalized to the expression level of GAPDH using the primer/probe combinations: 5′-CTGCCACCCAGAAGACTGTG-3′,5′-AGGCAGGGATGATGTTCTGG-3′, and 5′-FAM-CCCTCCGGGAAACTGTGGCG-3′. A t-test was applied to determine if differences between treatments were statistically significant.
| Gene name | Assay # Applied Biosystems |
|---|---|
| IL-8 | Hs00174103_m1 |
| CXCL1 (Gro-α) | Hs00605382_gH |
| CSF1 (GM-CSF) | Hs00174164_m1 |
| CSF2 (M-CSF) | Hs00171266_m1 |
| VEGFc | Hs00153458_m1 |
| CNN1 (Cyr61) | Hs00155479_m1 |
| CNN2 (CTGF) | Hs00170014_m1 |
| Integrin- β1 | Hs00559596_m1 |
| uPA | Hs00170182_m1 |
| PAI-1 | Hs00167155_m1 |
| NFKBIE | Hs00234431_m1 |
| PTX3 | Hs00173615_m1 |
| Glutamine-fructose-6-P-transaminase | Hs00192725_m1 |
Flow cytometry
U373 cells were flushed once with serum-free medium and once with Versene buffer (Invitrogen, Gibco). The cells supplemented with Versene buffer were detached from the culture flask by scraping. FACS buffer (phosphate-buffered saline containing 1% bovine serum albumin and 0.05% sodium azide) was added, and the cells were split in samples and control of 1× 106 cells and stored on ice. Cells were resuspended in FACS buffer at 4°C. Samples were incubated with 10 μg mL−1 monoclonal antibodies against TF (1F44A1, Novo Nordisk, Maalov, Denmark), PAR1 (ATAP-2) or PAR2 (SAM11) (Santa-Cruz Biotechnology, Santa Cruz, CA, USA) for 60 min at 4°C. Samples and controls were then washed and incubated with 50 μg mL−1 PE-conjugated goat antimouse IgG (Dako, Copenhagen, Denmark) in the dark for 30 min at 4°C, after which they were washed twice with FACS buffer and fixed for 2 h in the dark at 4°C in FACS buffer containing 0.5% paraformaldehyde and analyzed for fluorescence using a BD FACS CantoTM (Becton Dickinson, Brondby, Denmark).
Results
Genes identified as differentially regulated by FVIIa in MDA-MB-231 cells are similarly regulated by PAR2 agonist peptide
Previous studies have shown that MDA-MB-231 cells express high levels of functionally active TF, PAR1 and PAR2. To further elucidate the effect of TF–FVIIa-induced cell signaling, we characterized TF–FVIIa-induced gene expression profiles in MDA-MB-231 cells using cDNA micro-arrays consisting of approximately 8500 human genes and expressed sequence tags. Quiescent monolayers of MDA-MB-231 cells were exposed either to serum-free medium (control) or serum-free medium supplemented with 100 nm FVIIa for 1 and 6 h, and total RNA was harvested and used for micro-array analysis. Genes significantly (P < 0.01) differentially regulated more than 2-fold were considered regulated by FVIIa. Using these criteria, we identified 15 and 27 genes that were regulated by FVIIa after 1 and 6 h, respectively. Of these, three genes were regulated at both time points, resulting in a total of 39 FVIIa-regulated genes.
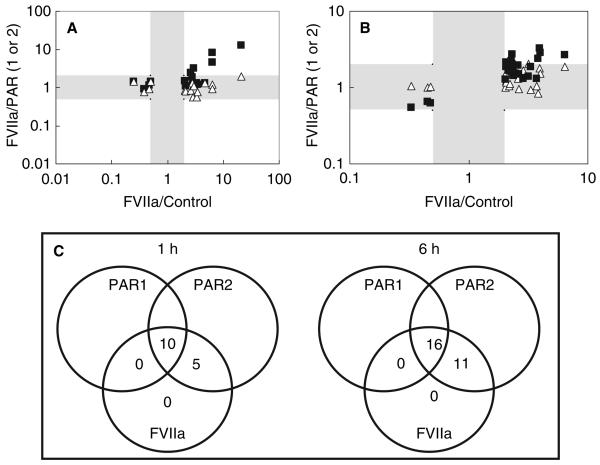
To analyze the mechanisms of FVIIa-induced intracellular signaling in MDA-MB-231 cells, we determined whether genes regulated by FVIIa were also regulated by selective stimulation of PAR1 or PAR2. For this, MDA-MB-231 cells were treated in parallel for 1 and 6 h with a selective agonist peptide for PAR1 (TFLLRNPNDK) or a selective agonist peptide for PAR2 (SLIGKV). The fold regulation of genes stimulated by FVIIa vs. control was compared with the fold regulations of FVIIa vs. PAR1 agonist or PAR2 agonist. Therefore, genes regulated by all three stimuli (FVIIa, PAR1- and PAR2 agonist) result in FVIIa/PAR1 and FVIIa/PAR2 ratios close to 1, whereas genes regulated by FVIIa, and not by either PAR1 or PAR2, result in FVIIa/PAR ratios above 2-fold. The results revealed that all 15 genes, which were regulated by FVIIa after 1 h, were also regulated by the PAR2 agonist. However, only 10 of the 15 genes were regulated by PAR1 agonist at this time point (Fig. 1A, Tables 1 and 2).
Fig. 1.
cDNA microarray analysis comparing transcriptional effects induced by factor VIIa (FVIIa) and selective protease-activated receptor 1 (PAR1) and PAR2 agonist peptides in MDA-MB-231 cells. Quiescent MDA-MB-231 cells were treated for 1 (A) or 6 h (B) with serum-free control media, or control media supplemented with 100 μm FVIIa, 10 μm TFLLRNPNDK (PAR1 agonist peptide) or 50 μm SLIGKV (PAR2 agonist peptide). Genes significantly (P < 0.01) differentially regulated more than 2-fold by FVIIa are included. The differential gene regulation ratio of FVIIa/PAR1 (squares) or the ratio of FVIIa/PAR2 (triangles) is plotted against the FVIIa/media control ratio. FVIIa/PAR1 and FVIIa/PAR2 ratios below 2-fold are to be found within the grey-scaled areas and represent genes regulated similarly by FVIIa and the PAR agonist peptides. Data points present in white-scaled areas with FVIIa/PAR1 or FVIIa/PAR2 ratios higher than 2-fold represent genes regulated by FVIIa but not the PAR1 agonist peptide (squares), or by FVIIa but not the PAR2 agonist peptide (triangles). Panel C is a schematic representation of cDNA microarray results comparing transcriptional effects induced by FVIIa and PAR1 and PAR2 agonist peptides.
Table 1.
Genes regulated by factor VIIa (FVIIa) (P < 0.01 and more than 2-fold) as well as both protease-activated receptor 1 (PAR1) and PAR2 agonist peptides in MDA-MB231 cells exposed to the agonist for 1 h
| Acc# | Annotation | FVIIa/ media 1 h |
FVIIa/ PAR1 1 h |
FVIIa/ PAR2 1 h |
Category | FVIIa regulation verified by qPCR |
|---|---|---|---|---|---|---|
| Y12084 | Cysteine rich protein 61 (Cyr61) (CNN1) | 3.48 | 1.22 | 0.76 | Cell adhesion, matrix modeling | Yes |
| X78947 | Connective tissue growth factor (CTGF) (CNN2) | 3.27 | 1.32 | 0.56 | Yes | |
| NM_001008701 | Latrophilin 1 (LPHN1) | 0.50 | 0.59 | 1.40 | Signaling | |
| AF035950 | BRCA1-associated protein 2 (BRAP2) | 2.88 | 1.25 | 0.55 | Transcriptional regulation | |
| AI971939 | Transcription factor 21 (Tcf21) | 0.25 | 1.04 | 1.42 | ||
| X63053 | Long pentraxin (PTX3) | 2.80 | 1.86 | 1.31 | Inflammation | Yes |
| BC017219 | Zinc finger protein 587 | 4.72 | 1.28 | 1.29 | Other | |
| AI741326 | Soluble liver antigen/liver pancreas antigen (SLA/LP) | 2.04 | 1.47 | 0.85 | ||
| NM_016396 | Carboxy-terminal domain. RNA polymerase II. polypeptide A small phosphatase like 2 (CTDSPL2) |
0.47 | 1.13 | 0.92 | ||
| AI400892 | Unknown. cDNA FLJ34852 fis. clone NT2NE2012113. | 0.38 | 0.90 | 0.77 | Unknown |
Table 2.
Genes regulated by factor VIIa (FVIIa) (P < 0.01 and more than 2-fold) as well as protease-activated receptor 2 (PAR2) agonist peptide in MDA-MB231 cells exposed to the agonist for 1 h
| Acc# | Annotation | FVIIa/ media 1 h |
FVIIa/ PAR1 1 h |
FVIIa/ PAR2 1 h |
Category | FVIIa regulation verified by qPCR |
|---|---|---|---|---|---|---|
| NM_001511 | Chemokine (C-X-C motif) ligand 1 (CxcL1). melanoma growth stimulating activity. alpha (Groα) | 20.65 | 12.73 | 1.95 | Chemokines cytokines, growth factors | Yes |
| M26383 | Interleukin 8 (IL-8) | 6.41 | 8.14 | 1.22 | Yes | |
| M11220 | Granulocyte-macrophage colony stimulating factor (GM-CSF) (csf2) | 6.39 | 4.51 | 0.91 | Yes | |
| NM_002211 | Integrin. beta 1 | 2.62 | 2.52 | 0.82 | Cell adhesion, matrix modeling | No |
| BC037420 | Baculoviral IAP (Inhibitor of apoptosis domain) repeat-containing 3 (Birc3) | 3.03 | 3.26 | 1.07 | Apoptosis |
A similar pattern was seen in cells treated for 6 h (Fig. 1B, Tables 3 and 4). Many of the FVIIa-regulated genes (16 out of 27) were also regulated by both PAR1 and PAR2 agonists (Table 3), and eight other genes were regulated by the PAR2 and FVIIa, but not by the PAR1 selective agonist (Table 4). Among the remaining three genes on the array, one appeared to be regulated by FVIIa through PAR1, whereas two appeared to be regulated by FVIIa independent of PAR1 and PAR2. However, the differential regulation was close to the cut-off value of 2-fold, and qPCR analysis (see the following section) of RNA samples from a separate experiment (data not shown) was used to confirm that the genes were weakly (approximately 2-fold) regulated by FVIIa and the PAR2 agonist, but not by the PAR1 agonist.
Table 3.
Genes regulated by factor VIIa (FVIIa) (P < 0.01 and more than 2-fold) as well as both protease-activated receptor 1 (PAR1) and PAR2 agonist peptides in MDA-MB231 cells exposed to the agonist for 6 h
| Acc# | Annotation | FVIIa/ media 6 h |
FVIIa/ PAR1 6 h |
FVIIa/ PAR2 6 h |
Category | FVIIa regulation verified by qPCR |
|---|---|---|---|---|---|---|
| M11220 | Granulocyte-macrophage colony stimulating factor (GM-CSF) (csf2) | 2.88 | 1.32 | 1.67 | Chemokines, cytokines and growth factors | Yes |
| D11143 | Urokinase-type plasminogen activator (uPa) | 2.64 | 1.50 | 0.96 | Proteases | Yes |
| M14083 | Serine (or cysteine) proteinase inhibitor. clade E. member 1 (Serpine1) (PAI-1) | 3.29 | 1.87 | 0.94 | protease inhibitors | Yes |
| L41816 | Cam kinase I | 2.58 | 1.96 | 1.30 | Signaling | |
| T04872 | Cyclin-dependent kinase 5. regulatory subunit 1 (p35). | 3.69 | 1.32 | 1.04 | Cell cycle control | |
| L26165 | Cyclin-dependent kinase inhibitor 1A (P21) | 2.05 | 1.88 | 1.22 | ||
| AI089791 | Cyclin-dependent kinase inhibitor 1c (p57. Kip2) | 0.33 | 0.55 | 1.05 | ||
| M69199 | Putative lymphocyte G0/G1 switch gene | 2.48 | 1.54 | 1.59 | ||
| AB011172 | Histone deacetylase 5 | 2.02 | 1.27 | 0.99 | Transcriptional regulation | |
| M55643 | Nuclear factor of kappa light chain gene enhancer in B-cells 1. p105 (Nfkb1) | 2.16 | 1.67 | 1.87 | ||
| NM_002698 | POU domain. class 2. transcription factor 2 (Pou2f2) | 2.06 | 1.96 | 1.56 | ||
| S78825 | Inhibitor of DNA binding 1 (Id1) | 0.45 | 0.66 | 0.99 | ||
| AI758821 | Programmed cell death 6 (Pdcd6) | 2.22 | 1.62 | 1.11 | Apoptosis | |
| AA829286 | Serum amyloid A1 (SAA1) | 2.35 | 1.81 | 1.58 | Inflammation | |
| NM_001093 | Acetyl-Coenzyme A carboxylase beta (ACACB) | 2.37 | 1.42 | 1.50 | Other | |
| NM_004772 | Unknown. chromosome 5 open reading frame 13 | 0.48 | 0.63 | 1.03 | Unknown |
Table 4.
Genes regulated by factor VIIa (FVIIa) (P < 0.01 and more than 2-fold) as well as protease-activated receptor 2 (PAR2) agonist peptide in MDA-MB231 cells exposed to the agonist for 6 h
| Acc# | Annotation | FVIIa/ media 6hrs |
FVIIa/ PAR1 6hrs |
FVIIa/ PAR2 6hrs |
Category | FVIIa regulation verified by qPCR |
|---|---|---|---|---|---|---|
| U43142 | Vascular endothelial growth factor C (Vegfc) | 2.07 | 2.12 | 1.16 | Chemokines, cytokines and growth factors | Yes |
| M11296 | Colony stimulating factor 1 (macrophage) (Csf1) | 2.27 | 2.22 | 2.10 | Yes | |
| AI343341 | Mitogen activated protein kinase kinase 3 (MAPKK3) | 2.19 | 2.01 | 1.04 | Signaling | |
| BC063609 | Nuclear factor of kappa light chain gene enhancer in B-cells inhibitor. epsilon (NFKBIE) | 2.27 | 2.22 | 2.10 | Transcriptional regulation | Yes |
| Z35085 | RB1-inducible coiled-coil 1 (Rb1 cc1) | 3.94 | 3.24 | 1.78 | ||
| BC037420 | Baculoviral IAP (Inhibitor of apoptosis domain) repeat-containing 3 (Birc3) | 4.02 | 2.89 | 1.52 | Apoptosis | |
| U37546 | Baculoviral IAP (Inhibitor of apoptosis protein 1) repeat-containing 2 (Birc2) | 2.31 | 2.73 | 1.62 | ||
| X63053 | Long pentraxin (PTX3) | 6.40 | 2.71 | 1.86 | Inflammation | Yes |
| AI539509 | Gem (nuclear organelle) associated protein 7 | 3.81 | 2.38 | 0.83 | Others | |
| AJ011896 | TNFAIP3 interacting protein 1 (Tnip1) | 2.31 | 2.05 | 1.45 | ||
| AB016789 | Glutamine fructose-6-phosphate transaminase 2 (Gfpt2) | 3.18 | 1.41 | 2.05 | Yes |
To examine whether the proteolytic activity was mandatory for FVIIa signaling, we treated MDA-MB-231 cells with active site blocked FVIIa (FFR-FVIIa). FFR-FVIIa induced only a few statistically significant transcriptional changes, which barely exceeded the 2-fold threshold set for differential expression. Analysis of signal intensities of these spots revealed very low levels for both control and FFR-FVIIa treated cells. Additionally, real-time qPCR analysis failed to confirm differential regulation of these genes (data not shown). This is consistent with the general experience that genes expressed at low levels are at an increased risk of being falsely scored as positives. We conclude that we were unable to find genes that are regulated by FFR-FVIIa. The data are therefore not at variance with a transcriptional FVIIa response mediated exclusively via proteolytic activation of PAR2.
Confirmation of FVIIa-induced differential gene expressions by real time qPCR analysis
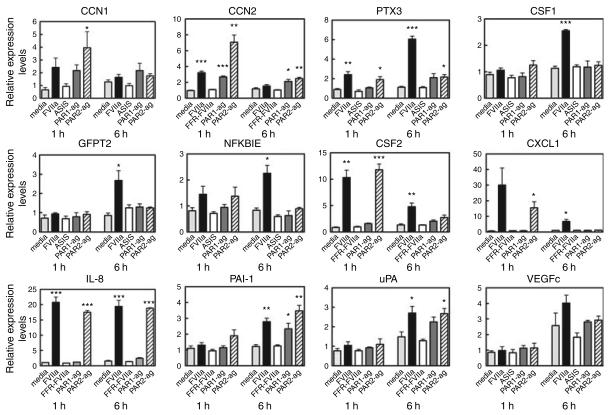
To validate the data obtained by cDNA microarray analysis, we subjected the array RNA samples to real-time qPCR analysis. Thirteen genes were selected for verification: CXCL1 (Groα), IL-8, CSF1 (M-CSF), CSF2 (GM-CSF), VEGFc, CCN1 (Cyr61), CCN2 (CTGF), integrin β1, uPA, PAI-1, NFKBIE, PTX3, and GFPT2. Except for one gene, integrin β1 (not shown), we confirmed the differential regulation by FVIIa of all genes selected (Fig. 2).
Fig. 2.
Validation of factor VIIa (FVIIa)-induced differential gene expression in MDA-MB-231 cells by real time qPCR analysis. Total RNA samples used for cDNA micro-array analysis were subjected to real-time qPCR analysis. Regulation of the mRNA level was measured relative to media control (mean ± SEM; n = 3). Statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001) was determined by t-test analysis.
To further confirm the results obtained by cDNA microarray analysis, six genes, IL8, CSF1, CCN1, CCN2, NFKBIE and GFPT2 were re-examined by qPCR analysis using RNA samples from an independent experiment. All six were differentially regulated by FVIIa (data not shown). According to the array analysis, two of the FVIIa-regulated genes (CSF1 and NFKBIE) appeared not to be regulated by either the PAR1 or the PAR2 selective agonist. However, when analyzed by real-time qPCR using RNA samples from an independent experiment, all genes were found to be regulated by the PAR2 agonist. Similarly, GFPT2 was found by real-time qPCR to be regulated by FVIIa and the PAR2 agonist. Moreover, IL-8 and IL-6 protein expression has been confirmed by enzyme-linked immunosorbent assay (ELISA) (data not shown). Overall, the cDNA microarray, real-time qPCR, and ELISA data are consistent with a model in which FVIIa signal through PAR2.
Concentration dependency of FVIIa-induced gene regulation
The cellular response to FVIIa stimulation measured by inositol 3 phosphate hydrolysis [6] or by IL-8 mRNA and protein levels [10] is reported to be saturable with an EC50 in the low nm range (< 10 nm). These observations were confirmed and expanded with dose–response data on the FVIIa-regulated genes, IL8, CXCL1 and CSF2 (unpubl. results). Saturation was obtained with an EC50 between 5 and 8 nm for all three genes suggesting that the gene response is more than 50% saturated at 10 nm FVIIa. To demonstrate physiological relevance and to avoid potential artefacts associated with supra-physiological concentrations, we designed a small series of experiments in which the agonists FVIIa and FXa were used at concentrations close to the plasma level of their respective zymogens, FVII and FX (Fig. 3). In addition, we verified by micro-array analysis (results not shown) that the general gene regulation pattern observed at 100 nm was also reproduced at 10 nm FVIIa.
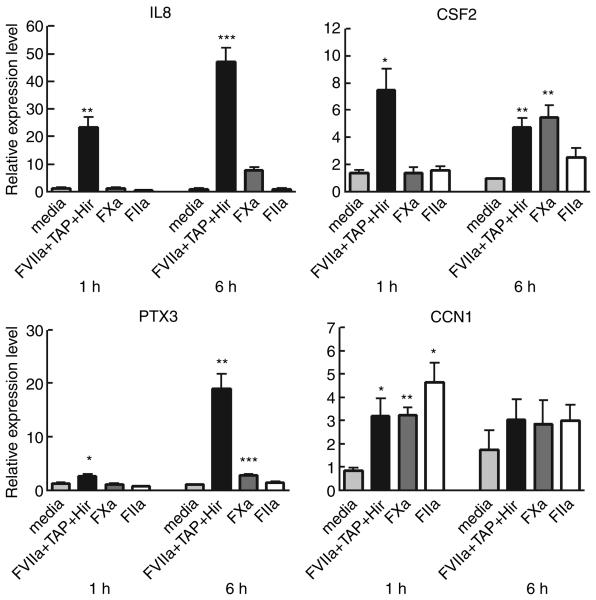
Fig. 3.
Differential gene regulation by factor VIIa (FVIIa), FXa and thrombin in MDA-MB-231 cells. Total RNA from MDA-MB-231 cells treated with control media, or control media supplemented with FVIIa (10 nm) + Tick anticoagulant protein (100 nm) + hirudin (25 U mL−1), FXa (100 nm), or thrombin (FIIa) (10 nm) for 1 or 6 h was isolated and subjected to real-time qPCR analysis. Regulation of the mRNA level was measured relative to media control (mean ± SEM; n = 3). Statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001) was determined by t-test analysis.
The transcriptional FVIIa response is not elicited by FXa or thrombin down-stream of FVIIa in the coagulation cascade
To exclude putative FVIIa-mediated down-stream activation of thrombin and FXa, we tested the effect of the specific inhibitors, TAP and hirudin on FVIIa-induced gene expression. We also investigated the ability of FXa and thrombin to induce transcriptional regulation of selected genes. Three FVIIa/PAR2-induced genes (IL8, CSF2, and PTX3) and two PAR1 and PAR2-induced genes (CCN1 and CCN2) were selected. The inhibitors of thrombin and FXa failed to abolish FVIIa-induced regulation of all genes tested (Fig. 3). Neither thrombin nor FXa induced the prominent PAR2-specific gene response seen with FVIIa. Thrombin, which is known to activate PAR1 and not PAR2, was devoid of effect on PAR2-regulated genes. Stimulation with 100 nm FXa did, however, induce a slight up-regulation of these genes at 6 h. This is in line with previous observations indicating PAR2 activation by FXa [17,18]. Thrombin and FXa stimulated the transcription of CCN1 and (not shown) CCN2 as expected from earlier studies using other cell model systems [19]. This implies that thrombin-regulated genes were observed only among FVIIa- and PAR2-induced genes which were also activated by the PAR1 agonists, and not among genes activated exclusively by FVIIa and the PAR2 peptide.
Genes regulated by PAR2 in MDA-MB-231 cells are regulated by PAR1 in U373 cells
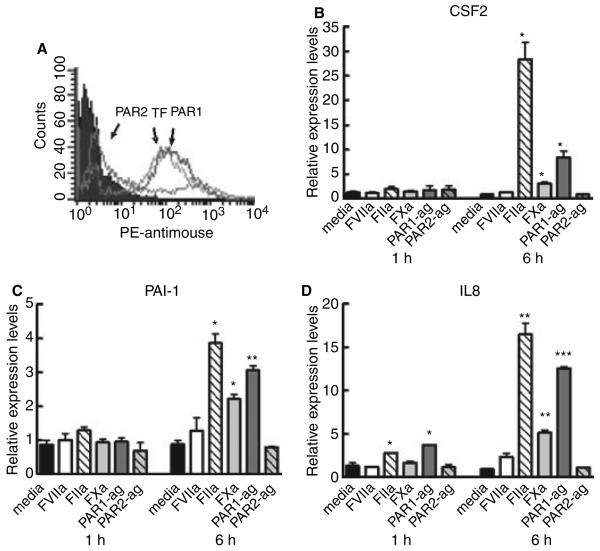
For screening of cell lines with high functional TF levels, we measured the IL-8 secretion in response to FVIIa and thrombin as an indicator of signal transduction. Interestingly, the human brain glioblastoma cell line, U373, was found to respond with enhanced IL-8 protein expression when treated with thrombin, but not with FVIIa. As compared with MDA-MB-231 cells, this cell line expressed relatively low antigen levels of PAR2 and TF (Fig. 4A). This was also reflected in a tenfold lower level of functional TF for U373 cells (0.14 nm FXa/min/106cells) as compared with MDA-MB-231 cells (1.9 nm FXa/min/106cells), possibly explaining the lack of FVIIa-induced effect. To analyze whether genes other than IL-8 were differentially expressed by thrombin in U373 cells, we performed real-time qPCR analysis of selected genes that were differentially expressed in MDA-MB-231 cells either by both PAR1 and PAR2 agonists or by the PAR2 agonist alone. Two genes, out of 13 genes analyzed, were below the detection level, whereas the remaining 11 genes were induced by thrombin and PAR1 agonist, including CSF2 and IL-8 that were shown to be specific to PAR2 activation in MDA-MB-231 cells (Fig. 4B–D). PAR2 agonist and FVIIa failed to up-regulate expression of any of the selected genes in U373 cells. These data indicate that the gene regulations elicited by FVIIa through PAR2 in MDA-MB-231 cells may occur in other cell lines through thrombin activation of PAR1.
Fig. 4.
Differential gene regulation in U373 cells. (A) Flow cytometry of U373 cells probed with antiprotease-activated receptor 1 (PAR1), anti-PAR2 or antitissue factor (TF). (B–D) Total RNA from U373 cells treated with: media; 50 nm FVIIa; 50 nm FXa; 50 nm thrombin (FIIa); 10 μm PAR1 agonist peptide or 50 μm PAR2 agonist peptide for 1 or 6 h was isolated and subjected to real-time qPCR analysis. Regulation of the mRNA level was measured relative to media control (mean ± SEM; n = 2). Statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001) was determined by t-test analysis.
Discussion
The present array analysis of MDA-MB-231 cells stimulated with FVIIa or with a selective PAR agonist (Fig. 1) showed that a total of 39 out of 8,500 genes were differentially regulated by FVIIa. Except for CCN1, CCN2 and IL-8, all genes, identified as FVIIa-regulated in the present study, are novel. All transcripts found to be FVIIa-regulated were similarly regulated by the PAR2 agonist. Furthermore, no FVIIa-regulated genes could be identified as specifically PAR1 regulated, as all PAR1-regulated genes were redundantly regulated also by PAR2. The data indicate that FVIIa regulates transcription exclusively via PAR2 in MDA-MB-231 cells, thus supporting recent reports suggesting that PAR2 activation is critical for FVIIa-induced cellular responses in those cells [10,20]. The present results define two groups of FVIIa-regulated genes: one group that appeared truly FVIIa specific and was activated solely through PAR2, and another group shared with thrombin that could be activated through PAR1 as well as PAR2. A caveat revealed by the present study is that FVIIa and thrombin, although acting on different PARs, regulate a set of overlapping genes, and are equally effective in eliciting the corresponding transcriptional response. Interestingly, we note that FVIIa in contrast to the PAR2 agonist, failed to induce intracellular Ca2+ release in MDA-MB-231 cells [10] and in other cell types that express both TF and PAR2 (unpublished data of the authors). Apart from this remarkable difference in Ca2+ signaling, there was little if any difference between the two PAR2 agonists in the transcriptional response pattern. The observation that the stimulation of gene transcription induced by FVIIa was not induced by FFR-FVIIa suggests that all genes identified were regulated as a consequence of FVIIa proteolytic activity. Furthermore, in our array of 8500 genes, none were found to be significantly regulated by FFR-FVIIa.
The striking difference in PAR-induced gene expression profiling observed between MDA-MB-231 and U373 cells is indicative of regulation of signaling on more than one level. The difference may partly be explained by a difference in the relative expression levels of TF and PARs (Fig. 4). In addition, cell-specific differences in the intracellular organization of the signaling network may account for the finding that genes regulated via PAR1 in U373 cells are specifically coupled to PAR2 activation in MDA-MB-231 cells. Cross activation of one PAR upon cleavage of the other and heterogeneous PAR clustering represent other possible explanations for the shunting of PAR signaling from one PAR to the other as was proposed to account for the distinct cellular responses obtained upon PAR1 cleavage with either thrombin or activated protein C[21]. Further studies are needed to elucidate the complex signaling networks that link PAR cleavage to gene transcription.
In addition to information about the role of PAR1 and PAR2 in FVIIa-induced cell signaling, the present data provide valuable insights into how FVIIa affects various cellular processes. Although compelling in vivo evidence is lacking, TF–FVIIa-induced cell signaling is thought to be important for normal physiology, particularly in relation to the host defense against tissue injury, and also to be implicated in the pathogenesis of inflammatory diseases and malignancy [6]. FVIIa has been shown to induce migration [10,22] and inhibit apoptosis [23,24]. TF–FVIIa signaling may also contribute to angiogenesis through a complex interplay between TF, PAR2 and integrins [25-27].
Several of the secreted proteins including chemokines, cytokines, and growth factors that were found to be regulated by FVIIa/PAR2 have been implicated in various aspects of cell proliferation, cell migration, cell adhesion, cell survival/apoptosis, and angiogenesis. Thus, IL-8 and CXCL1 have been described to be implicated in angiogenesis, metastasis, and tumor development [28,29]. CSF2 modulates growth, differentiation, and survival of macrophage, granulocyte, erythrocyte and megakaryocyte cells from bone marrow progenitors, as well as the functional activities of mature effector cells such as neutrophils, macrophages and dendritic cells. CSF1 is the primary regulator of the survival, proliferation, differentiation and function of mononuclear phagocytes. CSF2 like CSF1 regulates monocyte differentiation. The resulting CSF2-derived and CSF1-derived macrophages have, however, distinctly different features [30]. Both play important roles in innate immunity, angiogenesis, cancer, and inflammation. They are detrimental for the ability of macrophages to move to specific sites, increase matrix remodeling and induce angiogenesis and thus essential for normal physiological processes such as wound healing and inflammation. CSF1 and CSF2 are supposed to be essential components of the micro-environment produced in some tumors characterized by a high density of so called tumor-associated macrophages [31].
A possible ‘inflammatoid’ response, as a result of FVIIa/PAR2 stimulation of MDA-MB-231 cells, was evident from the data showing that a number of genes encoding inflammatory mediators such as CXCL1, CXCL8 (IL8), CSF2 (GM-CSF) and PTX3 are up-regulated already at 1 h. Later at 6 h, CSF1 (M-CSF), VEGFc, and acute-phase genes for serum amyloid A1 and PTX3 are also found to be up-regulated. In this context, it is interesting that PTX3 is presumed to function as a regulator of the innate immune response by its binding to complement C1q [32]. Furthermore, PTX3 is reported to up-regulate TF expression on endothelial cells and on activated monocytes [33]. The recent demonstration of PTX3 as a specific bFGF ligand/antagonist [34] suggests that PTX3 may also work as a modulator of angiogenesis. The inflammatory mediators CXCL1, CXCL8, CSF2, CSF1 and VEGFc have all been ascribed a role in angiogenesis. CSF1 [35] and VEGFc [36,37] are direct angiogenic factors promoting endothelial proliferation, migration and differentiation, whereas the inflammatory chemokines, CXCL1 and IL8 show both direct and indirect effects by stimulating endothelial proliferation and migration and by working as chemo-attractants towards monocytes/macrophages [28,38,39].
Finally, FVIIa also induces gene products involved in remodeling of the matrix during angiogenesis and tissue repair. These include matricellular proteins, CCN1 and CCN2 that are up-regulated at 1 h. The later products (at 6 h) comprise the serine protease, uPA, and its natural inhibitor, PAI-1. TF–FVIIa-induced expression of CCN1 and CCN2 is noteworthy. CCN1 has been shown to activate genes that play multiple and coordinated roles in the wound healing process, including angiogenesis, inflammation, and ECM remodeling [40]. Similarly, CCN2 has also been shown to induce gene expression that is relevant to wound healing and angiogenesis [41].
In conclusion, gene expression profile analysis of MDA-MB-231 cells exposed to FVIIa and to PAR1 and PAR2 selective peptide agonists show that FVIIa activates a specific genetic program and also suggest that this is mediated through activation of PAR2. The data show that FVIIa induces a set of genes whose products play a role in various steps of angiogenesis and inflammation. These data thus support the hypothesis that TF–FVIIa cell signaling may play an important role in inflammatory and angiogenic processes, and thus be implicated in the normal wound healing response to injury as well as in the pathogenesis of inflammatory diseases and cancer.
Acknowledgements
Tine Ø. Pedersen, Elke Gottfriedsen, Lone Langhoff, and Berit Lassen are thanked for excellent technical assistance. Kjeld Madsen, Novo Nordisk A/S is thanked for synthesis of PAR agonist peptides. Supported partly by grants from National Institutes of Health HL65550 and HL58869 (to L.V.M.R.).
Footnotes
Disclosure of Conflict of Interest
T. Albrektsen, B. B. Sørensen, G. M. Hjortø, J. Fleckner and L. C. Petersen are employees at Novo Nordisk A/S, whose product FVIIa is studied in this work. L. V. M. Rao states that he has no conflict of interest.
References
- 1.Osterud B, Bjorklid E. Sources of tissue factor. Semin Thromb Hemost. 2006;32:11–23. doi: 10.1055/s-2006-933336. [DOI] [PubMed] [Google Scholar]
- 2.Wilcox JN, Smith KM, Schwartz SM, Gordon D. Locallization of tissue factor in the normal vessel wall and in the atherosclerotic plaque. Proc Natl Acad Sci USA. 1989;86:2839–43. doi: 10.1073/pnas.86.8.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackman N. Role of tissue factor in hemostasis, thrombosis, and vascular development. Arterioscler Thromb Vasc Biol. 2004;24:1015–22. doi: 10.1161/01.ATV.0000130465.23430.74. [DOI] [PubMed] [Google Scholar]
- 4.Taubman MB, Fallon JT, Schecter AD, Giesen P, Mendlowitz M, Fyfe BS, Marmur JD, Nemerson Y. Tissue factor in the pathogenesis of atherosclerosis. Thromb Haemost. 1997;78:200–4. [PubMed] [Google Scholar]
- 5.Belting M, Ahamed J, Ruf W. Signaling of the tissue factor coagulation pathway in angiogenesis and cancer. Arterioscler Thromb Vasc Biol. 2005;25:1545–50. doi: 10.1161/01.ATV.0000171155.05809.bf. [DOI] [PubMed] [Google Scholar]
- 6.Rao LVM, Pendurthi UR. Tissue factor–factor VIIa signaling. Arterioscler Thromb Vasc Biol. 2005;25:47–56. doi: 10.1161/01.ATV.0000151624.45775.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callander NS, Varki N, Rao LVM. Immunohistochemical identification of tissue factor in solid tumors. Cancer. 1992;70:11941201. doi: 10.1002/1097-0142(19920901)70:5<1194::aid-cncr2820700528>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 8.Sorensen BB, Freskgard PO, Nielsen LS, Rao LVM, Ezban M, Petersen LC. Factor VIIa-induced p44/42 mitogen-activated protein kinase activation requires the proteolytic activity of factor VIIa and is independent of the tissue factor cytoplasmic domain. J Biol Chem. 1999;274:21349–54. doi: 10.1074/jbc.274.30.21349. [DOI] [PubMed] [Google Scholar]
- 9.Camerer E, Huang W, Coughlin SR. Tissue factor- and factor X-dependent activation of protease-activated receptor 2 by factor VIIa. Proc Natl Acad Sci USA. 2000;97:5255–60. doi: 10.1073/pnas.97.10.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hjortoe GM, Petersen LC, Albrektsen T, Sorensen BB, Norby PL, Mandal SK, Pendurthi UR, Rao LVM. Tissue factor–factor VIIaspecific up-regulation of IL-8 expression in MDA-MB-231 cells is mediated by PAR-2 and results in increased cell migration. Blood. 2004;103:3029–37. doi: 10.1182/blood-2003-10-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thim L, Bjoern S, Christensen M, Nicolaisen EM, Lund-Hansen T, Pedersen AH, Hedner U. Amino acid sequence and posttranslational modifications of human factor VIIa from plasma and transfected baby hamster kidney cells. Biochemistry. 1988;27:7785–93. doi: 10.1021/bi00420a030. [DOI] [PubMed] [Google Scholar]
- 12.Sorensen BB, Persson E, Freskgard PO, Kjalke M, Ezban M, Williams T, Rao LVM. Incorporation of an active-site inhibitor in factor viia alters the affinity for tissue factor. J Biol Chem. 1997;272:11863–8. doi: 10.1074/jbc.272.18.11863. [DOI] [PubMed] [Google Scholar]
- 13.Edwards D. Non-linear normalization and background correction in one-channel cDNA microarray studies. Bioinformatics. 2003;19:825–33. doi: 10.1093/bioinformatics/btg083. [DOI] [PubMed] [Google Scholar]
- 14.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 15.Gibson UEM, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 16.Heid CA, Stevens J, Livak KJ, Williams PM. Real-time quantitative pcr. Genome Res. 1996;6:986–94. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 17.Camerer E, Kataoka H, Kahn M, Lease K, Coughlin SR. Genetic evidence that protease-activated receptors mediate factor Xa signaling in endothelial cells. J Biol Chem. 2002;277:16081–7. doi: 10.1074/jbc.M108555200. [DOI] [PubMed] [Google Scholar]
- 18.Kawabata A, Kuroda R, Nakaya Y, Kawai K, Nishikawa H, Kawao N. Factor Xa-evoked relaxation in rat aorta: involvement of PAR- 2. Biochem Biophys Res Commun. 2001;282:432–5. doi: 10.1006/bbrc.2001.4597. [DOI] [PubMed] [Google Scholar]
- 19.Pendurthi UR, Allen KE, Ezban M, Rao LVM. Factor VIIa and thrombin induce the expression of Cyr61 and connective tissue growth factor, extracellular matrix signaling proteins that could act as possible downstream mediators in factor VIIa center dot tissue factor-induced signal transduction. J Biol Chem. 2000;275:14632–41. doi: 10.1074/jbc.275.19.14632. [DOI] [PubMed] [Google Scholar]
- 20.Morris DR, Ding Y, Ricks TK, Gullapalli A, Wolfe BL, Trejo J. Protease-activated receptor-2 is essential for factor VIIa and Xa-induced signaling, migration, and invasion of breast cancer cells. Cancer Res. 2006;66:307–14. doi: 10.1158/0008-5472.CAN-05-1735. [DOI] [PubMed] [Google Scholar]
- 21.Riewald M, Ruf W. Protease-activated receptor-1 signaling by activated protein C in cytokine-perturbed endothelial cells is distinct from thrombin signaling. J Biol Chem. 2005;280:19808–14. doi: 10.1074/jbc.M500747200. [DOI] [PubMed] [Google Scholar]
- 22.Siegbahn A, Johnell M, Rorsman C, Ezban M, Heldin CH, Ronnstrand L. Binding of factor VIIa to tissue factor on human fibroblasts leads to activation of phospholipase C and enhanced PDGF-BB- stimulated chemotaxis. Blood. 2000;96:3452–8. [PubMed] [Google Scholar]
- 23.Sorensen BB, Rao LVM, Tornehave D, Gammeltoft S, Petersen LC. Antiapoptotic effect of coagulation factor VIIa. Blood. 2003;102:1708–15. doi: 10.1182/blood-2003-01-0157. [DOI] [PubMed] [Google Scholar]
- 24.Versteeg HH, Spek CA, Richel DJ, Peppelenbosch MP. Coagulation factors VIIa and Xa inhibit apoptosis and anoikis. Oncogene. 2004;23:410–7. doi: 10.1038/sj.onc.1207066. [DOI] [PubMed] [Google Scholar]
- 25.Ahamed J, Ruf W. Protease-activated receptor 2-dependent phosphorylation of the tissue factor cytoplasmic domain. J Biol Chem. 2004;279:23038–44. doi: 10.1074/jbc.M401376200. [DOI] [PubMed] [Google Scholar]
- 26.Belting M, Dorrell MI, Sandgren S, Aguilar E, Ahamed J, Dorfleutner A, Carmeliet P, Mueller BM, Friedlander M, Ruf W. Regulation of angiogenesis by tissue factor cytoplasmic domain signaling. Nat Med. 2004;10:502–9. doi: 10.1038/nm1037. [DOI] [PubMed] [Google Scholar]
- 27.Dorfleutner A, Hintermann E, Tarui T, Takada Y, Ruf W. Cross-talk of integrin alpha 3 beta 1 and tissue factor in cell migration. Mol Biol Cell. 2004;15:4416–25. doi: 10.1091/mbc.E03-09-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belperio JA, Keane MP, Arenberg DA, Addison CL, Ehlert JE, Burdick MD, Strieter RM. CXC chemokines in angiogenesis. J Leukoc Biol. 2000;68:1–8. [PubMed] [Google Scholar]
- 29.Strieter RM, Belperio JA, Phillips RJ, Keane MP. CXC chemokines in angiogenesis of cancer. Semin Cancer Biol. 2004;14:195–200. doi: 10.1016/j.semcancer.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Barreda DR, Hanington PC, Belosevic M. Regulation of myeloid development and function by colony stimulating factors. Dev Comp Immunol. 2004;28:509–54. doi: 10.1016/j.dci.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 31.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71–8. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 32.Bottazzi B, Vouretcraviari V, Bastone A, Degioia L, Matteucci C, Peri G, Spreafico F, Pausa M, Dettorre C, Gianazza E, Tagliabue A, Salmona M, Tedesco F, Introna M. Multimer formation and ligand recognition by the long pentraxin ptx3 - similarities and differences with the short pentraxins c-reactive protein and serum amyloid-p component. J Biol Chem. 1997;272:32817–23. doi: 10.1074/jbc.272.52.32817. [DOI] [PubMed] [Google Scholar]
- 33.Napoleone E, Di-Santo A, Bastone A, Peri G, Mantovani A, de-Gaetano G, Donati MB, Lorenzet R. Long pentraxin PTX3 upregulates tissue factor expression in human endothelial cells - A novel link between vascular inflammation and clotting activation. Arterioscler Thromb Vasc Biol. 2002;22:782–7. doi: 10.1161/01.atv.0000012282.39306.64. [DOI] [PubMed] [Google Scholar]
- 34.Rusnati M, Camozzi M, Moroni E, Bottazzi B, Peri G, Indraccolo S, Amadori A, Mantovani A, Presta M. Selective recognition of fibroblast growth factor-2 by the long pentraxin PTX3 inhibits angiogenesis. Blood. 2004;104:92–9. doi: 10.1182/blood-2003-10-3433. [DOI] [PubMed] [Google Scholar]
- 35.Eubank TD, Galloway M, Montague CM, Waldman WJ, Marsh CB. M-CSF induces vascular endothelial growth factor production and angiogenic activity from human monocytes. J Immunol. 2003;171:2637–43. doi: 10.4049/jimmunol.171.5.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J, Gray A, Yuan J, Luoh SM, Avraham H, Wood WI. Vascular endothelial growth factor-related protein - a ligand and specific activator of the tyrosine kinase receptor flt4. Proc Natl Acad Sci USA. 1996;93:1988–92. doi: 10.1073/pnas.93.5.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enholm B, Jussila L, Karkkainen M, Alitalo K. Vascular endothelial growth factor-C: a growth factor for lymphatic and blood vascular endothelial cells. Trends Cardiovasc Med. 1998;8:292–7. doi: 10.1016/s1050-1738(98)00026-7. [DOI] [PubMed] [Google Scholar]
- 38.Desbaillets I, Diserens AC, Detribolet N, Hamou MF, Vanmeir EG. Up-regulation of interleukin-8 by oxygen-deprived cells in glioblastoma suggests a role in leukocyte activation, chemotaxis, and angiogenesis. J Exp Med. 1997;186:1201–12. doi: 10.1084/jem.186.8.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch AE, Polverini PJ, Kunkel SL, Harlow LA, Dipietro LA, Elner VM, Elner SG, Strieter RM. Interleukin-8 as a macrophage-derived mediator of angiogenesis. Science (Washington DC) 1992;258:1798–801. doi: 10.1126/science.1281554. [DOI] [PubMed] [Google Scholar]
- 40.Chen CC, Mo FE, Lau LF. The angiogenic factor Cyr61 activates a genetic program for wound healing in human skin fibroblasts. J Biol Chem. 2001;276:47329–37. doi: 10.1074/jbc.M107666200. [DOI] [PubMed] [Google Scholar]
- 41.Wang JF, Olson ME, Ball DK, Brigstock DR, Hart DA. Recombinant connective tissue growth factor modulates porcine skin fibroblast gene expression. Wound Repair Regen. 2003;11:220–9. doi: 10.1046/j.1524-475x.2003.11311.x. [DOI] [PubMed] [Google Scholar]