Abstract
The ability to undergo large-scale domain rearrangements is essential for the substrate binding function of periplasmic binding proteins (PBPs), which are indispensable for nutrient uptake in Gram-negative bacteria. Crystal structures indicate that PBPs typically adopt either an “open” unliganded configuration or a “closed” liganded one. However, it is not clear whether, as a general rule, PBPs remain open until ligand-induced interdomain closure, or are in equilibrium with a minor population of unliganded, closed species. Evidence for the latter has been recently reported on maltose-binding protein (MBP) in aqueous solution via paramagnetic relaxation enhancement (PRE), a technique able to probe lowly populated regions of conformational space. Here, we use PRE to study the unliganded open–closed transition of another PBP: glutamine-binding protein (GlnBP). Through a combination of domain structure knowledge, and intermolecular and concentration dependence PRE experiments, a set of surface residues was found involved in intermolecular interactions. Barring such residues, PRE data on ligand-free GlnBP, paramagnetically labeled at two sites (one at a time), could be appropriately explained by the unliganded, open crystal structure in that it both yielded a good PRE fit and was not significantly affected by PRE-based refinement. Thus, contrary to MBP, our data did not particularly suggest the coexistence of a minor closed conformer. Several possibilities were explored to explain the observed differences in such closely structurally related systems, among them, a particularly interesting one arises from close inspection of the interdomain “hinge” region of various PBPs: strong hydrogen bond interactions discourage large-scale interdomain dynamics.
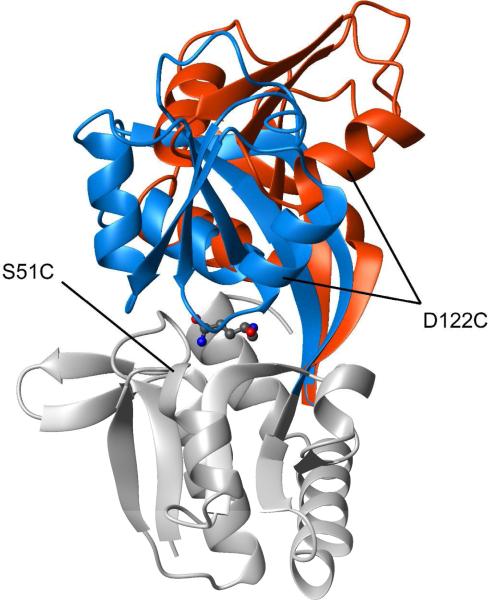
Found in all prokaryotic and eukaryotic species, ATP-binding cassette (ABC) systems actively transport solutes across cellular membranes, and are crucial to many important biological phenomena (for reviews see refs. (1, 2)). When involved in the uptake of nutrients in Gram-negative bacteria, ABC transporters generally rely on periplasmic binding proteins (PBPs) that bind the substrate with high-affinity and present it to the membrane-bound component of the system for translocation into the cytoplasm. Despite, when combined, PBPs can handle a wide variety of ligands, which include amino acids and short peptides, mono- and oligosaccharides, and inorganic and organic ions (3), members of the PBP family share a common three-dimensional architecture: two globular domains with a similar αβ fold, connected by one or more polypeptide linker segments (1, 4). High-resolution crystallographic studies have shown that, typically, ligand-free PBPs exhibit an “open” conformation with a solvent-accessible cleft between the domains. The ligand occupies this cleft and becomes trapped by a domain closure that involves significant rigid-body movement of the domains around the linker region that functions as a hinge (1, 4) (see Figure 1 for an example).
Figure 1.
Conformational change of GlnBP upon ligand binding. The crystal structures of unliganded (PDB ID 1GGG) and liganded (PDB ID 1WDN) GlnBP are superimposed via the large domain, colored grey (only that of liganded GlnBP is shown). The small domain and hinge region of unliganded and liganded GlnBP are colored red and blue, respectively. The L-Gln ligand is shown in “balls-and-sticks” representation. The mutation sites used for spin labeling are indicated. All molecular representations were generated with MOLMOL (19).
The existence of the aforementioned open–unliganded and closed–liganded states raises the question as to whether a closed conformation can be reached in the absence of ligand. Accessibility to a closed–unliganded form with the liganded conformation (or one very similar to it) would strongly suggest a population-shift or conformational selection mode of binding (5) where the interaction with the ligand stabilizes a preexisting, albeit disfavored, species. On the other hand, if such a closed conformer were forbidden, binding would proceed via induced-fit (6), where domain closure is a ligand-triggered event. Experimental evidence that indicates at least some PBPs are able to achieve a ligand-free open–closed transition stems from the crystal structures of two proteins in a closed, unliganded state: glucose/galactose-binding protein (GGBP) (7) and ChoX (8). Furthermore, nuclear magnetic resonance (NMR) studies based on paramagnetic relaxation enhancement (PRE) have recently afforded the structure determination of a minor (~5% population) unliganded, closed conformation of maltose-binding protein (MBP) in equilibrium with the major (~95% population) open form (9).
PRE has become a highly sensitive tool for probing large-scale dynamical processes in biomolecules and their complexes in solution, phenomena otherwise inaccessible by conventional biophysical techniques (for reviews see refs. (10, 11)). The magnetic dipolar interaction between a free electron on a paramagnetic center, and a nucleus, such as 1H, enhances the magnetic relaxation of the latter, giving rise to the PRE. For a paramagnet–nucleus distance r, the effect is proportional to r–6 and, due to the large magnetic moment of the free electron, is far-reaching. Additionally, in a system comprised of fast-exchanging species, the observed transverse PRE rate of nucleus i, Γ2,i, is given by the population-weighted average (12). For instance, in a two-state system:
| (1) |
Here, the contribution of the lowly populated (minor) species (with fractional population fminor; fmajor = 1 – fminor) can be significant, provided that its PRE is much larger than that of the major species (i.e., ) or, equivalently, that its paramagnet–nucleus distance is much shorter. This effect, which can be probed for all magnetic nuclei in the system, allows for the detection and characterization of the otherwise elusive minor species (10, 11).
The dynamical aspect of PRE introduced above can be exploited in the context of ligand-free domain rearrangement in a PBP by placing a paramagnetic label in the vicinity of the potential domain–domain interface generated upon domain closure. Due to the absence of ligand, the open conformation is expected to be favored (major species). However, the equilibrium with a small population of closed conformer may have a considerable influence on the observed average PREs (equation 1) as a result of the increased interdomain proximity in this minor species. Thus, interdomain PREs are particularly sensitive, as reflected in values larger than expected on the basis of a single open conformer. Indeed, this was the case for MBP, which prompted Tang et al. to postulate the coexistence of a minor closed form in order to successfully interpret the PRE data (9).
Here, we use the above-described PRE approach to investigate the open–closed transition of unliganded glutamine-binding protein (GlnBP), a 226-residue PBP involved in the active transport of L-glutamine across the cytoplasmic membrane of Escherichia coli, and closely related, in terms of fold and ligand binding, to mammalian and bacterial glutamate receptors (13). Although a PRE-based study has been previously performed on GlnBP (14), the focus was on the solution structure determination of the liganded conformation, under conditions unsuitable to probe interdomain dynamics. The latter has been the subject of several theoretical investigations (15-18), but little experimental research, hence the motivation behind the present work. Our results are interpreted in the context of the other PBPs that are able to reach a closed conformation in absence of ligand.
Materials and Methods
Sample Preparation
In addition to wild-type GlnBP, two mutants, namely S51C and D122C, were generated using the pJ133 plasmid (20) as a template and the QuikChange site-directed mutagenesis kit (Stratagene). Protein expression, purification, and mutant spin labeling were performed as detailed elsewhere (14). Briefly, using E. coli host cells, uniformly 15N-enriched or natural abundance protein was overexpressed in minimal medium respectively containing 15NH4Cl or 14NH4Cl as sole nitrogen source, followed by extraction by chloroform-shock (21), and purification via anion exchange (DEAE) and size exclusion (Superdex-75) columns (GE Healthcare) (20). In the case of cysteine mutant GlnBP (S51C or D122C), protein was concentrated, reduced for two hours in 20-fold molar excess of dithiothreitol (DTT), and desalted on a PD-10 column (GE Healthcare) using 0.1 M potassium phosphate buffer, pH 7.2 (NMR buffer). Nitroxide spin labeling was performed immediately after desalting by adding 20-fold molar excess of methanethiosulfonate spin label (MTSL; Toronto Research Chemicals Inc.), and incubating overnight at 4° C. Reverse-phase HPLC (C4 column; Vydac) was used to purify the unliganded spin-labeled protein (spin labeling confirmed by LC-MS), which was subsequently lyophilized, and re-dissolved overnight in NMR buffer to a final sample concentration ~0.5 mM (unless otherwise stated); when present L-Gln was in 3-fold molar excess. In the case of wild-type GlnBP, protein purification was performed as described above, except for the DTT incubation, desalting, and spin labeling steps, which were omitted. For the measurement of intermolecular PREs (see below), a mixed sample was prepared that contained 0.3 mM 15N-enriched wild-type GlnBP and 0.5 mM natural abundance GlnBP D122C derivatized with MTSL, both proteins ligand-free.
PRE Measurements
The transverse PRE rate of nucleus i, Γ2, i, is defined as the difference in its transverse relaxation rate, R2, between the paramagnetic and diamagnetic state of the protein. The latter was achieved by reduction of the nitroxide spin label with the addition of 10-fold molar excess of ascorbic acid to the paramagnetic NMR sample. Transverse PRE rates were measured for backbone 1HN protons using a two-time-point (0 and 15 ms) 15N–HSQC-based interleaved experiment (22), and their errors estimated as previously described (22). Measurements were carried out at 41 °C on a Bruker Avance 800-MHz spectrometer equipped with a cryoprobe and Z−pulsed field gradient. Spectra were processed with NMRPipe (23), and analyzed with PIPP (24) and home-written Python programs.
PRE Fit Based on the Crystal Conformations
Experimental PRE values were used to drive torsion-angle simulated annealing (SA) optimizations of the paramagnetic probe side chain, represented by a three-conformer ensemble (25). A total of four SA optimization types were performed, involving (i) unliganded GlnBP S51C, (ii) liganded GlnBP S51C, (iii) unliganded GlnBP D122C, and (iv) liganded GlnBP D122C. In each case, the corresponding PRE data set was used. The polypeptide backbone conformation was assumed to be that of the corresponding crystal structure, and held fixed during SA calculations. In contrast, all side chains were allowed to move, biased by a torsion-angle database potential (26) and van der Waals repulsions which, along with the PRE, were the major active energy terms. The correlation time for the internal motion of the paramagnetic probe, τi, was assumed to be negligible against the correlation time for the paramagnet–nucleus interaction, τc (τi << τc). The latter was optimized within an 8–15-ns range (25). The crystal structure models were “protonated” with REDUCE (27). Xplor-NIH (28, 29) was used for all SA calculations reported in this paper.
For each optimization type (i–iv above), the 10 structures (optimized probe side chains) with the lowest PRE energy, out of 200 independent SA runs, were selected and used to back-calculate PRE values. Goodness of fit between experimental and calculated PREs was assessed by a Q-factor (25):
| (2) |
where angular brackets indicate the average over the 10 selected optimizations, and summations extend over the set A, which may comprise either the entire PRE data set (leading to an “overall Q-factor”) or a subset of the total (e.g., intradomain, leading to an “intradomain Q-factor”). Similarly, an average root-mean-square deviation (rmsd) was taken as an additional figure of merit.
PRE-based Structural Refinement
Refinement of the backbone conformation of the open, unliganded crystal structure of GlnBP against the PRE data was achieved using a SA protocol based on that previously described (14). Briefly, starting from the open GlnBP crystal coordinates (PDB ID 1GGG), SA calculations were performed that treated the backbone of the two domains as rigid bodies, while giving the hinge and non-probe side chains full torsion-angle degrees of freedom. Both unliganded S51C and D122C PRE data sets were enforced simultaneously via a single hybrid GlnBP model spin-labeled at the two sites (25); the PRE data comprised the only experimentally derived energy terms. Each paramagnetic probe side chain was represented by a three-conformer ensemble (25), and constrained to the conformations previously optimized assuming the unliganded crystal model (see previous section) in order to avoid structural distortions due to probe flexibility (14). A total of 200 structures were computed, out of which the 10 with the lowest overall PRE energy were selected for analysis.
Generation of GlnBP Conformers with Various Degrees of Domain Closure
GlnBP models at different steps along the open→closed pathway, the ends respectively defined by the unliganded (PDB ID 1GGG) and liganded (PDB ID 1WDN) crystal backbone conformations (Figure 1), were generated via adiabatic mapping or “morphing” (30). Specifically, for each mutant, the unliganded and liganded structures with the lowest PRE energy after paramagnetic probe side chain optimization (see above) were selected as the two ends for the morphing process. The latter consisted in initially superimposing the two structures via the backbone N, Cα, and C’ atoms of the domain that contains the label. Subsequently, starting from the open conformation, an interpolation routine was iteratively run, that (i) evenly reduced the interatomic distances between the current structure and the closed conformer, and (ii) minimized the energy to ensure a correct covalent geometry. After each interpolation step, the resulting structure was output and labeled by the iteration number as it represents a measure of the progress along the open→closed pathway. For each mutant, a total of 100 models were generated, thus evenly sampling the space defined between the fully open (iteration 0) and fully closed (iteration 99) structures, populated by partially closed conformations (intermediate iterations). The morphing protocol was implemented in Xplor-NIH (28, 29), and closely followed that described elsewhere (31).
Hydrogen Bond Detection
Hydrogen bonds were calculated for the hinge region of several PBPs, using the crystal structure after the attachment of hydrogen atoms with the program REDUCE, which additionally performs a hydrogen bond optimization that includes sampling of the two Asn/Gln side chain amide group orientations, and rotation of OH and NH3 groups (27). In our hinge analysis, all possible hydrogen bond donor–acceptor combinations—which included both backbone and side chain groups as specified elsewhere (32)—were considered, and hydrogen bonds inferred on the basis of the N–H···O or O–H···O small angle (≥ 115°) and the H···O distance (≤ 2.5 Å) (33).
Results
Spin Labeling
GlnBP consists of two globular domains, termed large (residues 1–84 and 186–226) and small (residues 90–180), linked by a two-strand hinge (residues 85–89 and 181–185) (34, 35). Crystal structures of GlnBP have been previously determined in both unliganded (PDB ID 1GGG) (34) and liganded (PDB ID 1WDN) (35) states (Figure 1). Similar to other PBPs, a significant open→closed conformational change is observed upon ligand binding, in this case largely described by rotations of 41° in the ϕ angle of Gly89, and 34° in the ψ angle of Glu181, both residues located in the hinge region (Figure 1) (35).
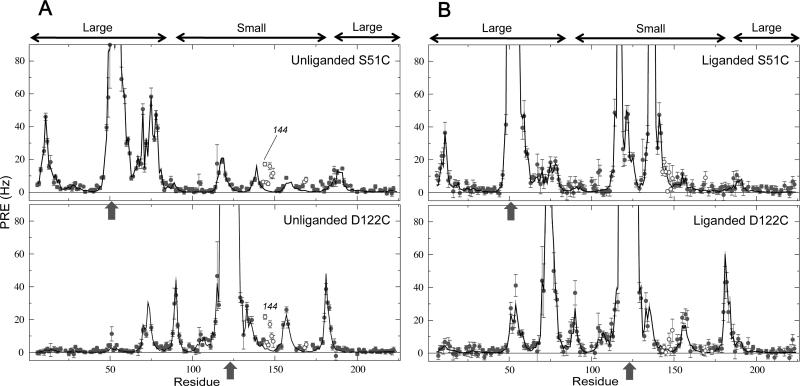
Site-directed nitroxide spin labeling was performed on GlnBP at two different positions (one at a time) by derivatizing with MTSL engineered cysteines S51C and D122C, located in the large and small domain, respectively (Figure 1). The chosen sites are near the domain–domain interface in the liganded structure, thus potentially useful for probing large-scale domain dynamics (9). In each case, mutation and spin labeling had little effect on the structure, as suggested by small and localized chemical shift changes in 15N–HSQC spectra, relative to wild-type GlnBP (true for both unliganded and liganded states). Furthermore, the S51C location has been successfully exploited for the PRE-based structure determination of liganded GlnBP in solution (14); the associated data set is used here as one of two controls (see below). The experimentally obtained transverse PRE rates of backbone 1HN protons for the S51C and D122C mutants, in both unliganded and liganded samples, are shown in Figure 2 as a function of the residue number.
Figure 2.
PRE profiles for unliganded and liganded GlnBP. In both, the unliganded (A) and liganded (B) cases, experimental transverse 1HN-PREs (circles) are shown against the calculated values (lines) obtained via ensemble paramagnetic probe optimization using the indicated PRE data set (S51C or D122C; single-headed arrows denote the mutation/paramagnetic labeling sites) and the corresponding crystal structure. Each calculated PRE value is the average over the 10 lowest-PRE energy results out of 200 independent optimizations. Double-headed arrows (top) indicate the large and small domains, respectively. Open circles involve residues 143–149, 168, 169, and 173, of which 144 is highlighted in the unliganded data sets (see text).
PRE Fit Assuming a Single GlnBP Conformer: the Crystal Structure
For each GlnBP mutant and ligand state (i.e., absence or presence of ligand), optimization of the conformational sampling space of the paramagnetic probe side chain (25) was performed against the associated PRE data set, assuming the backbone conformation of the protein to be that of the corresponding crystal structure. The results are summarized in Table 1, and Figures 2 and 3.
Table 1.
Goodness of Fit for the Overall PRE Data Sets and Their Intra-/Interdomain Partitions.a
| overall | intradomain | interdomain | |
|---|---|---|---|
| unliganded S51C | |||
| Q | 0.19 ± 0.01 | 0.17 ± 0.01 | 0.44 ± 0.00 |
| rmsd | 3.3 ± 0.1 | 3.8 ± 0.2 | 2.4 ± 0.0 |
| total PREsb | 145 | 83 | 54 |
| liganded S51C | |||
| Q | 0.21 ± 0.00 | 0.22 ± 0.00 | 0.19 ± 0.00 |
| rmsd | 2. 9 ± 0.0 | 2.7 ± 0.0 | 3.3 ± 0.1 |
| total PREsb | 153 | 86 | 57 |
| unliganded D122C | |||
| Q | 0.21 ± 0.00 | 0.17 ± 0.01 | 0.44 ± 0.00 |
| rmsd | 2.2 ± 0.0 | 2.6 ± 0.2 | 1.6 ± 0.0 |
| total PREsb | 150 | 48 | 92 |
| liganded D122C | |||
| Q | 0.27 ± 0.00 | 0.27 ± 0.01 | 0.26 ± 0.02 |
| rmsd | 3.9 ± 0.1 | 3.9 ± 0.2 | 3.2 ± 0.2 |
| total PREsb | 149 | 53 | 87 |
Statistics exclude residues 143–149, 168, 169, and 173 (see text).
Hinge residues are represented only in the overall data set. PREs associated to completely broadened, highly overlapped, or unassigned peaks are excluded.
Figure 3.
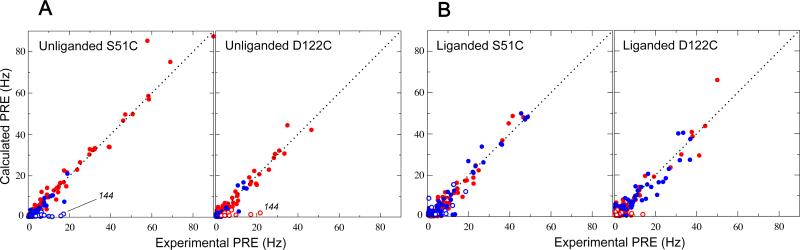
Correlations between calculated and experimental PRE values. Both the unliganded (A) and liganded (B) cases are shown. Each calculated PRE value is the average over the 10 lowest-PRE energy results out of 200 independent paramagnetic probe optimizations. Intradomain PREs are shown in red, interdomain in blue. Open circles involve residues 143–149, 168, 169, and 173, of which 144 is highlighted in the unliganded data sets (see text).
Despite being in a small proportion under ligand-free conditions, a closed conformer of a PBP in coexistence with a major open form may have a significant contribution to the measured PRE data (see equation 1) as a result of large interdomain PREs arising from the increased domain–domain proximity (9). A manifestation of the effect is the failure to fit interdomain PREs assuming the open conformation as the sole structural model (9). Close inspection of the unliganded data set on GlnBP S51C reveals a subset of PREs that cannot be properly fit by the ensemble probe optimization, comprising residues 143–149, 168, 169, and 173 (Figures 2A and 3A, open circles). These PREs are interdomain and larger than predicted from the open structure used in the optimizations, thus possibly suggesting the existence of a minor closed conformer. However, careful scrutiny of the unliganded PRE fits, particularly the profiles of Figure 2A, shows that the same residues that yield larger than expected interdomain PREs in the unliganded S51C data provide a very similar PRE pattern in the unliganded D122C mutant. In the latter case, however, such residues are in the same domain as the paramagnetic probe, thus giving rise to intradomain PREs that should not be significantly affected by a conformational equilibrium that involves rigid-body domain motions. The effect is reproducible across different sample preparations, and observed at protein concentrations as low as 0.25 mM (not shown). In addition, the PRE fit is not improved by increasing the number conformers used to represent the probe side chain during the ensemble probe optimizations (Supporting Information).
Figure 4 sheds light on the phenomenon. While residues 143–149, 168, 169, and 173 are non-contiguous in the amino acid sequence and are spread across various types of structural elements, such as α-helix, β-strand and loop regions, they comprise a solvent exposed patch on the small domain of GlnBP (Figure 4A). Two different experiments, each involving one of the two mutants, suggest that the larger than expected PREs observed for this patch (Figures 2A and 3A, open circles) result from intermolecular interactions that give rise to intermolecular PREs. First, the effect is concentration dependent, as suggested by Figure 4B which highlights the differences in PREs arising from a 0.5- and a 0.25-mM sample of unliganded spin-labeled GlnBP S51C. Second, a mixture of unliganded, 15N-enriched wild-type GlnBP and unliganded, natural isotopic abundance, spin-labeled GlnBP D122C—a sample that produces only intermolecular PREs since they are measured on the 15N-enriched protein that contains no paramagnetic label—shows the largest of its PREs on a residue within the patch (Figure 4C). Although not obvious for all such residues, the two experiments independently and clearly implicate residue 144 in intermolecular interactions. Interestingly enough, position 144, centrally located in the surface patch (Figure 4A), exhibits the largest PRE among its kind in the unliganded S51C and D122C data sets (Figures 2A and 3A). It should be noted that the small offset and noisy nature observed for the intermolecular PREs (Figure 4C) are possibly due to additional non-specific encounter complexes in this sample that bears the highest total GlnBP concentration (0.8 mM) (22).
Figure 4.
Intermolecular interactions on unliganded GlnBP. Residues 143–149, 168, 169, and 173 (magenta) form a continuous, solvent exposed surface on the small domain of unliganded, open GlnBP (A). For each residue, the PRE concentration dependence in unliganded, spin-labeled (i.e., MTSL-derivatized) GlnBP S51C is expressed in terms of the square of the PRE difference, (ΔΓ2)2, between two protein concentrations (0.5 and 0.25 mM), divided by the square of the average PRE error, σ2 (B). Intermolecular PREs arising from the interaction of wild-type (WT) 15N-enriched GlnBP and natural isotopic abundance, spin-labeled GlnBP D122C (both proteins unliganded) are shown (C). Residue 144 is indicated throughout.
The failure to fit PREs arising from the set of residues 143–149, 168, 169, and 173 as intradomain data (unliganded GlnBP D122C sample), in combination with the suggested involvement in intermolecular interactions, was used as criterion to exclude such residues from the following statistical analysis. An alternative possibility would have been to correct for intermolecular “artifacts” by using the PRE values extrapolated to zero concentration in a concentration dependence series. Indeed, use of the two concentration points implied in Figure 4B yields extrapolated PREs averaging 0.21 Hz for the most affected residues in the set (residues 144–147, and 149). Since such values are close to those predicted by ensemble probe optimization (Figures 2A and 3A), correcting their contribution to the PRE data sets would have little impact on the subsequent PRE fits; as a result, we opted to exclude all residues in the surface patch (Figure 4A) from consideration. With this in mind, a good agreement is achieved between experimental PREs from both unliganded S51C and D122C mutants and those calculated from the open crystal structure. This is visually appreciated in the profiles of Figure 2A and the correlation plots of Figure 3A, as well as quantitatively assessed by the low overall Q-factors of 0.19 and 0.21 for S51C and D122C data sets, respectively (Table 1).
The liganded PRE data serve as control since in the presence of an excess of ligand any existing equilibrium is expected to be heavily biased towards the closed, ligand-bound conformation: the major species. A minor population of open conformer would have a negligible effect on the observed, average PREs (see equation 1) since, in addition to being poorly weighted, the associated interdomain PREs are inherently small as a result of the large interdomain separation (i.e., in equation 1) (9). Consequently, the liganded data can be safely described by a single closed, ligand-bound species, as reflected by low overall Q-factors of 0.21 (intradomain, 0.22; interdomain, 0.19) and 0.27 (intradomain, 0.27; interdomain, 0.26) for the S51C and D122C data sets, respectively (Table 1, and Figures 2B and 3B).
The similitude between overall Q-factors for the unliganded data sets and their corresponding liganded controls suggests that the former can also be appropriately interpreted in terms of a single species, in this case with the open conformation. On the other hand, Q-factor statistics for the unliganded and liganded data sets differ when comparing the intra- and interdomain partitions. Specifically, while in the liganded controls the magnitudes of intra- and interdomain Q-factors are similar, in the unliganded data sets interdomain Q-factors are larger than the intradomain (0.44 vs. 0.17, coincidentally valid for both mutants; Table 1). At first glance, this might suggest a worse fit for the interdomain PREs in the unliganded state. However, larger interdomain Q-factors are more likely to reflect the fact that in the open structure experimental interdomain PRE values—which form the denominator of the Q-factor formula (equation 2)—are smaller than the intradomain as a result of larger paramagnet–1HN distances concomitant with the interdomain separation. This effective partitioning of the unliganded data into large (intra-) and small (interdomain) PREs breaks down, however, in the liganded case, where intra-and interdomain distances are comparable. (Indeed, it can be shown that the Q-factor distance dependence for the unliganded and liganded data sets are very similar; see Supporting Information.) Furthermore, the trend observed under the Q-factor disappears when using a figure of merit that is not normalized by the experimental values, such as the rmsd (Table 1). Q-factors are retained throughout this paper as the main goodness-of-fit measure in order to facilitate comparison with a growing body of literature that makes use of this statistic (10, 11).
PRE Fit Assuming a Single GlnBP Conformer: Refinement of the Crystal Structure
Since the unliganded crystal structure has been used thus far for fitting the unliganded PRE data sets, an alternative possibility to explain the above-discussed large interdomain Q-factors is a different domain arrangement in solution from that in the crystal. Such possibility can be readily tested by conjoined rigid-body/torsion-angle simulated annealing calculations (36, 37) that allow flexibility at the hinge. Using a protocol based on that successfully implemented for the calculation of the liganded GlnBP conformation in solution (14), the domains of the unliganded, open GlnBP crystal structure were allowed to move relative to each other, under the simultaneous influence of both unliganded S51C and D122C PRE data sets. Any possible strain associated with the large interdomain Q-factors listed in Table 1 would result in its relaxation through the hinge. The computed models, however, do not significantly deviate from the crystal structure (Supporting Information), and the Q-factor statistics remains essentially unchanged (Table 2).
Table 2.
Q-factor Statistics for the Refinement of the Unliganded GlnBP Crystal Structure Against the Unliganded S51C and D122C PRE Data Sets.a
| overall Q | intradomain Q | interdomain Q | |
|---|---|---|---|
| S51C | 0.18 ± 0.00 | 0.16 ± 0.00 | 0.42 ± 0.00 |
| D122C | 0.19 ± 0.01 | 0.16 ± 0.00 | 0.44 ± 0.00 |
Statistics exclude residues 143–149, 168, 169, and 173 (see text).
PRE Fit Assuming a Two-Conformer GlnBP System: Minor Species with the Closed, Liganded Conformation
In order to determine whether the PRE fit of the unliganded data sets could be further improved by postulating the presence of a minor closed species, the values respectively calculated from the unliganded and liganded crystal structures via ensemble paramagnetic probe optimization (i.e., calculated values appearing in Figures 2 and 3) were linearly combined using the following equation:
| (3) |
Equation 3 represents a particular instance of the fast-exchanging two-state system—described in broad terms by equation 1—where the conformations of the coexisting species are those of the unliganded and liganded GlnBP crystal structures, with fractional populations fopen and fclosed, respectively. Given the absence of ligand, it is implied that the closed conformation is that of the liganded structure, although devoid of the L-Gln ligand. Support for this model stems from GGBP and ChoX, to the best of our knowledge, the only two PBPs ever reported in a closed, unliganded crystal state, that, in addition, exhibit conformations highly superimposable to their liganded versions (Cα-rmsd of 0.3 and < 0.2 Å, respectively) (7, 8).
Figure 5 shows the Q-factor-based agreement between the experimental PREs from unliganded GlnBP S51C and D122C, and those calculated using equation 3, as a function of the population of the minor closed species. Expectedly, the interdomain Q-factor is the most affected by the closed species population, in particular that of the S51C data set, as a result of better positioning of the spin label to probe this specific domain motion (Figure 1). For both data sets, however, the trend is the same: the interdomain PRE fit remains almost unchanged for closed populations ≤ 3%, and worsens for populations ≥ 4%. Indeed, an F-test of additional term (38) indicates the fits of the overall data sets are not significantly improved by the assumption of the minor closed species—which are responsible for the “extra” term in eq. 3—as they yield F values up to 5.95 (S51C) and 6.55 (D122C), both above the acceptable probability F cutoff of 0.01.
Figure 5.
Effect of a minor population of closed GlnBP conformer on the fit of PREs arising from unliganded GlnBP S51C (full lines) and D122C (dashed lines). Overall, intra-, and interdomain Q-factors are indicated in black, red, and blue, respectively. The backbone coordinates for the closed conformer are those of the liganded GlnBP crystal structure (PDB ID 1WDN).
PRE Fit Assuming a Two-Conformer GlnBP System: Minor Species with Partially Closed Conformations
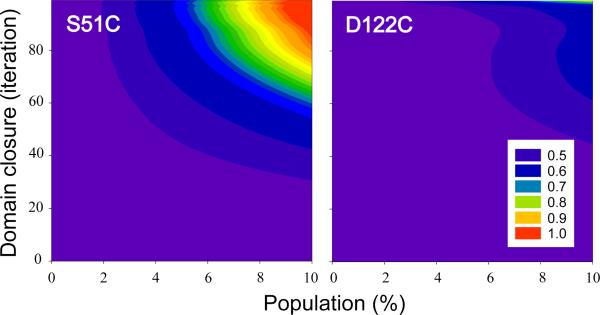
The fast-exchanging two-state system embodied by equation 3 was further explored by the postulation of minor GlnBP species with different degrees of domain closure, ranging from fully open to fully closed, as defined by the unliganded and liganded GlnBP crystal structures, respectively. By morphing these two extreme conformations via adiabatic mapping, a popular technique for the analysis of macromolecular flexibility (31, 39), partially closed GlnBP models were generated. For each GlnBP mutant, PREs calculated from the fully open structure (major species) and a morphed model (minor species) were combined via equation 3, and the agreement with the corresponding unliganded experimental data set was assessed by a Q-factor. Figure 6 displays the interdomain Q-factors for the unliganded S51C and D122C data sets as a function of the population and degree of domain closure of the minor species. Since, as detailed in Materials and Methods, the morphing protocol evenly samples the open→closed pathway by iteratively taking the current structure one step away from the starting unliganded conformation (iteration 0) and one step closer to the final liganded one (iteration 99), the iteration number is used to indicate the extent of domain closure. Thus, for example, the model generated at iteration 50 is approximately halfway in between the fully open and fully closed structures, and will be accordingly referred to as “50% closed”.
Figure 6.
Interdomain Q-factor surfaces for the fit of the unliganded GlnBP S51C (left) and D122C (right) PRE data sets as a function of the relative population and extent of domain closure of the minor species. Domain closure is expressed in terms of the iteration number of the morphing protocol that gradually converts the unliganded open structure (PDB ID 1GGG, iteration 0) into the liganded closed one (PDB ID 1WDN, iteration 99).
It is apparent from the Q-factor surfaces of Figure 6 that position 51 is more sensitive than 122 to probe this particular path in conformational space and, consequently, it shall be used as the basis for the following discussion. Here, it is assumed that Q-factor variations greater than 0.04, i.e., twice the value of the largest Q-factor error listed in Table 1, are significant. Thus, for example, minor species with ≤ 35% domain closure and populations of up to 10% could be accommodated by the unliganded data as the interdomain Q-factor is not considerably affected relative to that obtained assuming a single open species. Similarly, a 50% closed conformer with minor population ≤ 6% would also be consistent with the unliganded data. Beyond a domain closure of 75%, minor populations lower than ~3% would be possible. Across the entire surface, the interdomain Q-factor either has small variability, as in the above examples, or increases; that is, the unliganded PRE fit never improves by the assumption of the minor conformer.
Discussion
Implications of the Present GlnBP Study
The above results do not particularly support the existence of a minor closed GlnBP species in equilibrium with the major open form under ligand-free conditions, in that the open structure suitably predicted the observed experimental PRE values (Table 1, Figures 2A and 3A). Furthermore, neither the open GlnBP conformer nor the PRE fit was significantly affected by structural refinement against the PRE data (Table 2). These observations may owe to several reasons: (i) an unliganded, closed conformation is not accessible, or exists in a very low proportion under our experimental conditions; (ii) even when a closed conformation is favorable, its degree of domain closure is relatively small, thus yielding weak interdomain PREs with little impact on the observed, averaged values; (iii) the open–closed transition does not satisfy the fast exchange condition. Possibilities i and ii were tested under the assumption that the minor conformation lies somewhere in between the unliganded and liganded ones, allowing for the identification of relatively large regions of conformational/population space consistent with the PRE data, that is, regions where the interdomain Q-factor for the fit of the unliganded data sets remains constant (Figures 5 and 6). Those tests relied on the additional assumption of fast exchange in the PRE time-scale, where the interconversion between major and minor conformers is characterized by the exchange rate constant kex, with (12). Indeed, another possibility (option iii) is that such condition is not fulfilled by the system (e.g., at the limit of kex = 0, ). Assuming a minor species with the liganded conformation, the difference between the PREs calculated from the liganded and unliganded crystal structures for HN 117 of GlnBP S51C—site that exhibits the largest such difference in this mutant—helps estimate the fast exchange condition as kex >> 250 Hz. A possible reason why the fast exchange condition might not be met is given in the following section.
Fulfillment of any single one of the above three scenarios (along with any of their possible combinations) would explain the inability to detect a minor unliganded closed GlnBP conformer. Except for that assuming the inexistence of such species, the remaining options do not exclude interdomain dynamics, which has been recently suggested by a phosphorescence spectroscopy study (40). An additional, yet trivial, circumstance not considered above is that the selected labeling sites are inappropriate to probe the particular domain rearrangement involved in an otherwise detectable open–closed transition. This is not expected to be the case, however, if the closed structure were similar to the liganded one, as clearly suggested by Figure 5. In this regard, position 51 seems particularly suitable, as also indicated by its successful use in the PRE-based structure determination of ligand-bound GlnBP (14). Furthermore, since positions 51 and 122 do not directly oppose each other in the liganded structure, they seem complementary to probe a range of hinge-twist motions.
Comparison with other PBPs
The experimental strategy used here for the investigation of the ligand-free open–closed transition of GlnBP closely followed that of the seminal work by Tang et al. on MBP (9). Contrary to GlnBP, in absence of ligand the open MBP conformation failed to predict experimental PRE data, which prompted the structure determination of a coexisting minor closed conformer. Despite such species was found structurally different from the liganded conformation, the postulation of a minor species with the liganded conformation still yielded improved fit for the interdomain PRE data associated to one of the two paramagnetic labels on which Tang et al. relied (9). A similar approach on GlnBP, however, did not improve the PRE fit, resulting in significant interdomain Q-factor increases for the greater part of the population range considered (Figure 5).
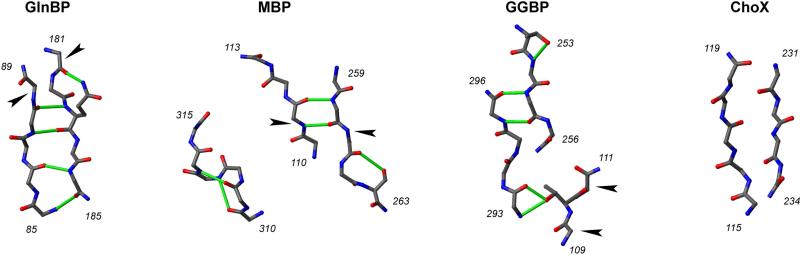
In addition to MBP, the other PBPs shown to be able to achieve a ligand-free closed conformation are GGBP and ChoX. This is thanks to crystallographic studies where the structures of the unusual closed–unliganded forms have been reported before those of the generally more common open–unliganded states (41, 42), and in the case of GGBP the reports are separated by twelve years. Figure 7 shows the hinge regions of GlnBP, MBP, GGBP, and ChoX. Such regions differ in the number of polypeptide linker segments, and the quantity and quality of hydrogen bonds. While ChoX has no hydrogen bonds within its hinge, GlnBP, MBP, and GGBP display a total of five. In GlnBP, hydrogen bonds comprise a compact network where four of them (i.e., all but one) directly connect the backbone of the two hinge segments, stabilizing a two-stranded antiparallel β-sheet. In contrast, MBP has a broken hydrogen bond network, with only two hydrogen bonds between the segments that support the major conformational change upon ligand binding. In the case of GGBP, while four hydrogen bonds tighten the whole hinge, they are divided among three different linker segments, and the conformational change focuses in a region relatively isolated from the rest of the hydrogen bond network.
Figure 7.
Hinge segments from the crystal structures of open–unliganded GlnBP (PDB ID 1GGG) (34), MBP (PDB ID 1OMP) (43), GGBP (PDB ID 2FW0) (41), and ChoX (PDB ID 3HCQ) (42). The hinges of GlnBP and ChoX have two segments, while those of MBP and GGBP have three; the residues flanking each segment are indicated. Backbone heavy atoms are shown, in addition to those of side chains involved in hydrogen bonding. Covalent bonds are colored according to the liked atoms (carbon, grey; oxygen, red; nitrogen, blue). Hydrogen bonds are colored green, and were detected as detailed in Materials and Methods. Arrowheads indicate regions reported to support the major conformational change upon ligand binding; such a specific region in ChoX has been neither reported nor observed here after backbone torsion-angle analysis (see text).
The hinge strands of GlnBP are stabilized by unusually strong hydrogen bond interactions (35), which has been suggested as a reason bending is concentrated at one end of the hinge, instead of the middle as in MBP (35). Furthermore, hydrogen bonds could also explain the reported location of major hinge change in GGBP (41) (see above), and why no such location can be detected by backbone torsion-angle analysis on the hinge of ChoX (not shown), where hydrogen bonds are absent. (When present, major hinge bending points are indicated in Figure 7.) In the context of unliganded open–closed conformational change that concerns us here, the suggested hinge stability in GlnBP could imply a high-energy transition state (e.g., with broken hydrogen bonds) that results in slow interconversion in the PRE time scale (see previous section), or even kinetic inaccessibility to a significantly closed state.
Yet another difference between GlnBP and the above-mentioned set of PBPs is ligand specificity. Whereas GlnBP binds L-Gln in a highly specific fashion (44), GGBP binds glucose and galactose (45), ChoX binds choline and acetylcholine (46), and MBP binds a series of maltodextrines (47). An interesting possibility is that a closed–unliganded conformation of GlnBP is inherently unstable, thus exerting more pressure on the ligand contribution to the stabilization of the complex.
Conclusions
Periplasmic binding proteins, essential for metabolite uptake, chemotaxis, and intercellular communication, represent small variations of a single basic structural motif. Long-standing speculations that PBPs undergo a constant ligand-free closing and opening around the hinge (1, 4, 48, 49) have found support in MBP, GGBP, and ChoX. The fact that the closed–unliganded structures of GGBP and ChoX are essentially the same as their liganded versions suggests a population-shift mode of binding. On the other hand, relative to the liganded conformation, the minor unliganded form of MBP is partially closed, and the domains are reoriented in order to relax lack of electrostatic and steric complementarity that arise in the absence of the ligand (9). Such minor conformer is assumed to facilitate binding, which implies an induced-fit mechanism starting from this intermediate species (9). In the case of GlnBP, the focus of this article, our results suggest that under ligand-free conditions a closed species is highly thermodynamically unfavorable (low population and/or partial closure) and/or that the open–closed transition is slow, possibly due to unusual hinge stability (PRE fast exchange condition unfulfilled and/or the open state is metastable). Although these possibilities are not enough to determine the mode of binding for GlnBP (population-shift vs. induced-fit), they highlight significant differences in the thermodynamic/dynamic behavior of these closely structurally related proteins.
Supplementary Material
Acknowledgment
We thank Dr. Duck-Yeon Lee (Protein Analysis Core facility of the NHLBI) for his assistance.
Abbreviations
- DTT
dithiothreitol
- HPLC
high-pressure liquid chromatography
- HSQC
heteronuclear single quantum correlation
- GGBP
glucose/galactose-binding protein
- GlnBP
glutamine-binding protein
- MBP
maltose-binding protein
- MTSL
methanethiosulfonate spin label
- NMR
nuclear magnetic resonance
- PBP
periplasmic binding protein
- PRE
paramagnetic relaxation enhancement
- rmsd
root-mean-square deviation
Footnotes
This work was supported by the NIH Grant GM-084614 to C.H., and the Intramural Research Program of the NIH, NHLBI, to N.T.
Supporting Information Available: A table with Q-factor statistics for the fit of the PRE data sets assuming a 6-conformer ensemble probe representation, plots of Q-factor dependence on the paramagnet–HN distance, and a figure with a family of structures and correlation plots generated from the refinement of the unliganded GlnBP crystal conformation against the unliganded PRE data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiology and Molecular Biology Reviews. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones PM, George AM. The ABC transporter structure and mechanism: perspectives on recent research. Cellular and Molecular Life Sciences. 2004;61:682–699. doi: 10.1007/s00018-003-3336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doeven MK, Kok J, Poolman B. Specificity and selectivity determinants of peptide transport in Lactococcus lactis and other microorganisms. Molecular Microbiology. 2005;57:640–649. doi: 10.1111/j.1365-2958.2005.04698.x. [DOI] [PubMed] [Google Scholar]
- 4.Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: Variation of common themes. Molecular Microbiology. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 5.Monod J, Wyman J, Changeux JP. On Nature of Allosteric Transitions - a Plausible Model. Journal of Molecular Biology. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 6.Koshland DE. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flocco MM, Mowbray SL. The 1.9 Angstrom X-Ray Structure of a Closed Unliganded Form of the Periplasmic Glucose/Galactose Receptor from Salmonella-Typhimurium. Journal of Biological Chemistry. 1994;269:8931–8936. [PubMed] [Google Scholar]
- 8.Oswald C, Smits SHJ, Hoing M, Sohn-Bosser L, Dupont L, Le Rudulier D, Schmitt L, Bremer E. Crystal Structures of the Choline/Acetylcholine Substrate-binding Protein ChoX from Sinorhizobium meliloti in the Liganded and Unliganded-Closed States. Journal of Biological Chemistry. 2008;283:32848–32859. doi: 10.1074/jbc.M806021200. [DOI] [PubMed] [Google Scholar]
- 9.Tang C, Schwieters CD, Clore GM. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449:1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 10.Clore GM. Visualizing lowly-populated regions of the free energy landscape of macromolecular complexes by paramagnetic relaxation enhancement. Molecular BioSystems. 2008;4:1058–1069. doi: 10.1039/b810232e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clore GM, Tang C, Iwahara J. Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement. Current Opinion in Structural Biology. 2007;17:603–616. doi: 10.1016/j.sbi.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 13.Lampinen M, Pentikainen O, Johnson MS, Keinanen K. AMPA receptors and bacterial periplasmic amino acid-binding proteins share the ionic mechanism of ligand recognition. Embo Journal. 1998;17:4704–4711. doi: 10.1093/emboj/17.16.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bermejo GA, Strub MP, Ho C, Tjandra N. Determination of the Solution Bound Conformation of an Amino Acid Binding Protein by NMR Paramagnetic Relaxation Enhancement: Use of a Single Flexible Paramagnetic Probe with Improved Estimation of Its Sampling Space. Journal of the American Chemical Society. 2009;131:9532–9537. doi: 10.1021/ja902436g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang A, Arinaminpathy Y, Sansom MSP, Biggin PC. Interdomain dynamics and ligand binding: molecular dynamics simulations of glutamine binding protein. Febs Letters. 2003;550:168–174. doi: 10.1016/s0014-5793(03)00866-4. [DOI] [PubMed] [Google Scholar]
- 16.Su JG, Jiao X, Sun TG, Li CH, Chen WZ, Wang CX. Analysis of domain movements in glutamine-binding protein with simple models. Biophysical Journal. 2007;92:1326–1335. doi: 10.1529/biophysj.106.086512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suhre K, Sanejouand YH. On the potential of normal-mode analysis for solving difficult molecular-replacement problems. Acta Crystallographica Section D-Biological Crystallography. 2004;60:796–799. doi: 10.1107/S0907444904001982. [DOI] [PubMed] [Google Scholar]
- 18.Sun TG, Hu JP, Li CH, Chen WZ, Wang CX. A molecular dynamics simulation study of glutamine-binding protein. Journal of Molecular Structure-Theochem. 2005;725:9–16. [Google Scholar]
- 19.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. Journal of Molecular Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 20.Shen QC, Simplaceanu V, Cottam PF, Ho C. Proton Nuclear Magnetic-Resonance Studies on Glutamine-Binding Protein from Escherichia-Coli - Formation of Intermolecular and Intramolecular Hydrogen-Bonds Upon Ligand-Binding. Journal of Molecular Biology. 1989;210:849–857. doi: 10.1016/0022-2836(89)90112-5. [DOI] [PubMed] [Google Scholar]
- 21.Ames GF, Prody C, Kustu S. Simple, Rapid, and Quantitative Release of Periplasmic Proteins by Chloroform. Journal of Bacteriology. 1984;160:1181–1183. doi: 10.1128/jb.160.3.1181-1183.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwahara J, Tang C, Clore GM. Practical aspects of H-1 transverse paramagnetic relaxation enhancement measurements on macromolecules. Journal of Magnetic Resonance. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. Journal of Biomolecular Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 24.Garrett DS, Powers R, Gronenborn AM, Clore GM. A Common-Sense Approach to Peak Picking in 2-Dimensional, 3-Dimensional, and 4-Dimensional Spectra Using Automatic Computer-Analysis of Contour Diagrams. Journal of Magnetic Resonance. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Iwahara J, Schwieters CD, Clore GM. Ensemble approach for NMR structure refinement against H-1 paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. Journal of the American Chemical Society. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 26.Clore GM, Kuszewski J. chi(1) Rotamer populations and angles of mobile surface side chains are accurately predicted by a torsion angle database potential of mean force. Journal of the American Chemical Society. 2002;124:2866–2867. doi: 10.1021/ja017712p. [DOI] [PubMed] [Google Scholar]
- 27.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: Using hydrogen atom contacts in the choice of side-chain amide orientation. Journal of Molecular Biology. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 28.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;48:47–62. [Google Scholar]
- 29.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. Journal of Magnetic Resonance. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 30.McCammon JA, Harvey SC. Dynamics of proteins and nucleic acids. Cambridge University Press; Cambridge; New York: 1987. [Google Scholar]
- 31.Krebs WG, Gerstein M. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Research. 2000;28:1665–1675. doi: 10.1093/nar/28.8.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker EN, Hubbard RE. Hydrogen bonding in globular proteins. Progress in Biophysics and Molecular Biology. 1984;44:97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 33.Lipsitz RS, Sharma Y, Brooks BR, Tjandra N. Hydrogen bonding in high-resolution protein structures: a new method to assess NMR protein geometry. Journal of the American Chemical Society. 2002;124:10621–10626. doi: 10.1021/ja020676p. [DOI] [PubMed] [Google Scholar]
- 34.Hsiao CD, Sun YJ, Rose J, Wang BC. The crystal structure of glutamine-binding protein from Escherichia coli. Journal of Molecular Biology. 1996;262:225–242. doi: 10.1006/jmbi.1996.0509. [DOI] [PubMed] [Google Scholar]
- 35.Sun YJ, Rose J, Wang BC, Hsiao CD. The structure of glutamine-binding protein complexed with glutamine at 1.94 angstrom resolution: Comparisons with other amino acid binding proteins. Journal of Molecular Biology. 1998;278:219–229. doi: 10.1006/jmbi.1998.1675. [DOI] [PubMed] [Google Scholar]
- 36.Clore GM, Bewley CA. Using conjoined rigid body/torsion angle simulated annealing to determine the relative orientation of covalently linked protein domains from dipolar couplings. Journal of Magnetic Resonance. 2002;154:329–335. doi: 10.1006/jmre.2001.2489. [DOI] [PubMed] [Google Scholar]
- 37.Schwieters CD, Clore GM. Internal coordinates for molecular dynamics and minimization in structure determination and refinement. Journal of Magnetic Resonance. 2001;152:288–302. doi: 10.1006/jmre.2001.2413. [DOI] [PubMed] [Google Scholar]
- 38.Bevington PR, Robinson DK. Data Reduction and Error Analysis for the Physical Sciences. 3rd ed. McGraw-Hill; Boston: 2003. [Google Scholar]
- 39.Echols N, Milburn D, Gerstein M. MolMovDB: analysis and visualization of conformational change and structural flexibility. Nucleic Acids Research. 2003;31:478–482. doi: 10.1093/nar/gkg104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D'auria S, Staiano M, Varriale A, Gonnelli M, Marabotti A, Rossi M, Strambini GB. The differences in the microenvironment of the two tryptophan residues of the glutamine-binding protein from Escherichia coli shed light on the binding properties and the structural dynamics of the protein. Proteins-Structure Function and Bioinformatics. 2008;71:743–750. doi: 10.1002/prot.21748. [DOI] [PubMed] [Google Scholar]
- 41.Borrok MJ, Kiessling LL, Forest KT. Conformational changes of glucose/galactose-binding protein illuminated by open, unliganded, and ultra-high-resolution ligand-bound structures. Protein Science. 2007;16:1032–1041. doi: 10.1110/ps.062707807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oswald C, Smits SH, Hoing M, Bremer E, Schmitt L. Structural analysis of the choline-binding protein ChoX in a semi-closed and ligand-free conformation. Biological Chemistry. 2009;390:1163–1170. doi: 10.1515/BC.2009.113. [DOI] [PubMed] [Google Scholar]
- 43.Sharff AJ, Rodseth LE, Spurlino JC, Quiocho FA. Crystallographic Evidence of a Large Ligand-Induced Hinge-Twist Motion between the 2 Domains of the Maltodextrin Binding-Protein Involved in Active-Transport and Chemotaxis. Biochemistry. 1992;31:10657–10663. doi: 10.1021/bi00159a003. [DOI] [PubMed] [Google Scholar]
- 44.Weiner JH, Heppel LA. Binding Protein for Glutamine and Its Relation to Active Transport in Escherichia-Coli. Journal of Biological Chemistry. 1971;246:6933. doi: 10.1016/0003-9861(71)90538-8. &. [DOI] [PubMed] [Google Scholar]
- 45.Anraku Y. Transport of sugars and amino acids in bacteria. II. Properties of galactose- and leucine-binding proteins. Journal of Biological Chemistry. 1968;243:3123–3127. [PubMed] [Google Scholar]
- 46.Dupont L, Garcia I, Poggi MC, Alloing G, Mandon K, Le Rudulier D. The Sinorhizobium meliloti ABC transporter Cho is highly specific for choline and expressed in bacteroids from Medicago sativa nodules. Journal of Bacteriology. 2004;186:5988–5996. doi: 10.1128/JB.186.18.5988-5996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kellerman O, Szmelcman S. Active-Transport of Maltose in Escherichia-Coli-K12 - Involvement of a Periplasmic Maltose Binding-Protein. European Journal of Biochemistry. 1974;47:139–149. doi: 10.1111/j.1432-1033.1974.tb03677.x. [DOI] [PubMed] [Google Scholar]
- 48.Mao B, Pear MR, McCammon JA, Quiocho FA. Hinge-bending in L-arabinose-binding protein. The “Venus's-flytrap” model. Journal of Biological Chemistry. 1982;257:1131–1133. [PubMed] [Google Scholar]
- 49.Sack JS, Saper MA, Quiocho FA. Periplasmic binding protein structure and function. Refined X-ray structures of the leucine/isoleucine/valine-binding protein and its complex with leucine. Journal of Molecular Biology. 1989;206:171–191. doi: 10.1016/0022-2836(89)90531-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.