Abstract
The degradation of intracellular components in lysosomes, generically known as autophagy, can occur through different pathways. This review discusses chaperone-mediated autophagy (CMA), a type of autophagy set apart from other autophagic pathways due to its selectivity and distinctive mechanism by which substrates reach the lysosomal lumen. CMA participates in quality control and provides energy to cells under persistently poor nutritional conditions. Alterations in CMA have recently been shown to underlie some severe human disorders for which the decline with age in the activity of this pathway becomes a major aggravating factor. Prevention of the age-dependent decline in CMA has major beneficial effects on cellular and organ homeostasis and function, revealing that CMA is an essential component of the anti-aging fight.
Autophagy: Cellular self-digestion with a purpose
We all probably invoke a very similar mental image in response to the word “lysosome”. I bet you just pictured a garbage container, didn’t you? This organelle has been associated with cellular cleaning almost since its description more than 50 years ago by Christian de Duve [1]. However, only recently has the importance of this cleaning function and its critical role in the maintenance of cellular homeostasis been appreciated in full. This improved understanding of the contribution of the lysosomal system to cellular quality control, along with the growing number of new functions identified for this organelle, have nowadays revitalized interest in lysosomes. These multiple functions are attained through a process known as autophagy, which refers to the lysosomal degradation of intracellular proteins, lipids and organelles. Once internalized in lysosomes, these structures are broken down by resident enzymes into their constituent building blocks that are then released and utilized by the cell to synthesize new cellular structures.
Most of the early studies on autophagy were performed in rat liver, tracking the degradation of the soluble pool of cytosolic protein or measuring changes in the size and volume of the lysosomal system [2-4]. However, major advances have taken place in the last 15 years, leading to a detailed dissection of the molecular components that participate in the execution and regulation of autophagy [5-7]. Genetic manipulation (knockout, knockdown or overexpression) of the genes required for autophagy (ATG) in different cellular and animal models has provided essential information on the consequences of changes in autophagic activity and has allowed the identification of autophagic failure in the pathogenesis of multiple human disorders, including among others, cancer, neurodegeneration, infectious diseases, myopathies and metabolic disorders [8].
Another consequence of these recent advances is the better understanding of the characteristics and differences among the autophagic pathways. Three different mechanisms for delivery of autophagic cargo to lysosomes co-exist in most mammalian cells: macroautophagy, microautophagy and chaperone-mediated autophagy (CMA) [6,8] (Box 1). This review focuses solely on chaperone-mediated autophagy because the recent links between this pathway and different human disorders and aging have awakened a previously unforeseen interest in this selective type of autophagy. I first review the current advances in our understanding of the mechanisms that mediate and regulate CMA and then comment on the different physiological functions attributed to this pathway and the consequences of its malfunctioning in disease and aging.
Box 1. Characteristics of the different autophagic pathways.
Three main autophagic pathways have been shown to co-exist in almost all mammalian cells (Figure I):
(a) Macroautophagy
In macroautophagy, whole regions of the cytosol are sequestered and delivered to lysosomes for degradation. Cargo sequestration occurs in the autophagosome, a double membrane vesicle that forms through the elongation and sealing of a de novo generated membrane [7]. This limiting membrane originates from a tightly controlled series of interactions between more than 10 different proteins, which resemble in many aspects the conjugation steps that mediate protein ubiquitinization. Formation of the limiting membrane also requires the interaction between a protein and a specific lipid molecule (phosphatidyl ethanolamine), also regulated by conjugating enzymes [7]. Once formed, the autophagosome receives the enzymes required for degradation of the sequestered cargo through fusion with lysosomes. Numerous reviews have summarized the recent findings on the coordinated orchestration of macroautophagy, the mechanisms that regulate this process, and its important and growing number of physiological functions [5-8].
(b) Microautophagy
A similar process of cytosolic sequestration takes place through microautophagy, but the limiting/sequestering membrane is the lysosomal membrane itself, which invaginates to form tubules or vesicles that pinch off into the lysosomal lumen [4]. Studies on microautophagy during recent years mainly focused on its characterization in yeast, where in addition to some genes shared with macroautophagy, a separate subset of genes also participates exclusively in this process [55]. However, microautophagy is still very poorly understood in mammals, and in fact, it is possible that this process actually takes place in late endosomes rather than in lysosomes and occurs through the formation of the multivesicular bodies in this compartment [56].
(c) Chaperone-mediated autophagy (CMA)
In contrast to macro- and microautophagy, delivery of cargo via CMA does not require formation of intermediate vesicle compartments, membrane fusion or membrane deformity of any kind [57,58]. Instead, the substrates for this type of autophagy are translocated from the cytosol into the lysosomal lumen directly across the membrane in a process mediated by a translocation protein complex that requires substrate unfolding [17,20]. Because of the particular characteristics of this delivery, only soluble proteins, but not complete organelles, can be degraded through CMA.
CMA: Selective degradation moves to the lysosome
Although an accepted concept nowadays, the idea that single proteins could be picked up from the cytosol and shuttled to the lysosome for degradation in a selective manner was rather provocative when first postulated in the mid 1980’s [9]. Selectivity was not a fit for lysosomal degradation, always perceived as an “in bulk” process. The initial idea originated from the observation that starvation in animals or serum removal in cultured cells beyond 8-10 hours accelerated the degradation in lysosomes of particular cytosolic proteins, but not others [9,10]. Tracking of individual proteins undergoing selective lysosomal degradation and analyzing their sequences led to the identification of both a targeting motif in all of the substrates for this pathway and a chaperone able to recognize this motif [11,12]. The unusual characteristics of CMA were not limited to its selectivity. In fact, as the mechanism for cargo delivery was revealed, it became evident that, in contrast to the other forms of autophagy, vesicle formation was not required in CMA. Instead, the substrate proteins were translocated across the lysosomal membrane. Selectivity and direct lysosomal translocation have thus become trademarks of CMA.
A tag and a chaperone are behind the selectivity of CMA
An essential requirement for a protein to become a CMA substrate is the presence of a pentapeptide motif biochemically related to KFERQ in its amino acid sequence [13] (Box 2). In fact, this motif, when mutated, prevents the degradation of proteins that otherwise would be degraded by this pathway. The recognition of this motif by a cytosolic chaperone leads to the delivery of the substrate protein to lysosomes. So far, HSC70, the constitutive member of the cytosolic family of chaperones of 70 kDa, is the only chaperone shown able to recognize the KFERQ-like motif [12] (Figure 1). HSC70 is a multifunctional chaperone that also contributes to the disassembly of clathrin from endocytic pits, folding of newly synthesized proteins, refolding of unfolded proteins and, as a partner of HSP90, to the stabilization of proteins in transient unstable conformations [14]. For most of these functions, HSC70 binds to stretches of hydrophobic residues in the substrates; however, to date, the KFERQ-like motif is the only region recognized by HSC70 in CMA substrates [12]. The interactions of particular co-chaperones know to modulate the ATPase activity of HSC70 might help decide between the different functions of this cytosolic chaperone. Some recent studies propose that the carboxyl terminus of HSP70-interacting protein (CHIP), another cytosolic chaperone that contributes to the degradation of cytosolic proteins by the proteasome, might also target proteins for lysosomal degradation [15]. However, future studies are required to determine whether this lysosomal degradation is indeed occurring via CMA.
Box 2. A tag for selective lysosomal degradation.
Like most targeting motifs, the CMA tag degenerates and allows for a series of amino acid combinations as far as they follow this rule: Q should be the flanking amino acid but could be located at the beginning or at the end of the sequence; there could be up to two of the allowed hydrophobic residues (I, F, L or V) or two of the allowed positive residues (R or K), but only one negative charge provided either by E or D [13]. In certain proteins, Q can be replaced with N, but this exchange does not work in all proteins, suggesting that the surrounding amino acid context might be important in this case [16].
The “pure” KFERQ motif is only present in ribonuclease A, the first protein identified as a CMA substrate. Sequence analysis revealed that KFERQ-like motifs are present in about 30% of cytosolic soluble proteins. The CMA motif is rarely present in proteins in organelles or membrane proteins. To date, only one membrane protein, the epidermal growth factor receptor, has been proposed to undergo lysosomal degradation in a manner dependent on the receptor for CMA [59]; however, the requirement for protein unfolding, essential to define this process as CMA, has not yet been determined for the epidermal growth factor receptor.
The location of the CMA tag in the protein is not important, although these motifs are usually hidden or covered. For the substrate to undergo CMA, it is necessary that the motif becomes accessible at some point to the chaperone. For example, some motifs located in the core of the protein will only be exposed upon unfolding [28]; other motifs placed in regions of protein/protein interaction will become accessible when the protein is no longer part of a complex [60]; lastly, in some enzymes the motifs are close to their catalytic site and only become accessible in the absence of substrate [16]. Post-translational modifications can, at least in theory, regulate when a protein becomes a CMA substrate. Incomplete motifs, missing one of the five amino acids required in the sequence, have been identified in different proteins. It is possible that modifications such as phosphorylation - when the negative residue is missing - or acetylation - when a positive residue would complete the sequence - might complete the CMA motif.
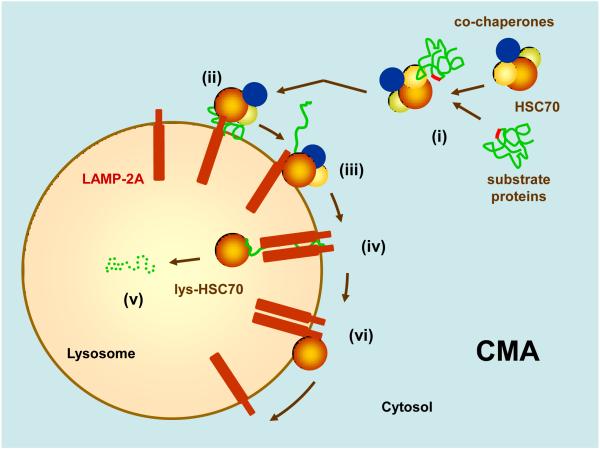
Figure 1. Hypothetical model of the steps and components of chaperone-mediated autophagy (CMA).
(i) Substrate proteins bearing a targeting motif are recognized by HSC70 and co-chaperones in the cytosol. (ii) The chaperone/substrate protein complex is delivered to the surface of lysosomes where it binds to the cytosolic tail of monomer of LAMP-2A. (iii) Substrate binding drives multimerization of LAMP-2A into a translocation complex. In between this and the next step, the substrate is unfolded by yet unknown mechanisms. (iv) The substrate crosses the lysosomal membrane assisted by a luminal form of HSC70. (v) Once inside the lysosome, the substrate is rapidly degraded (dotted structure). (vi) HSC70 promotes disassembly of LAMP-2A from the translocation unit, now devoid of substrate, to provide monomeric forms of LAMP-2A for a new cycle of substrate binding and translocation.
A novel way of crossing the lysosomal membrane: look mom, no vesicles!
Once the cytosolic chaperone binds CMA substrates, it delivers them to lysosomes. Although the steps resulting in the appearance of the cytosolic cargo protein in the lysosomal lumen are not completely elucidated, it is well-established that this process is clearly different from macro- and microautophagy in that delivery of substrates to the lysosomal lumen is saturable, it does not involve formation of vesiculations in the surface of the lysosomes, and it requires unfolding of the substrate protein [16,17]. The saturability results from the binding of each substrate to the lysosome-associated membrane protein type 2A (LAMP-2A) before translocation can occur [18]. In contrast with HSC70 that is often in excess in the cytosol, levels of LAMP-2A at the lysosomal membrane are limiting for CMA and hence, subject to tight regulation [19] (Box 3). LAMP-2A acts as a receptor for the substrates of this pathway, but its functions extend beyond that of a classic membrane receptor (Figure 2). Recent studies showed that LAMP-2A possesses a regulated lateral mobility that allows its assembly into a complex essential for substrate translocation [20]. However, instead of the typical stability of protein translocation systems in other organelles, the lysosomal CMA translocon is highly dynamic and undergoes continuous cycles of assembly/disassembly based on the availability of substrate proteins [20]. The presence of microdomains of discrete lipid composition at the lysosomal membrane also regulates the dynamics of LAMP-2A in lysosomes [21] (Figure 2).
Box 3. The unique characteristics of the CMA translocation system.
LAMP-2A is a single-span membrane protein at the lysosomal membrane required for CMA. It is likely that LAMP-2A participates in other cellular functions common to all LAMP-2 protein variants with which LAMP-2A shares a common luminal region [61]. However, LAMP-2A is the only one of the three LAMP-2 variants involved in CMA [25]. Although LAMP-2A was initially identified as a receptor for the substrates, current studies expanded the steps of CMA in which LAMP-2A participates:
a) LAMP-2A as a receptor for CMA substrates
The use of blocking antibodies revealed that substrate proteins bind to the lysosomal membrane through the cytosolic tail of LAMP-2A [18]. Although further investigation is required to determine the regions of substrate/receptor interaction, it is known that the KFREQ-like motif in the substrate is not utilized for binding to LAMP-2A and that a stretch of four positive residues in the cytosolic tail of the receptor is required for substrate binding [62].
b) LAMP-2A in the translocation complex
LAMP-2A can be detected at the lysosomal membrane in different multimeric states. Substrate binding occurs only to monomers of LAMP-2A, and this binding promotes the organization of LAMP-2A into a high multimeric complex of about 700 kDa, necessary for substrate translocation [20]. This lysosomal translocon undergoes continuous cycles of assembly/disassembly, where assembly is driven by binding of substrates to the lysosomal receptor and disassembly is actively mediated by HSC70 [20]. The disassembly function of HSC70 is only possible when substrate is no longer bound to it. Although the reasons for this dynamic nature of the translocation complex are not known, we speculate that it might be advantageous when rapid downregulation of this system is needed and might also be a safety mechanism to preserve the pH gradient characteristic of the lysosomal lumen.
c) Regulation of LAMP-2A levels
Conditions associated with enhanced CMA always require high LAMP-2A levels, which can be attained by transcriptional upregulation of the Lamp2 gene [28], but more often through changes in the degradation and lysosomal distribution of LAMP-2A [19]. Thus, LAMP-2A is subject to a regulated degradation in specific lipid microdomain regions of the lysosomal membrane [21]. Degradation is initiated through dual cleavage by a pair of lysosomal proteases, cathepsin A and a yet unidentified metalloprotease, followed by the release of the truncated form of LAMP-2A in the lumen and its complete degradation by resident hydrolases [50]. Enhanced recruitment of LAMP-2A to these areas of degradation at the lysosomal membrane reduces levels of this receptor and downregulates CMA [21]. In addition, a portion of intact LAMP-2A is present in the lysosomal lumen and can be recruited to the lysosomal membrane when upregulation of CMA is needed [19].
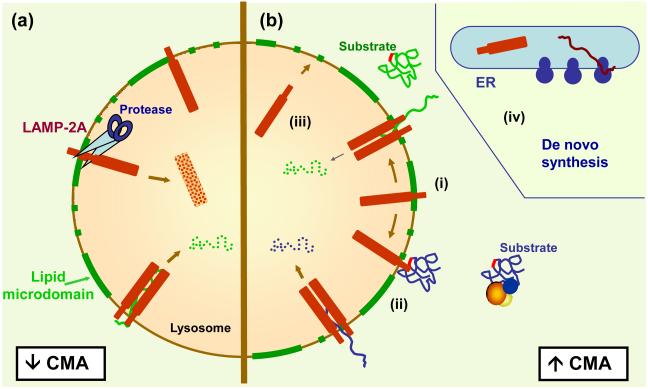
Figure 2. Regulation of the dynamics of LAMP-2A in lysosomes.
(a) Under basal conditions, a percentage of LAMP-2A molecules at the lysosomal membrane are continuously turned over through their recruitment to lipid microdomains (green) where the proteases required for cleavage reside. (b) Activation of CMA is associated with (i) mobilization of LAMP-2A outside of the lipid microdomains to prevent degradation and result in a net increase of LAMP-2A. (ii) LAMP-2A undergoes assembly and disassembly to mediate substrate binding and uptake. Increased LAMP-2A levels can also be attained by (iii) recruiting a percentage of full size LAMP-2A that normally resides in the lysosomal lumen or (iv) through de novo synthesis of LAMP-2A in the ER.
Substrate translocation also requires two forms of HSC70 associated to lysosomes, one located on the cytosolic side of the lysosomal membrane, likely involved in substrate unfolding, and a second one present inside lysosomes (Figure 2). The steps and chaperones involved in substrate unfolding are not known. Chimeras of CMA substrates with a protein unable to unfold can bind to the lysosomal membrane but do not translocate, suggesting that unfolding occurs after binding but before translocation [17]. Blocking antibodies against luminal HSC70 prevented substrate translocation, supporting that this chaperone might act by “pulling” the substrates inside the lysosomal lumen as they cross the membrane [22]. Luminal HSC70 is necessary for CMA, and in fact, the subset of lysosomes able to perform CMA is often identified in cells by the presence of HSC70 in their lumen [23].
HSP90 also associates to lysosomes and affects CMA activity, but rather than participating in the translocation of substrate proteins, the form of HSP90 bound to the luminal side of the membrane is required to stabilize LAMP-2A while transitioning from the monomeric to the multimeric state [20] (Box 3).
Physiological functions of CMA
The first function proposed for CMA was to provide an alternative source of amino acids under conditions of prolonged starvation [9,10,24]. Macroautophagy is activated in most cells during the first hours of starvation, reaches its maximum activity at about 6 hours and then gradually declines [25,26]. CMA is activated when starvation persists beyond 6-8 hours, reaches its maximum activity at about 24 hours and remains activated for at least 3 days [24] (Figure 3). This switch from bulk degradation to a more selective proteolysis might allow cells to avoid degrading proteins and structures essential for cell survival under these compromised conditions, while at the same time degrading proteins of lesser importance to obtain the amino acids required for cell maintenance and survival. CMA and macroautophagy can compensate for each other in this function, at least temporally. For example, RNA interference against LAMP-2A, which blocks CMA results in constitutive activation of macroautophagy and vice versa; however, both pathways are non-redundant, and the lack of the defective pathway becomes evident in response to an additional stressor [25,27].
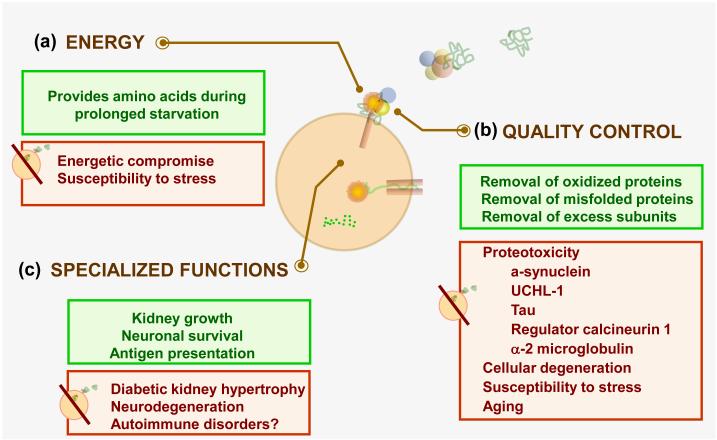
Figure 3. Pathophysiology of CMA.
CMA fulfills two general functions shared with other autophagic pathways - providing an alternative source of (a) energy and (b) performing quality control. (c) In addition, some specialized functions have also been proposed for CMA which are often cell type dependent. Some of the physiological consequences of each of those CMA functions (green) and the cellular alterations resulting from CMA dysfunction (red) are summarized.
CMA, as any other type of autophagy, also participates in cellular quality control (Figure 3). CMA of substrate proteins increases when they are oxidized, misfolded or truncated [28]. Exposure of cultured cells or animals to different oxidants or pro-oxidant compounds enhances rates of CMA, confirming its function in removing oxidized proteins in vivo [28]. Interestingly, increased levels of oxidized proteins might also contribute to keeping CMA upregulated during prolonged starvation, because the increased levels of circulating ketone bodies in these conditions promotes protein oxidation [29]. Blockage of CMA in cultured cells increases their sensitivity to multiple cellular stressors, supporting the notion that activation of CMA is an essential component of the cellular response to stress [25,28].
CMA has specialized functions in particular cell types, often related to the degradation of a specific protein or group of proteins in these cells (Figure 3). In professional antigen presenting cells, CMA participates in the presentation of cytoplasmic peptides by class II molecules contributing to organism autoimmunity [30]. In neurons, CMA promotes neuronal survival by modulating the intracellular levels of the transcription factor myocyte enhancer factor D [31]. Modulation of kidney growth is a specialized function attributed to CMA through the degradation of a specific protein, the transcription factor Pax2, in renal tubular cells [32].
Further understanding of the physiological functions of CMA should be gained once an animal model with selective blockage of this pathway becomes available. A mouse knock-out for the Lamp2 gene has been developed already [33]. However, because all isoforms of the LAMP-2 protein (LAMP-2A, B and C) are absent in this mouse model, the phenotype is of complex interpretation. For example, the alterations in macroautophagy and in lysosomal biogenesis observed in Lamp2 null mice [33] and after knocking-out or knocking-down all LAMP-2 isoforms in cultured cells [34,35], cannot be reproduced by blocking only LAMP-2A in cultured cells [25].
CMA and aging
The finding that CMA activity decreases with age [36], the identification of the main defect responsible for this functional decline [37], and the recent rescue of the CMA age-related defect in whole animals [38] all provide strong support for the contribution of CMA failure to different aspects of aging [39].
The decrease in CMA activity with age was first described even before the molecular components for this pathway were characterized. Early studies in cultured primary fibroblasts revealed that the pool of cytosolic proteins whose lysosomal degradation was accelerated upon serum removal was degraded at slower rates as cells become senescent [36]. This decrease was later confirmed in lysosomes from various organs of old rodents which displayed lower translocation of well-characterized CMA substrate proteins [37]. A systematic comparative study revealed that chaperone recognition and targeting of CMA substrates and their degradation once in the lumen are preserved with age [37]. However, binding and translocation of substrate proteins by lysosomes is drastically reduced in older organisms due to a gradual decrease in the lysosomal levels of LAMP-2A [37]. This decrease does not result from age-related changes in the transcriptional activation of the Lamp2 gene, changes in its splicing toward one of its protein variants, or problems in trafficking of this protein from Golgi to its final location in lysosomes [40]. Instead, the stability of the CMA receptor is markedly compromised once it reaches the lysosome in old organisms because a large percentage of lysosomal LAMP-2A remains in the lysosomal lumen where it undergoes massive unregulated degradation [40]. The decrease in LAMP-2A levels is initially compensated by a gradual increase in the percentage of lysosomes able to perform CMA, i.e. those containing HSC70 in their lumen, through still unknown mechanisms [37]. However, eventually, this compensation is not enough, and failure of this pathway becomes evident.
Because age-dependent decreases in LAMP-2A levels seem universal, our group attempted to preserve LAMP-2A levels in mice until later in life as a possible mechanism to keep CMA active. We developed a bi-transgenic mouse model expressing an additional copy of LAMP-2A in liver that can be activated in middle life to supplement the reduced levels of endogenous protein [38]. CMA activity is preserved in these animals when they are old at rates comparable to those in young mice. This results in improved cellular homeostasis (lower content of oxidized and aggregated proteins and improved activity of other quality control systems), enhanced resistance to different stressors, and preserved organ function [38]. These results support that, at least in the liver, preventing the age-dependent decrease in CMA activity slows down aging. Future studies in mouse models with regulatable LAMP-2A expression in other organs should help determine whether aging of CMA can be prevented in all cell types and if preservation of CMA in the whole organism until late in life prolongs healthspan (disease-free time). A recent study reported abnormal upregulation of macroautophagy in mice with accelerated aging that might contribute to their systemic degeneration [41]. Whether or not CMA is also upregulated during accelerated aging remains to be determined.
CMA and disease
Recently, malfunctioning of CMA has been connected to a growing number of human disorders [42]. The clue to the discovery that CMA activity was faulty in these pathologies has often been the detection of abnormally high levels of a CMA substrate protein in the affected cells (Figure 3).
CMA and neurodegeneration
The first connection between CMA and neurodegenerative diseases was established in Parkinson’s disease (PD) because alpha-synuclein, the protein that accumulates in protein inclusions in PD-affected neurons, was identified as a bona fide CMA substrate [43]. Alpha-synuclein can be degraded by the ubiquitin proteasome system, CMA and macroautophagy [43,44]. However, mutant and post-translationally modified forms of alpha-synuclein, identified in PD patients, are targeted to lysosomes through their CMA-motif but are unable to cross the lysosomal membrane. Instead, pathogenic alpha-synucleins remain tightly bound to the CMA receptor, preventing the translocation of other CMA substrates into lysosomes [43,45]. The specific reasons why pathogenic forms of alpha-synuclein cannot cross the lysosomal membrane remain unknown, but they have been found to form irreversible oligomers on the surface of lysosomes which likely interfere with lysosomal functions. Consequently, the homeostatic problem in PD is twofold, involving first, the inability to properly turnover alpha-synuclein and second, the associated blockage of CMA which renders cells more susceptible to multiple stressors. Furthermore, not only pathogenic forms of alpha-synuclein, but even high levels (overexpression) of the wild type form of this protein has recently been found to hinder the degradation of neuronal proteins by CMA [31]. This finding is relevant for disease because a triplication of the alpha-synuclein gene has also been identified in some patients with familial PD. The connections between CMA and PD have further expanded with the discovery that mutant forms of the ubiquitin carboxyl-terminal esterase L1 (UCHL-1), another pathogenic protein in PD, interact abnormally with LAMP-2A leading to CMA blockage [46].
Interference of the CMA translocating machinery by pathogenic proteins extends beyond PD to other neurodegenerative diseases. A mutant cleaved form of Tau, the protein that organizes into toxic fibrillar structures in Alzheimer’s disease (AD), is also delivered to lysosomes via CMA, and similar to the abnormal alpha-synucleins, mutant Tau fails to translocate properly [47]. Instead, the C-terminus of Tau reaches the lysosomal lumen and undergoes a dual cleavage by cathepsin D which generates a highly amyloidogenic fragment. Persistence of Tau fibers on the surface of lysosomes diminishes the stability of the lysosomal membrane and favors leakage of lysosomal enzymes into the cytosol [47]. A second connection between CMA and AD has been recently proposed based on the finding that the regulator of calcineurin 1, a protein also implicated in the pathogenesis of this disease, undergoes CMA [48]. In contrast to the other pathogenic proteins described above, translocation of the regulator of calcineurin 1 and its degradation in lysosomes via CMA occurs efficiently. However, it is possible that reduced degradation of this protein, as a result of the described changes in CMA activity with age, might explain, at least in part, the aggravating effect that aging has in AD. Similarly, the age-dependent decline in CMA activity could be an aggravating factor in most of these neurodegenerative disorders [39].
CMA and lysosomal storage disorders
Lysosomal storage disorders (LSD) are a group of genetic diseases in which the lack of enzymatic activity in lysosomes leads to massive accumulation of the substrate of that enzyme, resulting in lysosomal engorgement [49]. The critical function of lysosomes as the terminal station where extracellular and intracellular components are delivered for degradation supports that all types of autophagy, including CMA, are secondarily altered to some extent in LSD. However, there are some LSD that directly affect CMA components. Abnormally high rates of CMA have been found in cells from patients with galactosialidosis, a LSD caused by mutations in the gene of protective protein/cathepsin A that acts as a chaperone for several glycosidases in lysosomes. Consequently, lysosomes from the affected cells accumulate oligosaccharides and glycopeptides. CMA is upregulated in this disease because protective protein/cathepsin A is required for the regulated cleavage of LAMP-2A [50].
Defective CMA has also been identified in patients with mucolipidosis type IV, an inherited LSD characterized by severe neurologic and ophthalmologic abnormalities caused by mutations in the transient receptor potential mucolipin 1 (TRPML1) gene. The primary function of TRPML1 remains unknown, but a possible role in pH maintenance has been proposed based on its ability to transport chloride molecules. The observed changes in CMA activity in the affected cells could be secondary to the changes in pH; however, because their lysosomes contain lower amounts of cytosolic chaperones, it is also possible that TRPML1 might participate in lysosomal docking of CMA chaperones [51].
Lastly, CMA is also likely to be altered in Danon’s disease, a type of vacuolar myopathy resulting from mutations in the LAMP2 gene [52]. However, the severe abnormalities observed in the lysosomal system in cells from these patients (accumulation of autophagic vacuoles and altered lysosomal biogenesis) - functions probably governed by the other LAMP-2 protein variants - prevents accurate measurement of CMA in these cells. Studies in livers of Lamp2 null mice, which reproduce Danon’s disease pathology, revealed a pronounced decrease in the degradation of long-lived proteins, supporting the observed severe alterations of the lysosomal system [33]. In contrast, proteolysis was normal in fibroblasts from embryos of these animals [34], suggesting that the functional defect in the lysosomal system may only become evident after birth.
CMA and kidney pathology
CMA activity is markedly high in kidney [10,53], and the first connection of CMA with disease was established in this organ. Study of a form of chemically induced hyaline droplet nephropathy revealed that CMA activity increased in the early stages of the disease to prevent the accumulation of alpha-2-microglobulin, a bona fide CMA substrate and the protein altered by the various chemical compounds that trigger this kidney condition [53]. However, as the disease progresses, CMA is not sufficient to accommodate the high levels of altered alpha-2-microglobulin which then starts to accumulate in kidney cells in the form of hyaline droplets. This kidney lesion often evolves to chronic progressive nephropathy and carcinogenesis.
Alterations in the degradation of selective proteins by CMA have also been linked to the pathogenesis of renal hypertrophy, characteristic of the diabetic kidney [54]. Decreased CMA activity under these conditions leads to high levels of the nuclear transcription factor Pax2, essential in regulating cellular growth in the kidney [54]. The reasons for the reduced levels of LAMP-2A observed in the diabetic kidney remain unknown.
Other pathologies
Altered macroautophagy has been described in infectious diseases and cancer. In light of the described contribution of CMA to antigen presentation, it is possible that CMA dysfunction underlies the basis of some immunoreactive and autoimmune disorders, but this aspect remains unexplored. We are also investigating now a possible relationship of CMA activity and oncogenesis. It is anticipated that the current development of easier methods to measure CMA in intact cells - without requiring the isolation of lysosomes - should allow the identification of abnormal CMA activity in more pathological conditions.
Concluding remarks and future perspectives
During the last decade a new appreciation for lysosomes and their multiple functions has emerged. In a short time, the mental image of this organelle has evolved from that of a mere “garbage disposal” to that of a sophisticated and active “recycling center”, a life-saver “energy boost for rainy days” and a complex “control panel” where cellular levels of specific proteins are regulated and the quality of proteins and organelles is controlled. The revival of the lysosomal system has lead to the realization that cargo delivery is not always “in bulk” and instead, can take place in a very selective manner. CMA is still the “champion” in lysosomal selectivity for the degradation of soluble proteins. This selectivity makes CMA suitable for removal of damaged or altered proteins or for the recycling of nonessential components to guarantee cellular survival when nutrients are scarce. These critical functions could explain why failure of CMA is associated with aging and age-related disorders. Despite the long list of remaining questions regarding this pathway (Box 4), the more we learn about CMA, the more this type of autophagy reveals itself as unique. From the selective motif in the substrates, the dual functions of the chaperones involved in this pathway, to the transient nature of the translocation machinery, there is always a new surprise waiting at the next curve of the CMA road.
Box 4. Outstanding questions.
The recent advances in our understanding of CMA and the identification of new components and functions for this autophagic pathway leave us now with a long list of unanswered questions:
How does HSC70 decide whether cargo proteins should be repaired or degraded by the proteasome or by CMA?
What is the role of the different co-chaperones known to associate with the HSC70/substrate complex?
Where does the force that drives translocation originate from?
What are the other components of the translocation machinery?
Why does LAMP-2A stability decrease with age?
What mediates the cross-talk between CMA and other autophagic pathways?
What determines the differences in levels of basal CMA among cell types?
Are basal and inducible CMA subject to the same regulation?
What signaling mechanisms contribute to the regulation of CMA?
More questions will arise as we learn about conditions and pathologies associated with altered CMA, although at the same time, these types of diseases are helping the field attain a better understanding of what the physiological functions of this pathway are as well as the consequences of a faulty CMA.
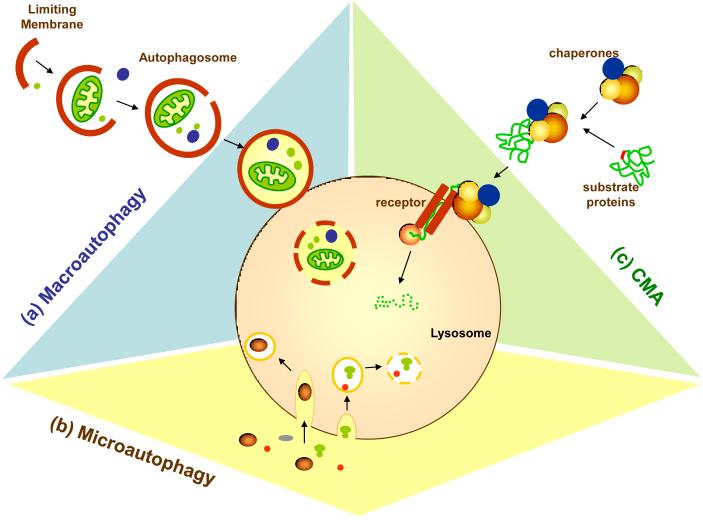
Figure I of Box 1. Types of autophagy in mammalian cells.
Three different main types of autophagy have been described in mammalian cells: (a) Macroautophagy, (b) Microautophagy, and (c) Chaperone-mediated autophagy (CMA).
Acknowledgements
I thank my numerous colleagues in the field of autophagy who through their animated discussions have helped shape this review. I am in particular debt to Dr. Susmita Kaushik and Ms. Samantha Orenstein for critically reading the manuscript. Work in our laboratory is supported by NIH grants from NIA (AG021904, AG031782), NIDKK (DK041918), NINDS (NS038370), a Glenn Foundation Award and a Hirsch/Weill-Caulier Career Scientist Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference list
- 1.De Duve C, Wattiaux R. Functions of lysosomes. Ann Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. Review. [DOI] [PubMed] [Google Scholar]
- 2.Mortimore G, Pösö A. Lysosomal pathways in hepatic protein degradation: Regulatory roles for amino acids. Fed Proc. 1984;43:1289–1294. [PubMed] [Google Scholar]
- 3.Mortimore GE, Poso AR. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annual Review of Nutrition. 1987;7:539–564. doi: 10.1146/annurev.nu.07.070187.002543. [DOI] [PubMed] [Google Scholar]
- 4.Ahlberg J, Glaumann H. Uptake--microautophagy--and degradation of exogenous proteins by isolated rat liver lysosomes. Effects of pH, ATP, and inhibitors of proteolysis. Exp Mol Pathol. 1985;42:78–88. doi: 10.1016/0014-4800(85)90020-6. [DOI] [PubMed] [Google Scholar]
- 5.Abeliovich H, et al. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Backer J, et al. Regulation of catabolism of microinjected ribonuclease A requires the amino-terminal 20 amino acids. Proc Natl Acad Sci USA. 1983;80:2166–2170. doi: 10.1073/pnas.80.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wing S, et al. Proteins containing peptide sequences related to KFERQ are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. Biochem J. 1991;275:165–169. doi: 10.1042/bj2750165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang H, Dice J. Peptide sequences that target proteins for enhanced degradation during serum withdrawal. J Biol Chem. 1988;263:6797–6803. [PubMed] [Google Scholar]
- 12.Chiang H, et al. A role for a 70 kDa heat shock protein in lysosomal degradation of intracellular protein. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 13.Dice J. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 14.Hohfeld J, et al. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Reports. 2001;2:885–890. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin Y, et al. The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 16.Cuervo AM, et al. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J Biol Chem. 1994;269:26374–26380. [PubMed] [Google Scholar]
- 17.Salvador N, et al. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. Journal of Biological Chemistry. 2000;275:27447–27456. doi: 10.1074/jbc.M001394200. [DOI] [PubMed] [Google Scholar]
- 18.Cuervo A, Dice J. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 19.Cuervo A, Dice J. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 20.Bandyopadhyay U, et al. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaushik S, et al. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agarraberes F, et al. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–834. doi: 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuervo AM, et al. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 24.Cuervo A, et al. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 25.Massey AC, et al. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Nat Acad Sci USA. 2006;103:5905–5910. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finn P, et al. Effects of small molecules on chaperone-mediated autophagy. Autophagy. 2005;1:141–145. doi: 10.4161/auto.1.3.2000. [DOI] [PubMed] [Google Scholar]
- 27.Kaushik S, et al. Constitutive Activation of Chaperone-mediated Autophagy in Cells with Impaired Macroautophagy. Mol Biol Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiffin R, et al. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn PF, Dice JF. Ketone bodies stimulate chaperone-mediated autophagy. J Biol Chem. 2005;280:25864–25870. doi: 10.1074/jbc.M502456200. [DOI] [PubMed] [Google Scholar]
- 30.Zhou D, et al. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 2005;22:571–581. doi: 10.1016/j.immuni.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Yang Q, et al. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franch H, et al. A mechanism regulating proteolysis of specific proteins during renal tubular cell growth. J Biol Chem. 2001;276:19126–19131. doi: 10.1074/jbc.M101777200. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka Y, et al. Accumulation of autophagic vacuoles and cardiomyopathy in Lamp-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 34.Eskelinen E, et al. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol Biol Cell. 2004;15:3132–3145. doi: 10.1091/mbc.E04-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortunato F, Kroemer G. Impaired autophagosome-lysosome fusion in the pathogenesis of pancreatitis. Autophagy. 2009;5:850–853. doi: 10.4161/auto.8839. [DOI] [PubMed] [Google Scholar]
- 36.Dice J. Altered degradation of proteins microinjected into senescent human fibroblasts. J Biol Chem. 1982;257:14624–14627. [PubMed] [Google Scholar]
- 37.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 38.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat. Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiffin R, et al. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci. 2007;120:782–791. doi: 10.1242/jcs.001073. [DOI] [PubMed] [Google Scholar]
- 41.Marino G, et al. Premature aging in mice activates a systemic metabolic response involving autophagy induction. Hum Mol Genet. 2008;17:2196–2211. doi: 10.1093/hmg/ddn120. [DOI] [PubMed] [Google Scholar]
- 42.Massey A, et al. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- 43.Cuervo AM, et al. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 44.Vogiatzi T, et al. Wild type alpha-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. J Biol Chem. 2008;283:23542–23556. doi: 10.1074/jbc.M801992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez-Vicente M, et al. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kabuta T, et al. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem. 2008;283:23731–23738. doi: 10.1074/jbc.M801918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, et al. Tau Fragmentation, Aggregation and Clearance: the Dual Role of Lysosomal Processing. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp367. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H, et al. Degradation of regulator of calcineurin 1 (RCAN1) is mediated by both chaperone-mediated autophagy and ubiquitin proteasome pathways. FASEB J. 2009 doi: 10.1096/fj.09-134296. In press. [DOI] [PubMed] [Google Scholar]
- 49.Wenger D, et al. Lysosomal storage disorders: diagnostic dilemmas and prospects for therapy. Genet Med. 2002;4:412–419. doi: 10.1097/00125817-200211000-00003. [DOI] [PubMed] [Google Scholar]
- 50.Cuervo AM, et al. Cathepsin A regulates chaperone-mediated autophagy through cleavage of the lysosomal receptor. EMBO J. 2003;22:12–19. doi: 10.1093/emboj/cdg002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venugopal B, et al. Chaperone-mediated autophagy is defective in mucolipidosis type IV. J Cell Physiol. 2009;219:344–353. doi: 10.1002/jcp.21676. [DOI] [PubMed] [Google Scholar]
- 52.Nishino I, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 53.Cuervo A, et al. Direct lysosomal uptake of alpha2-microglobulin contributes to chemically induced nephropathy. Kidney Int. 1999;55:529–545. doi: 10.1046/j.1523-1755.1999.00268.x. [DOI] [PubMed] [Google Scholar]
- 54.Sooparb S, Price SR, Shaoguang J, Franch HA. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 2004;65:2135–2144. doi: 10.1111/j.1523-1755.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 55.Tuttle DL, Dunn WA., Jr. Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. Journal of Cell Science. 1995;108:25–35. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- 56.Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 2009;16:70–78. doi: 10.1038/cdd.2008.168. [DOI] [PubMed] [Google Scholar]
- 57.Dice J. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 58.Massey A, et al. Pathophysiology of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2420–2434. doi: 10.1016/j.biocel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 59.Shen S, et al. Cyclodepsipeptide toxin promotes the degradation of Hsp90 client proteins through chaperone-mediated autophagy. J Cell Biol. 2009;185:629–639. doi: 10.1083/jcb.200810183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cuervo AM, et al. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 61.Eskelinen EL, et al. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–145. doi: 10.1016/s0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 62.Cuervo A, Dice J. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000;113:4441–4450. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]