Abstract
The inability to forecast outcomes for malignant mesothelioma prevents clinicians from providing aggressive multimodality therapy to the most appropriate individuals who may benefit from such an approach. We investigated whether specific microRNAs (miRs) could segregate a largely surgically-treated group of mesotheliomas into good or bad prognosis categories. A training set of 44 and a test set of 98 mesothelioma tumors were analyzed by a custom microRNA platform, along with 9 mesothelioma cell lines and 3 normal mesothelial lines. Functional implications as well as downstream targets of potential prognostic microRNAs were investigated. In both the training and test sets, hsa-miR-29c* was an independent prognostic factor for time to progression as well as survival after surgical cytoreduction. The miR was expressed at higher levels in epithelial mesothelioma, and the level of this miR could segregate patients with this histology into groups with differing prognosis. Increased expression of hsa-miR-29c* predicted a more favorable prognosis, and overexpression of the miR in mesothelioma cell lines resulted in significantly decreased proliferation, migration, invasion, and colony formation. Moreover, major epigenetic regulation of mesothelioma is mediated by hsa-miR-29c* and was demonstrated through downregulation of DNA methyltransferases as well as upregulation of demethylating genes. A single microRNA has the potential to be a prognostic biomarker in mesothelioma, and validation of these findings as well as investigation of its downstream targets may give insight for potential therapies in the future.
Introduction
Malignant Pleural Mesothelioma (MPM) is a lethal, asbestos related cancer with numerous genomic abnormalities(1). MicroRNAs are short (17–22 nucleotides) non-coding RNAs that regulate gene expression by inhibition of translation, and play a major role in oncogenesis(2). Their exceptional tissue specificity has made them potent biomarkers for diagnosing the tissue source of metastatic cancers(3, 4), and microRNAs have also been reported as prognostic markers for a multitude of cancers including ovarian(5), pancreatic(6), lung(7), and breast(8). We have identified a member of microRNA family 29, hsa-miR-29c*, as an independent predictor of time to progression as well as overall survival for MPM using a custom microRNA microarray, with validation by qRT-PCR. Functional assays reveal profound changes in cell proliferation, invasion, migration, and colony formation after hsa-miR-29c* cell line transfection, and we postulate that miR-29c* is integral for the balance between methylation and demethylation events in MPM. The Rosetta Genomics microRNA platform was used since it was the most technologically advanced at the time of study inception, containing probes for ~900 microRNAs which could differentiate microRNA family members differing by only one base. In addition, a highly sensitive qRT-PCR for each microRNA was available which was so specific that one mismatch would result in a complete signal loss.
Methods
Mesothelioma tumors
One hundred and forty-two snap frozen, immunohistochemically-proven (positive staining for WT1 [Wilms Tumor Antigen], cytokeratins, calretinin, and the absence of staining for CEA [carcinoembryonic antigen] and B72.3) MPM surgical specimens were collected from 1990–2005. Age, sex, stage, histology, time to progression from surgery (TTP), and time to death from surgery were recorded and current as of September 2008 (Table 1).
Table I.
Patient Demographics
| Training Set (n=37) | Test Set (n=92) | |
|---|---|---|
| Age, years (range) | 63.1 (43–78) | 61.6 (34–87) |
| Sex | 9F/28M (76%) | 16F/76M (83%) |
| Stage | I/II:10 (27%); III/IV: 27 | I/II: 29 (32%); III/IV:63 |
| Histology | 23 Epithelial (62%); 14 non-Epithelial | 58E (63%);34 non-Epithelial |
| Cytoreductive Surgery | 34 Yes(92%); 3 No | 83 Yes (90%);9 No |
RNA extraction
Total RNA was extracted from the tumors and from MPM cell lines HP1, HP3, H2373, H2452, H2591, H2595, H2596, and H2461(9), and tert immortalized mesothelial cell line LP9 (10), primary mesothelial cell culture NYU-590.2 (passage 3), and SV40 transformed mesothelial cell line Met5A . For each tumor sample approximately 0.5cm3 in dimension was used for RNA extraction. Total RNA was extracted using the miRvana miRNA isolation kit (Ambion) according to the manufacturer’s instructions. Briefly, the sample was homogenized in a denaturing lysis solution followed by an acid-phenol:chloroform extraction. Finally, the sample was purified on a glass-fiber filter. Total RNA quantity and quality was checked by spectrophotometer (Nanodrop ND-1000). Of the tumor specimens, 129/142 (37/44 training set, 92/98 test set, 91%) satisfied quality assurance for microarray analyses.
MicroRNA microarray
Custom microRNA microarrays were prepared as described previously(3). Briefly, ~900 DNA oligonucleotide probes representing microRNAs were spotted in triplicate on coated microarray slides (Nexterion® Slide E, Schott, Mainz, Germany).
Microarray data analysis and statistics
Recent publications have reported median TTP and survival after cytoreductive surgery as 8–12 months, and 12–20 months, respectively(11, 12). Therefore, in this report, a good prognosis group was defined as a TTP greater than or equal to 12 months or survival from the time of surgery greater than or equal to 15 months. Univariate survival analysis was performed by the Kaplan Meier method and evaluated by log-rank, with significance at p less than 0.05. Only microRNAs with a median signal higher than signal background levels were compared using the Mann-Whitney non-parametric test. Corrections for multiple comparisons were performed using the Benjamini-Hochberg “False Discovery Rate” (FDR) method(13). Group separation (bad or good) was based on the median value of microRNA expression for the group.
qRT-PCR
16 samples, 8 with good prognosis and 8 with poor prognosis were selected for validation using quantitative real-time PCR (qRT-PCR) analysis. The qRT-PCR analysis was performed for all microRNAs besides mir 29c* which came out as differential in the microarray experiments and for 5 microRNAs that were chosen for normalization as described below. We defined a microRNA as differential if it met one of the following criteria: (i) p < 0.05 and fold change between the medians > 1.5 when comparing the good and bad TTP prognosis groups in either the training or test set; or (ii) if the microRNA had the same criteria when considering only the 16 samples chosen for qRT-PCR analysis; or (iii)a non significant p-value but a fold change > 1.5 at a very high expression level (accommodating potential saturation) in either the training or test set. MicroRNAs which bore strong sequence similarity to others previously selected by these criteria were excluded. These criteria resulted in 7 differential microRNAs: hsa-miR-451, hsa-miR-99a, hsa-miR-150, hsa-miR-29c*, hsa-miR-199b-5p, hsa-miR-210, and hsa-miR-221. In addition, 5 microRNAs (hsa-miR-181a, hsa-let-7c, hsa-miR-193a-5p, hsa-miR-27b, and hsa-miR-339-5p) which had very low fold change in all datasets, fairly low dispersion and spanning expression levels 500–50000 fluorescence units, were chosen as normalizers. MicroRNA amounts were quantified using a qRT-PCR method recently described(4). The cycle threshold (CT, the PCR cycle at which probe signal reaches the threshold) was determined for each well. Each CT was normalized by subtracting the mean of all 12 microRNAs for that sample and scaled so that the mean expression for each sample was a constant. To allow comparison with results from the microarray, each value received was subtracted from 50. This 50-CT (50CT) expression for each microRNA for each patient was compared with the signal obtained by the microarray method. Linear regression for the microRNA readings over all patients was used to model 50CT by microarray values. Using this model, the threshold for the separation between high and low expression samples was transferred from microarray to qRT-PCR readings and used for Kaplan-Meier analysis.
In vitro Functional Analyses of miR-29c*
miRNA-oligonucleotides & transfection
Pre-miR™ miRNA mimic (miR-29c*) and scrambled oligonucleotides (Ambion, Austin, TX) were complexed with Lipofectamine 2000 for transfection into H2595 and HP-1. Forty-eight hours later, cells were trypsinized, counted and assayed for proliferation, soft agar colony formation, wound closure, and matrigel invasion in triplicate experiments.
Stem-loop RT-PCR
Stem-loop technology(14) in its end-point PCR modification(15) was used to validate miR-29c* delivery to the cells. We designed a stem-loop RT primer to anneal to the 3′-end of miR-29c* and ensure extension of the product size during the PCR stage: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGAACACCAGG-3′. In the PCR stage, we used a miR-29c*-specific forward primer 5′-TGACCGATTTCTCCTGG-3′ and a stem-loop-specific oligonucleotide 5′-GTGCAGGGTCCGAGGT-3′ as a reverse primer. RT-PCR was performed on total miR-containing RNA isolated from cultured MM cells using pulsed RT and PCR protocols.
Analysis of Potential Targets of mir-29c*
MiRanda™ was used to predict microRNA target sites (16). RNAs from an additional 30 tumor specimens and matching normal peritoneum (easily harvested from the abdomen during MPM operations as normal mesothelium without contamination by tumor cells) to tumors were used for differential gene expression of DNA methyltransferase 3A (DNMT3A) using the Affymetrix U133 2.0 Plus platform. (17). A separate set of 7 matched tumor-normal MPM pairs were used to validate the Affymetrix DNMT3A data by qRT-PCR using specially designed primer pairs. The effect of hsa-mir-29c*overexpression on the DNMTs, C1QTNF1, C1QTNF8, and adiponectin from RNA derived from the transfection experiments was assessed by qRT-PCR using specially designed primer pairs (See Supplementary Material Table 1).
Statistical analysis
All assays were performed in triplicate and statistical evaluations performed using the SPSS software package (bivariate correlations-Pearson coefficient and T-tests). All values in the text and figures represent the means ± SD.
Results
Mesothelioma Tumor Training Set: Time to Progression
Median TTP in the training set was 7 months. In a multivariate analysis for the training set, stage was the only independent demographic or clinical predictor of time to progression (Hazard Ratio 3.8, p=0.0099, CI: 1.3–10.2).
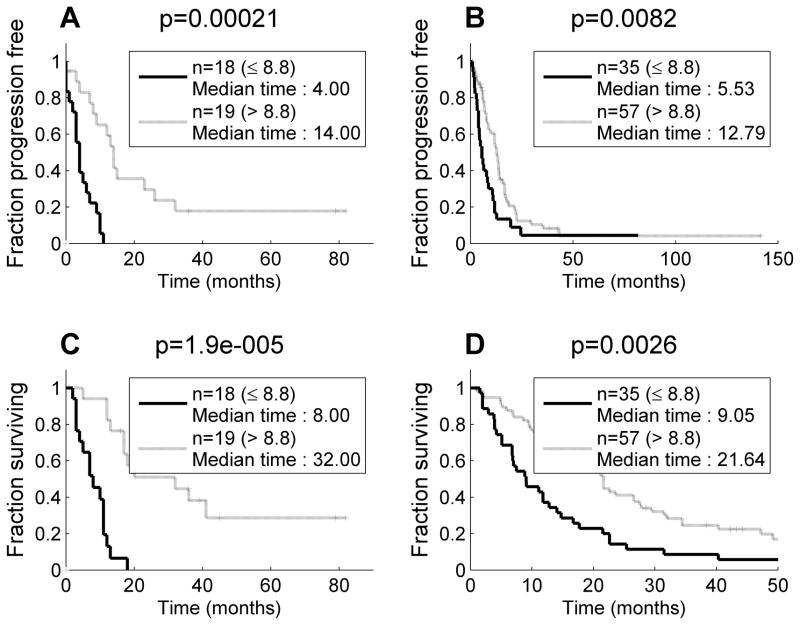
Only hsa-miR-29c* had significantly different expression (p = 0.00036, fold change 1.8) after adjusting for the false discovery rate (FDR) for the training set comparing good prognosis patients (TTP ≥ 12 months, n=10) to those with TTP < 12 months (n=24). When median expression of hsa-miR-29c* (8.8) was used as a cutoff, two groups with significantly different TTP were seen (4 months vs. 14 months, p=0.0002) (Figure 1A). Elevated hsa-miR-29c* was associated with an increased TTP.
Figure 1.
Elevated hsa-miR-29c* was associated with a significantly longer time to progression and survival time (A) Training set: time to progression (B) Training set: survival time. (C) Test set: time to progression (D) Test set: survival time
In the training set, hsa-miR-29c* expression did not differ between Stage I/II and Stage III/IV. Within each of these groups however, the expression levels of hsa-miR-29c* maintained an association with progression-free survival which was significant for Stages I/II (p=0.0031) with a trend for samples of Stages III/IV (p=0.068).
Mesothelioma Tumor Training Set: Survival
The median survival for the training set was 12 months. In multivariate analysis, stage was the only independent demographic or clinical predictor of survival (Hazard Ratio 2.6, p=0.04, CI:1.0–6.5). Hsa-miR-29c* was the best discriminator (p=0.006, fold change 1.7) for survival of the training set. When median expression of hsa-miR-29c* was used as the cut-off point, two distinct groups of MPM patients with significantly different survival curves were seen (median survival of 8 months vs. 32 months, p=0.000019, Figure 1B).
Mesothelioma Tumor Test Set: Time to Progression
The training set and the test set (Table I) were similar with regard to sex, age, histology, stage, and treatment options. Median TTP in the test set was 9.8 months. In multivariate analysis, stage was the only independent predictor of TTP (Hazard Ratio 3.8, p=0.000012, CI: 2.1–7.1) and survival (Hazard Ratio 3.1, p=0.00016, 1.7–5.5).
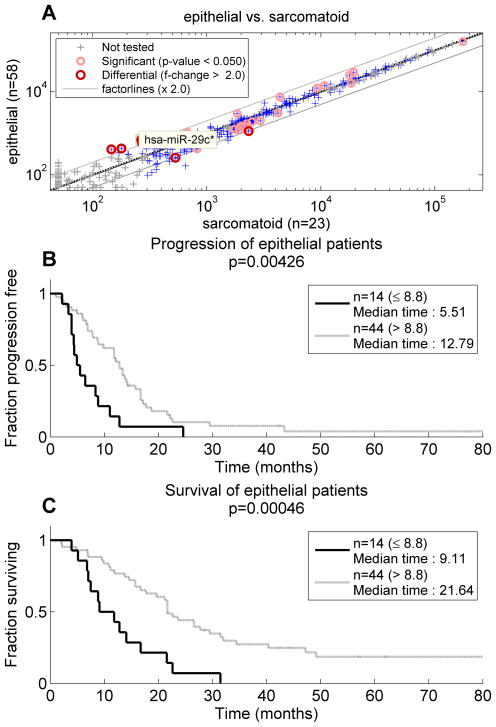
When microRNAs were investigated in the test set comparing good (TTP ≥ 12 months, n=33) to bad prognosis (TTP < 12 months, n=50) patients, hsa-miR-29c* was again significant, surpassing FDR=0.1 with p= 7.5E-005. Other microRNAs were also differential in the test set including hsa-miR-451, hsa-miR-99a, hsa-miR-150, hsa-miR-199b-5p, hsa-miR-210, and hsa-miR-221, but none of these 6 were differential in the training set. When median expression of hsa-miR-29c* from the training set (8.8) was used as a cutoff (Figure 1C), two groups in the test set with significantly different TTP were seen (5.5 months vs 12.8 months, p=0.008). In the test set, hsa-miR-29c* was elevated in epithelial tumors (Figure 2A, fold change =2.63; p=0.0005). Although histology itself was not a predictor of time to progression (p=0.69), expression of hsa-miR-29c* could separate the epithelial group of mesotheliomas into two categories with significantly different TTP (Figure 2B, p=0.0043) using the predetermined threshold for hsa-miR-29c* expression.
Figure 2.
Histology and mesothelioma prognosis influenced by mir-29c*. (A) hsa-miR-29c* was differentially expressed between epithelial and biphasic or sarcomatoid cases with higher levels in the former (B) hsa-mir-29c* could subclassify epithelial mesotheliomas into those with short versus long time to progression.
(C) hsa-mir-29c* could subclassify epithelial mesotheliomas into those with short versus long survival times.
Mesothelioma tumor Test Set: Survival
The median overall survival for the test set was 16 months, and due to the larger size of the test set, a significant difference in survival (p=0.03) favoring epithelial histology patients was seen. Hsa-miR-29c* upregulation was associated with good prognosis (survival ≥15 months, p=0.0014, hazard ratio 1.83). The threshold of expression (8.8) determined in the training set separated the patients in the test set into two groups with median survival times of 9.1 months vs. 21.6 months (p=0.0026, Figure 1D). Moreover, as in the TTP analysis, hsa-miR-29c* could separate the epithelial mesotheliomas into a good prognosis group (median survival, 21.6 months) and a poor prognosis group (median survival, 9.1 months, p=0.00046, (Figure 2C.)
In a Cox multivariate model of stage, gender, lymph node involvement, histological type and hsa-miR-29c* expression for the test set, we found that stage, hsa-miR-29c* expression, and lymph node status were independent predictors of survival (p<0.0001, p=0.0187, and 0.0001, respectively), while gender and histological type were not predictive (p>0.05). When all 129 patients in the analysis were considered in Cox multivariate analysis, stage, hsa-miR-29c* expression, and lymph node status remained the only independent predictors of survival (p<0.0001, p< 0.0001, and p<0.0001, respectively).
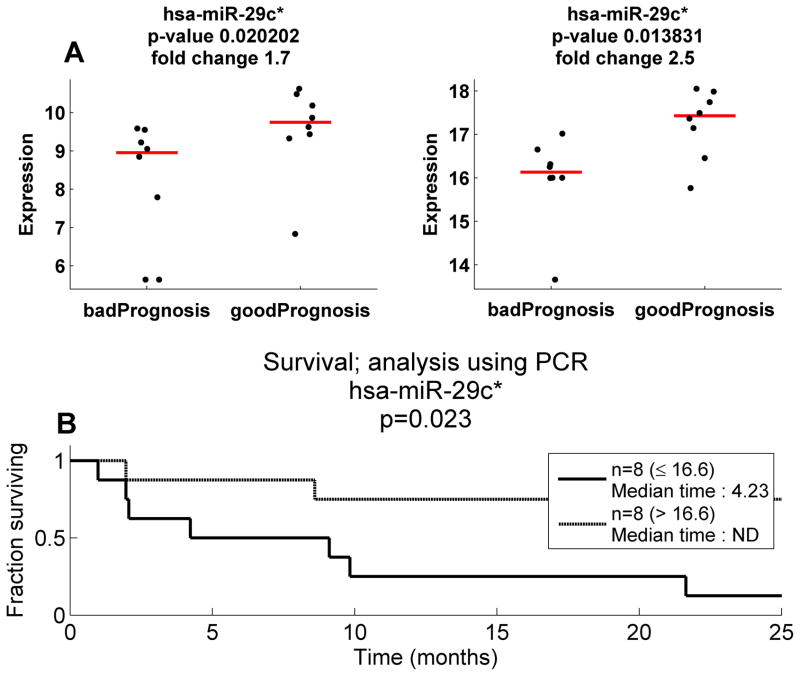
PCR Validation of hsa-miR-29c* expression
PCR validation of microarray expression was achieved for all differential and normalizer microRNAs evaluated with r values ranging from 0.46 to 0.79. However, none of these miRs with the exception of hsa-mir-29c* were capable of separating good versus bad prognosis by both PCR and microarray. Hsa-miR-29c* was significantly higher in the good prognosis group for both the custom array and the PCR (Figure 3A), and the correlation coefficient between microarray expression and PCR was 0.649, and using the mathematical regression line, the training/test set separation expression of 8.8 correlated with a 50-CT measurement of 16.6. The qRT-PCR cutoff divided the 16 MPMs into a good and bad prognosis group (median survivals, 4.2 months vs >10 years, P=0.02) (Figure 3B). These data, validating hsa-mir-29c* as the only common prognostic microRNA in both the training set and the test set, as well as its ability to achieve prognostic significance on both platforms, determine that hsa-mir-29c* is the sole candidate prognostic microRNA in these investigations of MPM.
Figure 3.
Validation of prognostic potential of hsa-miR-29c* using qRT-PCR (A) left panel reveals microarray expression values for hsa-mir-29c* in 16 mesotheliomas compared to expression measured by qRT-PCR (right panel). In both cases, elevated hsa-mir-29c* was associated with good prognosis. (B) qRT-PCR for hsa-mir-29c* separating the validation individuals significantly by their survival at a cut-off comparable to the consistent cutoff used for the microarray data.
Hsa-mir-29c* and Asbestos History
For the entire set of 129 patients, the mean expression value of hsa-mir-29c* for 24 patients without exposure (9.2) was not significantly different from those with a history of asbestos exposure (8.7), and there was an insignificant trend for prolonged survival for unexposed patients compared to patients with exposure to asbestos (median survivals, 27 months vs. 13 months, p=0.11). When asbestos exposure was stratified by expression of mir29c*, however, elevated expression of the mir (at a cutoff of 8.8) was associated with significantly prolonged survival in both exposed and unexposed MPM patients (exposed individuals, 20 vs 9 months, p=0.00028; unexposed individuals, 31 vs 11 months, p = 0.0012). These data reinforce that high expression of has-mir-29c* is independently associated with a better prognosis in MPM, irregardless of asbestos exposure.
MicroRNA 29 family analysis in Mesothelial Cell Lines
Seven members of the mir-29 family were analyzed for their expression in nine MPM and three mesothelial cell lines. Low levels of mir-29c* were detected in the three normal mesothelial cells. The actual levels of measured mir-29c* were 1.8 fold higher in the normal cell lines compared to MPM (p=0.05).
Biological Characterization of miR-29c*
The hsa-miR 29c* mimic was transfected into cell lines H2595 and HP1. Hsa-miR-29c* overexpression was associated with decreased proliferation, decreased migration, as well as a significant decrease in the invasive capacity of both cell lines (Figure 4). Colony formation was significantly inhibited with transfection of miR- 29c* resulting in smaller and fewer colonies, with the most impressive results recorded with H-2595. These data are compatible with the clinical findings that MPMs with higher expression of miR-29c* are less aggressive than those with low expression of the miR.
Figure 4.
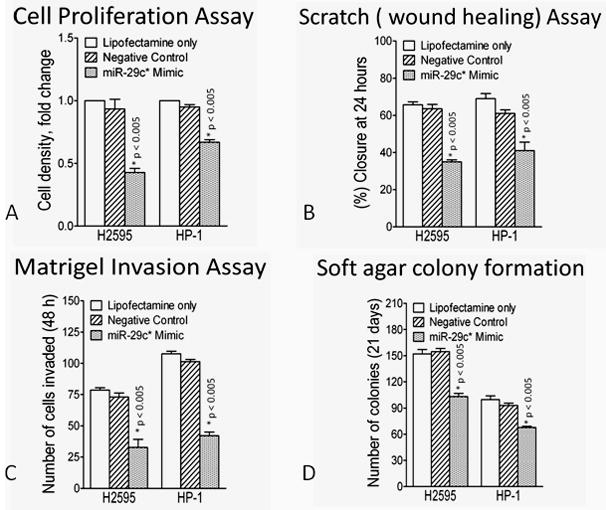
Functional consequences of hsa-miR-29c* overexpression in mesothelioma cell lines compared to lipofectamine alone or irrelevant (scrambled) miR. (A) hsa-miR-29c* overexpression resulted in a significant decrease in cell proliferation (B) hsa-miR-29c* overexpression resulted in a significant decrease in cellular migration (C) hsa-miR-29c* overexpression resulted in a significant decrease in invasion (D) hsa-miR-29c* overexpression resulted in a significant decrease in soft agar colony formation
Targets of miR-29c*
Previous data have associated the miR-29c family with methyltransferases(18). Analysis of our U133 Plus 2 expression arrays revealed that DNMT3A was elevated significantly in MPMs (Figure 5A) and these data were validated in matched normal tumor pairs from our archives (Figure 5B). When miR-29c* mimic was transfected into H2595 and HP1, both cell lines exhibited significantly decreased expression of DNMT3A compared to controls (Figure 5C).
Figure 5.
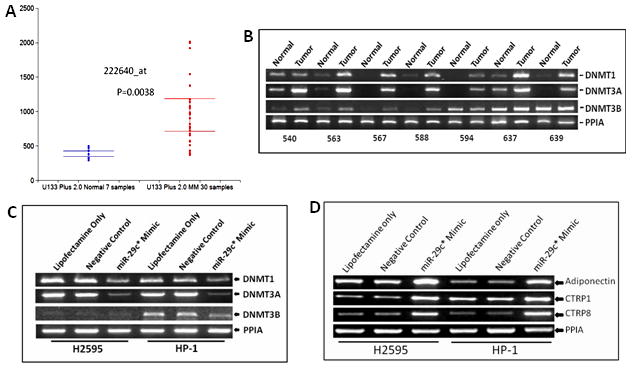
hsa-miR-29c* regulates epigenetic gene expression in mesothelioma (A) DNA methyltransferase 1 expression measured by U133Plus 2 Affymetrix microarray platform is elevated in mesothelioma compared to normal mesothelium from the peritoneum (B) RT-PCR validates overexpression of all of the DNMTs in mesothelioma compared to matching normal peritoneum. 18/21 specimen revealed overexpression of a DNA methyltransferase in the tumor. (C) Transfection of hsa-miR-29c* resulted in decreased expression of all DNA methyltransferases in the two cell lines.. H2595 did not have expression of DNMT3B (D) Transfection of hsa-miR-29c* upregulated Adiponectin, C1QTNF1 and C1QTNF8, three genes associated with active gene demethylation. C1QTNF8 was thus validated as a target of hsa-miR-29c*.
One of the key targets for hsa-mir-29c* by MiRanda™ with the highest significance (p=1×10−6) was C1q and TNF related protein 8 (CTRP8). This molecule belongs to a family of proteins characterized by a common TNF alpha–like globular domain. A member of this proteinfamily includes adiponectin, a major insulin-sensitizing multimeric hormone. Liu et al(19) reported that adiponectin regulates methyltransferases by decreasing the methylation of CpG islands located within the proximal promoter region of WIF1 by inhibition of specificity protein 1 (Sp1) transcription factor and its downstream target DNA methyltransferase 1. We postulated that mir-29c* may control methylation-demethylation mechanisms in MPM and investigated the effect of overexpression of mir-29c* on adiponectin, CTRP1 and CTRP8 levels. As seen in Figure 4D, mir-29c* overexpression increased the levels of all three of these genes, further linking this microRNA to genes controlling methylation in MPM.
Discussion
We report here that the presence of a single microRNA has significant prognostic implications for MPM. Using a proprietary microarray capable of analyzing over 900 microRNAs, hsa-miR-29c* was not only able to separate MPM patients by time to progression after surgery, but also stratified these patients by their survival. This microRNA is preferentially expressed in epithelial MPM, and further stratifies this group into favorable and unavorable cohorts. Moreover, the prognostic impact on survival of the microRNA was independent of histology in a multivariate analysis. Validation of miR-29c* was accomplished by microarray in an independent test set and by using qRT-PCR. The results of the in vitro functional assays in which the miR is expressed in MM cell lines are entirely consistent with the clinical findings that the expression of this miR decreases the tumors proliferative, migration and invasive potential.
Few papers describe the expression or role of microRNAs in MPM. Guled et al(20) were the first to profile 17 snap frozen mesothelioma specimens along with pericardium as “control” mesothelium. Twelve microRNAs were exclusively expressed in mesotheliomas. Insightful discussion pointed to important MiRanda™ predicted targets including those known to be involved with MPM pathogenesis (HGF, neurofibromatosis gene 1, and jun). We did not find any of the microRNAs cited as significant by the Guled group; however, this is not surprising given the difference in the “control” samples. Most recently, Busacca (21) reported differences in miR expression between two mesothelioma cell lines and a tert transformed mesothelial cell culture, as well as in 24 mesothelioma specimens. This elegant study was characterized by validation of the candidate microRNAs, and, similar to Guled, used informatics algorithms to predict targets of the miRs which may be relevant for MPM. This group reported that the expression of miR-17-5p, miR-21, miR-29a, miR-30c, miR-30e-5p, miR-106a and miR-143 was significantly associated with the histopathological subtypes and that the reduced expression of two microRNAs (miR-17-5p and miR-30c) correlated with better survival of patients with sarcomatoid subtype.
Downregulation of the miR-29 family has been reported in chronic lymphocytic leukemia(22), lung cancer(18, 23), and nasopharyngeal cancer, and a possible role of miR-29 as a tumor suppressor has been hypothesized. We have validated the findings of Fabbri et al that the 29c family of miRs has a direct effect on DNMTs (18) and demonstrated that this family of miRs also influences a demethylation pathway involving adiponectin, CTRP1, and CTRP8. We have noted that mir-29c* levels are elevated in epithelial mesothelioma compared to sarcomatoid, and it is well known that epithelial histology is associated with longer survival in MPM. Moreover, mir-29c* was noted to have higher expression in mesothelial cell lines than in MPM cell lines, implying that the microRNA is associated with a more differentiated phenotype. We also were able to demonstrate that the levels of the microRNA were able to subcategorize epithelial histology into good risk and bad risk cohorts, implying that lower levels of the microRNA would potentially impart a more “non-epithelial” like natural history for the patient. Since the mir seems to be associated with epigenetic surveillance, one could ask whether this epithelial/non-epithelial survival difference is on the basis of methylation “homeostasis.” In the literature, there has been no convincing evidence that the degree of methylation differs between epithelial and non-epithelial histology. If this were correct, then, given an equal degree of methylation, one would speculate that the ability of epithelial histology to have greater epigenetic control of methylation and demethylation (i.e. greater expression of has-mir-29c*) may indeed provide survival advantage for this histotype over the others with lower expression of the mir. Moreover, epithelial mesenchymal transition (EMT) has been under study in mesothelioma(24), and non-epithelial mesotheliomas exhibit a more mesenchymal-type molecular phenotyping with poorer survivals. In fact, the mir-29 family itself has been associated with EMT in limited investigations (23).
Our findings that elevated levels of miR-29c* are associated with greater survival and longer times to progression in mesothelioma are compatible with this miR having an effect on gene methylation(25–28) and other pathways that are known to be upregulated in MPM. These pathways include, among others, the PI3 Kinase pathway(29, 30), NfKB(31–33), apoptosis(34), and immunity(35). In myoblasts, miR-29 is repressed by NF-kB, and in primary rhabdomyosarcomas that possess impaired differentiation, miR-29 is epigenetically silenced by an activated NF-kB pathway. Reconstitution of miR-29 in this system inhibits tumor growth and stimulates differentiation, suggesting that miR-29 acts as a tumor suppressor through its promyogenic function. Since NF-kB is known to be elevated in MPM and potentially contribute to chemoresistance, these results in rhadomyosarcoma could imply that a similar NF-kB miR-29 regulatory circuit may be present. Apoptotic mechanisms in MPM are evident with the reports of significant differences in abnormal expression of apoptosis proteins between epithelial and sarcomatoid MPM including overexpression of MCL1, an anti-apoptotic member of the Bcl-2 family of proteins, in 92% of the specimens(36). Pertinent to the present discussion, the mir 29 family appears to regulate MCL1 since forced expression of mir-29b in cholangiocarcinoma cells reduced Mcl-1 protein expression and sensitized the cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) cytotoxicity. Aberrant immune surveillance mechanisms have also been described in mesothelioma including those involving co-stimulatory molecules of the B7 family (B7-1 and B7-2) which play a key role in antigen-specific T-cell activation via their interaction with the counter-receptor CD28(37, 38). Human B7-H3 (also named as CD276) is a member of the B7/CD28 immunoglobulin superfamily, and specifically inhibits natural killer cells and T cells. Most recently, B7-H3 protein expression was found to be inversely correlated with miR-29 levels in cell lines and tumor tissues, and knock-in and knockdown of miR-29a results in down-and up-regulation, respectively, of B7-H3 protein expression(39). Further study, specifically of the ability hsa-mir-29c* to control B7-H3 protein expression could have implications in immune escape by mesothelioma..
Many of the aforementioned pathways are directly associated with asbestos fiber exposure on mesothelial cells(40, 41). One of the primary methods for genetic damage by asbestos involves promoter methylation, and various genes have been found to be methylated in mesothelioma at varying frequencies(42). Data are not consistent regarding the effect of asbestos exposure and the prognosis of MPM (43–46), and in the present study, there was neither a difference in survival nor mean expression of hsa-mir-29c* between MPM patients with or without a history of asbestos exposure. Nonetheless, high expression of the microRNA was able to significantly increase survival within both the asbestos and non-asbestos exposed cohorts. One could hypothesize that since the degree of methylation may be the least in individuals with no history of asbestos exposure, the efficacy of the microRNA could be the greatest since the degree of methylation is lowest. Validation of these prognostic findings in larger cohorts of MPM patients with precise asbestos exposure, or ideally, with pateints who have had lung fiber analysis will be necessary in the future.
These data point to important control of carcinogenic mechanisms in MPM by selected microRNAs, with the ability of such microRNAs to prognosticate patients with surgically cytoreducible MPM. Investigations which decipher the complexity and integration of miR-29c* with other pathways involved in MPM should be explored because of the potential therapeutic implications these studies might reveal. Our findings must not only be validated in other surgical cohorts, but in patients who are treated in non-surgical ways (i.e. chemotherapy, targeted therapies) to see whether hsa-mir-29c* retains prognostic (and possibly predictive) power as well. A significant advantage of microRNA exploration is that the studies can be performed in formalin fixed paraffin embedded tissues, thus simplifying specimen procurement in the future.
Supplementary Material
Acknowledgments
This project was supported by an NCI/NIH EDRN Biomarker Discovery Laboratory Grant UO1CA1111295 to Dr. Pass, philanthropy from Belluck and Fox, LPA and from the Stephen Banner Lung Cancer Foundation. The work represents a collaboration between NYU Langone Medical Center, Rosetta Genomics, and other participating academic centers. Dr. Pass is Vice Chairman of the Medical Advisory Board for Rosetta Genomics and receives consultation fees for his services. MicroRNA microarrays and PCR validations were performed by Rosetta Genomics, along with bioinformatics analyses. Stem loop amplifications of hsa-mir-29c*, all functional assays, U133Plus2 analyses, and resulting PCR validations were performed by the Thoracic Surgery Laboratory, NYU Langone Medical Center.
Reference List
- 1.Kaufman AJ, Pass HI. Current concepts in malignant pleural mesothelioma. Expert Rev Anticancer Ther. 2008;8:293–303. doi: 10.1586/14737140.8.2.293. [DOI] [PubMed] [Google Scholar]
- 2.Bentwich I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005;579:5904–10. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 3.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–9. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 4.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eitan R, Kushnir M, Lithwick-Yanai G, David MB, Hoshen M, Glezerman M, et al. Tumor microRNA expression patterns associated with resistance to platinum based chemotherapy and survival in ovarian cancer patients. Gynecol Oncol. 2009;114:253–9. doi: 10.1016/j.ygyno.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 6.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–84. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 7.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Lowery AJ, Miller N, Devaney A, McNeill RE, Davoren PA, Lemetre C, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009;11:R27. doi: 10.1186/bcr2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pass HI, Stevens EJ, Oie H, Tsokos MG, Abati AD, Fetsch PA, et al. Characteristics of nine newly derived mesothelioma cell lines. Ann Thorac Surg. 1995;59:835–44. doi: 10.1016/0003-4975(95)00045-m. [DOI] [PubMed] [Google Scholar]
- 10.Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–47. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanov SV, Miller J, Lucito R, Tang C, Ivanova AV, Pei J, et al. Genomic events associated with progression of pleural malignant mesothelioma. Int J Cancer. 2009;124:589–99. doi: 10.1002/ijc.23949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weder W, Stahel RA, Bernhard J, Bodis S, Vogt P, Ballabeni P, et al. Multicenter trial of neo-adjuvant chemotherapy followed by extrapleural pneumonectomy in malignant pleural mesothelioma. Ann Oncol. 2007;18:1196–202. doi: 10.1093/annonc/mdm093. [DOI] [PubMed] [Google Scholar]
- 13.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 14.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov SV, Miller J, Lucito R, Tang C, Ivanova AV, Pei J, et al. Genomic events associated with progression of pleural malignant mesothelioma. Int J Cancer. 2009;124:589–99. doi: 10.1002/ijc.23949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Lam JB, Chow KH, Xu A, Lam KS, Moon RT, et al. Adiponectin stimulates Wnt inhibitory factor-1 expression through epigenetic regulations involving the transcription factor specificity protein 1. Carcinogenesis. 2008;29:2195–202. doi: 10.1093/carcin/bgn194. [DOI] [PubMed] [Google Scholar]
- 20.Guled M, Lahti L, Lindholm PM, Salmenkivi K, Bagwan I, Nicholson AG, et al. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma -A miRNA microarray analysis. Genes Chromosomes Cancer. 2009;48:615–23. doi: 10.1002/gcc.20669. [DOI] [PubMed] [Google Scholar]
- 21.Busacca S, Germano S, De CL, Rinaldi M, Comoglio F, Favero F, et al. MicroRNA Signature of Malignant Mesothelioma with Potential Diagnostic and Prognostic Implications. Am J Respir Cell Mol Biol. 2009 doi: 10.1165/rcmb.2009-0060OC. [DOI] [PubMed] [Google Scholar]
- 22.Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–3. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 23.Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009;10:400–5. doi: 10.1038/embor.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schramm A, Opitz I, Thies S, Seifert B, Moch H, Weder W, et al. Prognostic significance of epithelial-mesenchymal transition in malignant pleural mesothelioma. Eur J Cardiothorac Surg. 2009 doi: 10.1016/j.ejcts.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 25.Christensen BC, Houseman EA, Godleski JJ, Marsit CJ, Longacker JL, Roelofs CR, et al. Epigenetic profiles distinguish pleural mesothelioma from normal pleura and predict lung asbestos burden and clinical outcome. Cancer Res. 2009;69:227–34. doi: 10.1158/0008-5472.CAN-08-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen BC, Godleski JJ, Roelofs CR, Longacker JL, Bueno R, Sugarbaker DJ, et al. Asbestos burden predicts survival in pleural mesothelioma. Environ Health Perspect. 2008;116:723–6. doi: 10.1289/ehp.11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsou JA, Galler JS, Wali A, Ye W, Siegmund KD, Groshen S, et al. DNA methylation profile of 28 potential marker loci in malignant mesothelioma. Lung Cancer. 2007;58:220–30. doi: 10.1016/j.lungcan.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer JR, Ohnmacht U, Rieger N, Zemaitis M, Stoffregen C, Kostrzewa M, et al. Promoter methylation of RASSF1A, RARbeta and DAPK predict poor prognosis of patients with malignant mesothelioma. Lung Cancer. 2006;54:109–16. doi: 10.1016/j.lungcan.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 29.Cole GW, Jr, Alleva AM, Zuo JT, Sehgal SS, Yeow WS, Schrump DS, et al. Suppression of pro-metastasis phenotypes expression in malignant pleural mesothelioma by the PI3K inhibitor LY294002 or the MEK inhibitor UO126. Anticancer Res. 2006;26:809–21. [PubMed] [Google Scholar]
- 30.Graziani I, Eliasz S, De Marco MA, Chen Y, Pass HI, De May RM, et al. Opposite effects of Notch-1 and Notch-2 on mesothelioma cell survival under hypoxia are exerted through the Akt pathway. Cancer Res. 2008;68:9678–85. doi: 10.1158/0008-5472.CAN-08-0969. [DOI] [PubMed] [Google Scholar]
- 31.Amatori S, Papalini F, Lazzarini R, Donati B, Bagaloni I, Rippo MR, et al. Decitabine, differently from DNMT1 silencing, exerts its antiproliferative activity through p21 upregulation in malignant pleural mesothelioma (MPM) cells. Lung Cancer. 2009 doi: 10.1016/j.lungcan.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 32.Sartore-Bianchi A, Gasparri F, Galvani A, Nici L, Darnowski JW, Barbone D, et al. Bortezomib inhibits nuclear factor-kappaB dependent survival and has potent in vivo activity in mesothelioma. Clin Cancer Res. 2007;13:5942–51. doi: 10.1158/1078-0432.CCR-07-0536. [DOI] [PubMed] [Google Scholar]
- 33.Yang H, Bocchetta M, Kroczynska B, Elmishad AG, Chen Y, Liu Z, et al. TNF-alpha inhibits asbestos-induced cytotoxicity via a NF-kappaB-dependent pathway, a possible mechanism for asbestos-induced oncogenesis. Proc Natl Acad Sci U S A. 2006;103:10397–402. doi: 10.1073/pnas.0604008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villanova F, Procopio A, Rippo MR. Malignant mesothelioma resistance to apoptosis: recent discoveries and their implication for effective therapeutic strategies. Curr Med Chem. 2008;15:631–41. doi: 10.2174/092986708783885273. [DOI] [PubMed] [Google Scholar]
- 35.Hegmans JP, Hemmes A, Hammad H, Boon L, Hoogsteden HC, Lambrecht BN. Mesothelioma environment comprises cytokines and T-regulatory cells that suppress immune responses. Eur Respir J. 2006;27:1086–95. doi: 10.1183/09031936.06.00135305. [DOI] [PubMed] [Google Scholar]
- 36.O’Kane SL, Pound RJ, Campbell A, Chaudhuri N, Lind MJ, Cawkwell L. Expression of bcl-2 family members in malignant pleural mesothelioma. Acta Oncol. 2006;45:449–53. doi: 10.1080/02841860500468927. [DOI] [PubMed] [Google Scholar]
- 37.Fox SA, Loh S, Thean AL, Garlepp MJ. Identification of differentially expressed genes in murine mesothelioma cell lines of differing tumorigenicity using suppression subtractive hybridization. Biochim Biophys Acta. 2004;1688:237–44. doi: 10.1016/j.bbadis.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 38.Leong CC, Marley JV, Loh S, Milech N, Robinson BW, Garlepp MJ. Transfection of the gene for B7-1 but not B7-2 can induce immunity to murine malignant mesothelioma. Int J Cancer. 1997;71:476–82. doi: 10.1002/(sici)1097-0215(19970502)71:3<476::aid-ijc28>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 39.Xu H, Cheung IY, Guo HF, Cheung NK. MicroRNA miR-29 modulates expression of immunoinhibitory molecule B7-H3: potential implications for immune based therapy of human solid tumors. Cancer Res. 2009;69:6275–81. doi: 10.1158/0008-5472.CAN-08-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H, Testa JR, Carbone M. Mesothelioma epidemiology, carcinogenesis, and pathogenesis. Curr Treat Options Oncol. 2008;9:147–57. doi: 10.1007/s11864-008-0067-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toyokuni S. Mechanisms of asbestos-induced carcinogenesis. Nagoya J Med Sci. 2009;71:1–10. [PMC free article] [PubMed] [Google Scholar]
- 42.Christensen BC, Houseman EA, Godleski JJ, Marsit CJ, Longacker JL, Roelofs CR, et al. Epigenetic profiles distinguish pleural mesothelioma from normal pleura and predict lung asbestos burden and clinical outcome. Cancer Res. 2009;69:227–34. doi: 10.1158/0008-5472.CAN-08-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toyooka S, Kishimoto T, Date H. Advances in the molecular biology of malignant mesothelioma. Acta Med Okayama. 2008;62:1–7. doi: 10.18926/AMO/30990. [DOI] [PubMed] [Google Scholar]
- 44.Christensen BC, Godleski JJ, Marsit CJ, Houseman EA, Lopez-Fagundo CY, Longacker JL, et al. Asbestos exposure predicts cell cycle control gene promoter methylation in pleural mesothelioma. Carcinogenesis. 2008;29:1555–9. doi: 10.1093/carcin/bgn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsou JA, Galler JS, Wali A, Ye W, Siegmund KD, Groshen S, et al. DNA methylation profile of 28 potential marker loci in malignant mesothelioma. Lung Cancer. 2007;58:220–30. doi: 10.1016/j.lungcan.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki M, Toyooka S, Shivapurkar N, Shigematsu H, Miyajima K, Takahashi T, et al. Aberrant methylation profile of human malignant mesotheliomas and its relationship to SV40 infection. Oncogene. 2005;24:1302–8. doi: 10.1038/sj.onc.1208263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.