Abstract
Purpose
Epigenetic alterations including changes to cellular DNA methylation levels contribute to carcinogenesis and may serve as powerful biomarkers of the disease. This investigation sought to determine whether hypomethylation at the long interspersed nuclear elements (LINE1), reflective of the level of global DNA methylation, in peripheral blood-derived DNA is associated with increased risk of bladder cancer.
Experimental Design
LINE1 methylation was measured from blood-derived DNA obtained from participants of a population-based incident case control study of bladder cancer in New Hampshire. Bisulfite-modified DNA was pyrosequenced to determine LINE1 methylation status; a total of 285 cases and 465 controls were evaluated for methylation.
Results
Being in the lowest LINE1 methylation decile was associated with a 1.8-fold increased risk of bladder cancer (95% CI, 1.12-2.90) in models controlling for gender, age and smoking, and the association was stronger in women than in men (ORs = 2.48, 95% CI 1.19-5.17 in women and 1.47, 95% CI 0.79-2.74 in men). Amongst controls, women were more likely to have lower LINE1 methylation than men (p-value 0.04), and levels of arsenic in the 90th percentile were associated with reduced LINE1 methylation (p-value 0.04).
Conclusions
LINE1 hypomethylation may be an important biomarker of bladder cancer risk, especially amongst women.
Keywords: Bladder Cancer, Epidemiology, Gender Differences
Introduction
Bladder cancer is the ninth most incident form of cancer in the United States, with an estimated 70, 980 new cases expected to occur in 2009 (1). High rates of bladder cancer are especially widespread in New England, with women in this area experiencing some of the highest bladder-cancer mortality rates in the U.S (2). Incidence rates of bladder cancer have been stable in men in the last twenty years but have been increasing in women at a rate of 0.2% a year; despite this, men are nearly four times more at risk of developing bladder cancer than women (1, 3). Furthermore, bladder cancer is thought to be a heterogeneous disease with different characteristics related to the histopathologic presentation of the disease primarily as non-invasive versus invasive, with the latter contributing to the vast majority of disease mortality(4). It is also believed that different molecular mechanisms may contribute to the heterogeneity of this disease, calling for more research to clarify the differences between pathological pathways (4).
Genetics, dietary factors, occupational exposures, toxicants and lifestyle factors play an important role in bladder cancer initiation and development, and the major known risk factors for bladder cancer include smoking, aromatic hydrocarbons, water chlorination byproducts, and inorganic arsenic (3, 5-6). However, the effects of some of these risk factors on bladder cancer, particularly arsenic, has only been noted in certain populations (7). Ingestion of high levels of arsenic has been associated with bladder cancer mortality in certain regions of the world, most notably in Argentina, Chile and Taiwan (7). Despite this, exposure to lower levels of arsenic has been found largely in susceptible subgroups (5). A recent New Hampshire study found the association between levels of arsenic higher than 0.330 mcg/g and an increased risk of bladder cancer to hold true only in smokers; confirming results reported in two other studies, one conducted in Utah and the other in Finland (5, 8, 9).
Epigenetics is an emerging area of epidemiologic interest, seemingly important for understanding both disease susceptibility and etiology (10, 11). Much work has characterized promoter region hypermethylation of cancer-related genes in bladder tumors, reporting associations between these alterations and exposures causal for the disease (12-14). Past research has also shown tumor cells to be hypomethylated, on a global level, in comparison to normal cells (15, 16). Examination of the DNA methylation pattern in repetitive elements, such as long interspersed nuclear elements (LINE1), has been used as a surrogate of overall global methylation levels (17, 18). At the same time, epigenetic alterations of LINE1 may also be of interest in that they may initiate the clonal expansion of pre-malignant cells in early stages of cancer development, may contribute to genomic instability and may lead to structural changes in chromosomes (10, 11, 19, 20). DNA hypomethylation may also lead to gene overexpression, occurring when genes become hypomethylated, and to the activation transposable elements which may result in genetic mutations (20-22). In addition, DNA methylation may be altered by dietary availability of methyl groups; specifically with two of the enzyme's methylenetetrahydrofolate reductase single nucleotide polymorphisms, MTHFR-677T/T and 1298C/C, having been associated in previous work with lower global methylation in lymphocytic DNA (23).
There is emerging evidence that changes to the global or repetitive element methylation status in DNA derived from peripheral blood cells can serve as a biomarker of exposure and disease. Recent reports suggest that exposure to carcinogens and pollutants, specifically including particulate matter and benzene, are associated with global hypomethylation detectable in peripheral blood (24-26). Further, global DNA hypomethylation in blood, measured using a radioactive methyl-acceptor assay, has been associated with bladder cancer (27) and gastric cancer (28), while altered methylation of the LINE1 region has been linked to head and neck cancer risk (18).
In this study we sought to determine what factors are associated with LINE1 hypomethylation, whether LINE1 hypomethylation is associated with increased risk of bladder cancer and if any risk factors for the disease modulate or modify this association.
Materials and Methods
Subjects
A description of the study design appears in earlier reports (5, 29). Briefly, cases were selected from the New Hampshire State Department of Health and Human Services' Cancer Registry and included only New Hampshire residents aged 25-74 years old with a first diagnosis of bladder cancer ranging from July 1, 1994 to June 30, 1998. Of the N=618 potential cases eligible for this study, n=10 (2%) were not able to be contacted due to denial of contact by their physicians, n=63 (10%) were reported as deceased by a household member or physician, n=3 (<1%) did not return telephone calls about participation in the study, n=75 (12%) refused to participate, and n=8 (1%) were too ill to take part. Finally, N=459 or 85% of the eligible cases were interviewed for this study. A standardized review was conducted by a single study pathologist (A.R.S.) to verify the diagnosis and histopathology of the cases. For the analyses presented here, the case group was restricted to cases having LINE1 methylation data and excluded cases that were diagnosed as carcinoma in situ; this included a total of 285 cases, whose characteristics are presented in Table 1. For efficiency purposes, the same control group used in a study of non-melanoma skin cancer conducted from July 1, 1993 to June 30, 1995 was used and additional controls were selected afterwards from the sources described below (30). All controls less than 65 years of age were selected from records obtained from the New Hampshire Department of Transportation and controls older than 65 years of age were chosen from records obtained from the Health Care Financing Administration's Medicare Program. Controls were then frequency matched to cases by age (25-34, 35-44, 45-54, 55-64, 65-69, 70-74) and gender and were randomly assigned a reference date comparable to the cases' diagnosis date. Of the 990 potential controls eligible for this study, n=18 (2%) were reported as deceased by a member of the household, n=17 (2%) did not return telephone calls about participation in the study (2%), n=261 (26%) refused to participate, and n =29 (3%) were mentally incompetent or too ill to take part. Finally, N=665 or 70% of the eligible controls were interviewed for this study. For the analyses presented here, the control group was restricted to the 465 controls having LINE1 methylation data, whose characteristics are presented in Table 1. No significant differences in the demographic or risk factor variables examined were found between subjects with and without methylation data and to ensure comparability of cases and controls, all subjects were asked if they currently held a driver's license or a Medicare enrollment card.
Table 1.
Selected Characteristics of bladder cancer cases and controls with LINE1 methylation data
| Variable | Total N=750 (%) |
Controls N=465 (62.0%) |
Cases N=285 (38.0%) |
|---|---|---|---|
| Sex (N=750) | |||
| Male | 521 (69.5%) | 297 (63.9%) | 224 (78.6%) |
| Female | 229 (30.5%) | 168 (36.1%) | 61 (21.4%) |
| Smoking status (N=750) | |||
| Never | 194 (25.9%) | 140 (30.1%) | 54 (19.0%) |
| Former | 382 (50.9%) | 240 (51.6%) | 142 (49.8%) |
| Current | 174 (23.2%) | 85 (18.3%) | 89 (31.2%) |
| Carcinoma Severity (N=730) | |||
| Non-invasive | 194 (68.1%) | -- | 194 (68.1%) |
| Invasive | 71 (24.9%) | -- | 71 (24.9%) |
| Missing | 20 (7.0%) | -- | 20 (7.0%) |
| Toenail arsenic (N=733) | |||
| 0.01 to <0.06: 0-25% | 179 (23.9%) | 110 (23.7%) | 69 (24.2%) |
| 0.059 to <0.09: 25.1-50% | 201 (26.8%) | 120 (25.8%) | 81 (28.4%) |
| 0.09 to <0.13: 50.1-75% | 180 (24.0%) | 116 (25.0%) | 64 (22.5%) |
| 0.13 to <0.20: 75.1-90% | 104 (13.9%) | 69 (14.8%) | 35 (12.3%) |
| 0.20 to 2.48: 90.1-100% | 69 (9.2%) | 40 (8.6%) | 29 (10.2%) |
| Missing | 17 (2.3%) | 10 (2.2%) | 7 (2.5%) |
| Age | |||
| Mean (+/-SD) | 62.2 (10) | 61.7 (10.4) | 63.1 (9.4) |
Personal Interview and Arsenic measurement
Consenting subjects underwent a detailed in-person interview, usually at their home, which assessed sociodemographic information, occupational history, detailed information about the use of tobacco products (such as history of cigarette smoking) and medical history prior to the reference or diagnosis date. Subjects were requested to save a toenail clipping specimen prior to the interview or were sent a self-addressed envelope to send in their sample after the interview. Toenail samples, which reflect arsenic burden in the body from all sources of exposure, were analyzed for arsenic using instrumental neutron activation analysis at the University of Missouri's Research Reactor Center (30). All procedures and study materials were approved by the appropriate Institutional Review Boards.
DNA extraction, modification and quantification of LINE1 Methylation and Genotyping Assay
DNA was extracted from peripheral blood buffy coats using the QIAmp DNA mini kit according to the manufacturere's protocol (Qiagen, Valencia, CA). DNA (1 μg) was subjected to sodium bisulfite modification using the EZ DNA Methylation Kit (Zymo Research, Orange, CA) following the manufacturer's protocol. LINE1 region methylation extent was determined through quantitiative bisulfite Pyrosequencing (31), following the protocol of Bollati et al (26), which examines the cytosine methylation status at 4 CpG sites in the LINE1 region. All PCR reactions, performed using Qiagen Hot Star Taq polymerase, included a no template control, unmodified DNA control, and 7 standardized percent methylation controls (0%, 15%, 25%, 45%, 65%, 75%, and 100%) derived from Qiagen EpiTect® PCR control DNA set samples. Each sequencing reaction used 10μl of PCR product and was run according to instrument/manufacturer protocols (Qiagen) on a PyroMark™MD System (Qiagen). Three PCR amplifications were performed on each sample and 2 Pyrosequencing runs were performed from each PCR reaction, resulting in 6 replicates for each individual to assess repeat measure variability. The standard error of the averaged individual repeats was found to be the same as the standard error for each replicate so the average measure of LINE1 methylation across the 4 CpG sites for the 6 replicates was used for each individual. The following polymorphisms were assessed in this study: methylenetetrahydrofolate reductase (MTHFR)-677 C>T and MTHFR-1298 C>T and both were genotyped using the 5′ nuclease TaqMan allelic discrimination assay on the ABI 7900HT (Applied Biosystems) as previously described (32).
Statistical Methods
In order to assess any dose dependence of LINE1 methylation extent on bladder cancer risk, LINE1 methylation was coded into deciles, based on the distribution in controls, to determine more precisely the amount of methylation which would increase the risk of bladder cancer. As an increase in the OR for disease was present only in the lowest decile, LINE1 methylation was coded as a binary outcome comparing the lowest decile with the referent including the remaining deciles of LINE1. Age was coded as a continuous variable with N=750, Mean=62.24, Standard deviation=10.04, Minimum=25, and Maximum=74. Arsenic was classified using percentiles of the control distribution, based again on our previous research: 0.009-0.059 (<=25%-used asthe referent group), 0.060-0.086 (25.1-50%), 0.087-0.126 (50.1-75%), 0.127-0.193 (75.1-90%), 0.194-0.277 (90.1-95%), 0.278-0.330 (95.1-97%) and 0.331-2.484 (97.1-100%) (4). Smoking was evaluated using variables measuring intensity of smoking, duration of smoking and their combination, as well as the status (never, former, current) amongst subjects. Smoking was used as a categorical variable defined as never, former, or current smokers as has been done previously in examinations in this and other populations (5, 27, 33). To explore the influence of MTHFR 677 and MTHFR 1298 genotypes on LINE1 methylation and bladder cancer risk, these variables were coded using wt/wt as the referent and were compared to wt/var and var/var (18). In univariate analyses, the relationship between amount of methylation and case control status and other factors was evaluated with χ2 statistics and t-tests using a p-value less than 0.05 as statistically significant. To investigate the association between the level of LINE1 methylation and bladder cancer, unconditional and stratified logistic models were used and included smoking, age and gender (the latter were matching variables in the original study). To examine the association between LINE1 methylation and risk of either noninvasive or invasive bladder cancer, we performed the analysis stratified by tumor stage. Similarly, to examine the differential association of LINE1 methylation with risk of bladder cancer between men and women, we performed analysis stratified by gender. These models were adjusted for gender and smoking status. To examine the relationship between variables including gender, age, arsenic and smoking with the amount of LINE1 methylation, logistic models were used with low LINE1 methylation defined as being in the first decile as a binary outcome. Selenium, education, MTHFR 677 and MTHFR 1298 were examined as possible predictors of bladder cancer risk and as possible important predictors of LINE1; however, these variables were not found to significantly influence methylation or bladder cancer risk, and did not change the estimates of the effects of the other variable in the model, and thus were dropped from further models. The interaction between smoking and LINE1 methylation was examined with the likelihood ratio test. Data were analyzed by use of SAS statistical software, version 9.1.
Results
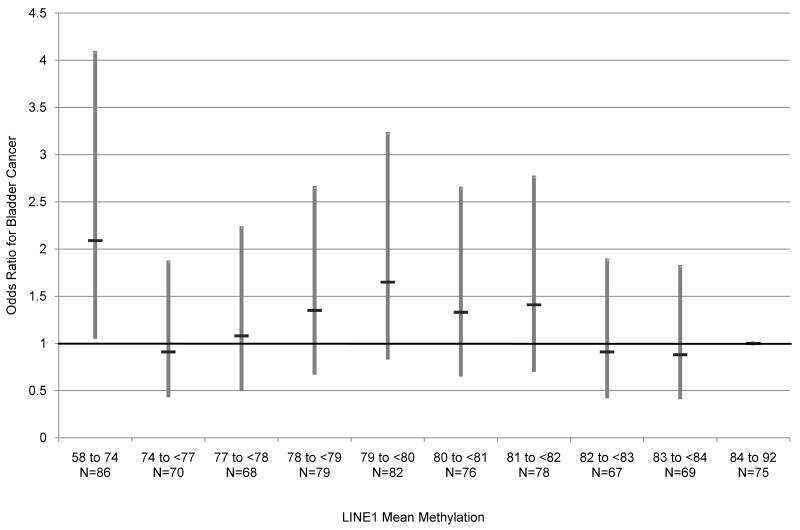
LINE1 methylation levels were determined for 465 controls and 285 cases, and overall ranged from 57.9% and 92.0%. The characteristics of controls and cases with LINE1 methylation data are shown in Table 1. MTHFR codon 677 and 1298 genotypes, mean toenail arsenic levels and age were not significantly different in cases and controls while smoking status and gender were significantly different in the two groups. Figure 1 shows the relationship of LINE1 methylation extent with bladder cancer risk, grouping LINE1 methylation data into deciles defined by levels in the controls. These data demonstrate that a significant estimate of disease risk exists only in the lowest decile of LINE1 methylation. The risk estimate, for being in the lowest decile was an odds ratio of 2.08 (95% CI, 1.07-4.07) compared to the referent highest LINE1 methylation decile, and adjusted for age, gender, and smoking status. Based upon this figure, we further defined LINE1 as a binary variable with the lowest decile grouping the remaining deciles as the referent.
Figure 1.
Modeling of LINE1 methylation deciles and corresponding risk of bladder cancer. LINE1 methylation decile bins are represented on the x-axis, and corresponding risk, modeled as odds ratios represented on the y-axis, of bladder cancer adjusting for smoking, age and gender using logistic regression. The black horizontal line depicts the odds ratio point estimate at each LINE1 methylation decile, compared to the referent highest decile. The gray bar represents the range of the 95% upper and lower confidence intervals. Below each decile bin on the x-axis are the number of observations present at each decile LINE1 methylation level. The deciles were coded based on the distribution in controls and range from 67 to 86 observations.
Table 2 presents the association between LINE1 methylation extent in the lowest decile compared to all others as referent, adjusting for gender, age and smoking status using logistic regression. A 1.80-fold increased risk of bladder cancer (95% CI, 1.12-2.90) was observed among those with LINE1 methylation extent in the lowest decile. As previous reports have suggested an interaction between global DNA methylation and smoking (27), we examined the interaction of LINE1 methylation with smoking status and found no significant association (data not shown). Since bladder cancer presents in a dichotomous fashion as non-invasive or invasive disease and these forms of bladder cancer have significant pathological differences, we performed stratified analysis by invasive status (4). Amongst non-invasive cancers, being in the lowest LINE1 methylation decile confers an almost two fold risk of bladder cancer (OR 1.94, 95% CI, 1.17-3.22) while the risk is non-significant amongst invasive cancers, in models adjusted for smoking, age, and gender. We then stratified our model by gender, adjusting for age and smoking status in each stratum. Amongst females, being in the lowest LINE1 methylation decile confers a significant almost 2.5-fold increased risk of bladder cancer (95% CI 1.19-5.17) while the risk was non-significant amongst males (OR = 1.47, 95% CI = 0.79-2.74).
Table 2.
Association of LINE1 methylation and risk of bladder cancer: main effects model and models further stratified by invasiveness and gender
| Variable | N | Odds Ratio (95% CI) | p-value |
|---|---|---|---|
| All subjects (N=465 controls and N=285 cases)* | |||
| LINE1 Mean Methylation | |||
| 57.89 to <74.25-10% | 86 | 1.80 (1.12, 2.90) | 0.02 |
| 74.25 to 91.96-90% | 664 | Ref | |
| Smoking status | |||
| Never smoker | 194 | Ref | |
| Former smoker | 382 | 1.31 (0.89, 1.94) | 0.17 |
| Current smoker | 174 | 2.46 (1.59, 3.83) | <0.0001 |
| Non-invasive cases (N=465 controls and N=194 cases)* | |||
| LINE1 Mean Methylation | |||
| 57.89 to <74.25-10% | 31 | 1.94 (1.17, 3.22) | 0.01 |
| 74.25 to 91.96-90% | 163 | Ref | |
| Smoking status | |||
| Never smoker | 34 | Ref | |
| Former smoker | 103 | 1.58 (1.00, 2.49) | 0.05 |
| Current smoker | 57 | 2.43 (1.46, 4.03) | 0.00 |
| Invasive cases (N=465 controls and N=71 cases)* | |||
| LINE1 Mean Methylation | |||
| 57.89 to <74.25-10% | 6 | 0.88 (0.36, 2.18) | 0.78 |
| 74.25 to 91.96-90% | 65 | Ref | |
| Smoking status | |||
| Never smoker | 14 | Ref | |
| Former smoker | 32 | 1.12 (0.57, 2.20) | 0.74 |
| Current smoker | 25 | 2.60 (1.28, 5.29) | 0.01 |
| Males (N=297 controls and N=224 cases)** | |||
| LINE1 Mean Methylation | |||
| 57.89 to <74.25-10% | 45 | 1.47 (0.79, 2.74) | 0.22 |
| 74.25 to 91.96-90% | 476 | Ref | |
| Females (N=168 controls and N=61 cases)** | |||
| LINE1 Mean Methylation | |||
| 57.89 to <74.25-10% | 41 | 2.48 (1.19, 5.17) | 0.02 |
| 74.25 to 91.96-90% | 188 | Ref | |
All models were adjusted for gender and age
All models were adjusted for age and smoking status
Given the risk associated with low methylation, we sought to identify predictors of hypomethylation in healthy controls. We utilized the binary LINE1 methylation variable as the dependent variable, and examined the association between this variable and known risk factors of bladder cancer (including gender, age, smoking status and toenail arsenic levels) stratified by case status in Table 3. Women were more likely to be in the lowest LINE1 methylation decile than men (male controls: OR 0.49, 95% CI 0.25-0.97; males cases: OR 0.38, 95% CI 0.18-0.81). The lowest decile of LINE1 methylation had a higher percentage of individuals with high toenail arsenic concentrations. A trend in ORs for low LINE1 methylation from the lowest to the highest toenail arsenic levels was statistically significant (p<0.01), with the 75th to 90th percentile (0.13 to <0.20 μg/g arsenic) having an OR for low LINE1 methylation of 3.68 (95% CI 1.17-11.58), and those with greater than the 90th percentile of arsenic exposure (>0.20 μg/g arsenic) having an OR for LINE1 methylation of 3.68 (95% CI 1.04-13.02).
Table 3.
Predictors of the risk of being in the lowest tenth decile of LINE1 methylation
| Variable | N=465 | Controls Odds Ratio (95% CI) | p-value | N=285 | Cases Odds Ratio (95% CI) | p-value |
|---|---|---|---|---|---|---|
| Sex | ||||||
| Male | 297 | 0.49 (0.25, 0.97) | 0.04 | 224 | 0.38 (0.18, 0.81) | 0.01 |
| Female | 168 | Ref | 61 | Ref | ||
| Reference Age | 465 | 1.03 (0.99, 1.06) | 0.11 | 285 | 1.00 (0.97, 1.04) | 0.97 |
| Smoking status | ||||||
| Never smoker | 140 | Ref | 54 | Ref | ||
| Former smoker | 240 | 0.67 (0.31, 1.42) | 0.29 | 142 | 0.75 (0.29, 1.93) | 0.55 |
| Current smoker | 85 | 1.01 (0.42, 2.43) | 0.99 | 89 | 1.20 (0.46, 3.14) | 0.71 |
| mth677 | ||||||
| Homozygous wild type | 190 | Ref | 116 | Ref | ||
| Heterozygous | 197 | 0.80 (0.35, 1.81) | 0.59 | 136 | 0.53 (0.23, 1.19) | 0.12 |
| Homozygous variant | 62 | 2.08 (0.76, 5.70) | 0.15 | 30 | 0.68 (0.20, 2.29) | 0.53 |
| mth1298 | ||||||
| Homozygous wild type | 229 | Ref | 136 | Ref | ||
| Heterozygous | 175 | 0.78 (0.35, 1.75) | 0.54 | 118 | 0.58 (0.26, 1.31) | 0.19 |
| Homozygous variant | 47 | 0.88 (0.24, 3.24) | 0.85 | 29 | 0.29 (0.07, 1.31) | 0.11 |
| Toenail arsenic | ||||||
| 0.01 to <0.06: 0-25% | 110 | Ref | 69 | Ref | ||
| 0.059 to <0.09: 25.1-50% | 120 | 1.62 (0.55, 4.76) | 0.38 | 81 | 0.69 (0.24, 1.96) | 0.49 |
| 0.09 to <0.13: 50.1-75% | 116 | 2.20 (0.77, 6.30) | 0.14 | 64 | 1.21 (0.43, 3.41) | 0.72 |
| 0.13 to <0.20: 75.1-90% | 69 | 3.68 (1.17, 11.58) | 0.03 | 35 | 2.72 (0.88, 8.39) | 0.08 |
| 0.20 to 2.48: 90.1-100% | 40 | 3.68 (1.04, 13.02) | 0.04 | 29 | 1.26 (0.36, 4.41) | 0.72 |
Discussion
In models controlling for smoking status, gender and age, we have found a significant 1.8-fold increased risk of bladder cancer among subjects with mean LINE1 methylation levels in the lowest decile of methylation, ranging from 57.89 to <74.25%, as compared to subjects with >74.25% LINE1 methylation. This suggests that LINE1 hypomethylation is an independent risk factor for bladder cancer and that it may be an excellent biomarker. Our results are consistent with a previous report that also noted a significantly increased risk of bladder cancer amongst patients with lower global DNA methylation levels, as assessed by the radioactive methyl-incorporation assay (27). Interestingly, unlike that report we did not observe a dose-dependent association of LINE1 methylation extent with bladder cancer risk, but rather saw only an association with disease risk at the lowest level, a result consistent with our previous work examining LINE1 methylation in head and neck cancer (18).
We also observed that the risk of non-invasive bladder cancer was increased almost two fold for subjects in the lowest LINE1 methylation decile, while there was no significant association in invasive cases. This is of interest, because it suggests that LINE1 methylation may be a specific biomarker of noninvasive disease. The mechanism responsible for this apparent distinction is unclear but it is consistent with the fact that there are well known differences in this disease associated with an invasive pathology (4).
In examining the predictors of lowest tenth decile of LINE1 methylation we found that women were greater than two-fold significantly more likely to have reduced LINE1 methylation extent. This finding is consistent with a study which found that females had statistically lower methylation levels than males in LINE1 and Alu repeats measured from whole blood (34) as well as with our previous work in head and neck cancer (18). Two other studies did not find a significant association between LINE1 methylation and gender in leucocytes and in adenomas; however, these studies did not use a large number of subjects and may have been underpowered to observe this association (17, 23). Our gender-stratified analyses further suggest that the risk for bladder cancer conferred by low LINE1 methylation extent is significant only among women, and is consistent with Moore et al. (27) who observed a similar increased risk of bladder cancer among women with reduced overall global DNA methylation levels.
Our finding of reduced LINE1 methylation in relation to arsenic exposure is consistent with experimental data showing that arsenic and its inorganic salt, arsenite, induce DNA hypomethylation in cultures of human lung and rat hepatocytes, as well as in rat livers through feeding studies (35-37). A study in Bangladesh, with endemic, high level arsenic exposures, observed that reduced DNA methylation was a risk factor for arsenic-associated skin lesions (38) but found a positive relationship between urinary arsenic concentrations and global DNA methylation in peripheral blood leucocytes (using the radioactive methyl-incorporation assay) (39). This different finding may be related to the different measures of DNA methylation and the far greater exposures in Bangladesh compared with ours. Although the methylation extent of the LINE1 region has been correlated with overall cellular 5-methycytosine content (40, 41), and thus serve as a surrogate marker, changes in the extent of methylation of these elements may lead to specific functional consequences on their expression. Expression of these elements can lead to their retrotransposition to various regions of the genome, possibly leading to downstream effects on other genes in these cells (42). It is unclear if the changes in methylation extent being measured lead to changes in expression of specific functional LINE1 elements. Going forward, it will be important to delineate the biological nature of this association, as LINE1 measurement may be, in some cases, in the causal pathway for disease.
We did not find an interaction between LINE1 methylation and smoking status, unlike previous reports examining methylation by smoking intensity (27). The non-significant interaction in our model could be attributable to differential smoking patterns between the populations.
Strengths of this study included the population-based nature of the study, as well as the use of quantitative pyrosequencing for LINE1 methylation determination and detailed exposure assessment including the use of toenail analysis of arsenic concentrations. Limitations of this study included the exclusion of cases and controls without methylation data which could lead to a selection bias if included and excluded cases and controls differ from one another. However, we did not note any differences between included and excluded cases and controls in any of our demographic information and use of tobacco products. Another limitation of this study was the low number of invasive cases available; a larger study is necessary to further understand the impact of global hypomethylation on the risk of development of non-invasive as compared to invasive bladder cancer. Another limitation would be the poor age matching done in this study; however we do not think this impacted the results as age is not found to be significant in any of our analyses. Further research on hormonal influences on global methylation and risk of bladder cancer is also warranted related to our findings of differential risk amongst women.
In summary, this study adds to the growing literature on the role of epigenetics in human cancer etiology, and suggests LINE1 hypomethylation as a risk factor for bladder cancer, with potential utility as a disease biomarker. In addition, this study shows an increased risk of LINE1 hypomethylation with high levels of arsenic, raising important questions about the nature of the causal pathway for alterations in DNA repeat methylation to contribute to cancer risk. These results also suggest that women demonstrate lower levels of LINE1 methylation, and may suggest distinct etiologies of bladder cancer between men and women, which may also aid in explaining the disparate incidence of this disease in men and women. Further work is needed to better clarify how exposures important to the disease play a role in LINE1 region methylation, and to clarify the specific subgroups where this factor may be most predictive of disease risk.
Acknowledgments
Funding: This work was supported by the Flight Attendant Medical Research Institute (YCSA 052341 to C.J.M.); and the National Institutes of Health (R01CA121147, K07CA102327, P50CA097257, and P42ES007373 and R01CA57494).
Footnotes
Statement of Translational Relevance: Bladder cancer is a common, exposure-related disease which contributes significantly to morbidity and health care costs in the United States. Although tobacco use and occupational exposures have been linked to bladder cancer, much of the etiology of this disease remains unexplained. We have identified a novel epigenetic marker of bladder cancer which has the potential for utility as a screening strategy for this disease, and may aid in providing a more comprehensive understanding of the etiology of bladder cancer.
Disclosure of Potential Conflicts of Interest: The authors declared no conflicts of interest.
References
- 1.American Cancer Society. Cancer facts and Figures, 2009. Atlanta, Ga: American Cancer Society; 2009. [Google Scholar]
- 2.Brown LM, Zahm SH, Hoover RN, Fraumeni JF., Jr High bladder cancer mortality in rural New England (United States): an etiologic study. Cancer Causes Control. 1995;6:361–8. doi: 10.1007/BF00051412. [DOI] [PubMed] [Google Scholar]
- 3.Parkin MP. The global burden of urinary bladder cancer. Scand J Urol Nephrol. 2008;42:12–20. doi: 10.1080/03008880802285032. [DOI] [PubMed] [Google Scholar]
- 4.Dinney CP, McConkey DJ, Millikan RE, et al. Focus on bladder cancer. Cancer Cell. 2004;6:111–6. doi: 10.1016/j.ccr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Karagas MR, Tosteson TD, Morris JS, et al. Incidence of transitional cell carcinoma of the bladder and arsenic exposure in New Hampshire. Cancer Causes Control. 2004;15:465–72. doi: 10.1023/B:CACO.0000036452.55199.a3. [DOI] [PubMed] [Google Scholar]
- 6.Boffetta P. Human cancer from environmental pollutants: The epidemiological evidence. Mutat Res. 2006;608:157–62. doi: 10.1016/j.mrgentox.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Abernathy CO, Liu YP, Longfellow D, et al. Arsenic: health effects, mechanisms of actions, and research issues. Environ Health Perspect. 1999;107:593–7. doi: 10.1289/ehp.99107593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bates MN, Smith AH, Cantor KP. Case–control study of bladder cancer and arsenic in drinking water. Am J Epidemiol. 1995;141:523–530. doi: 10.1093/oxfordjournals.aje.a117467. [DOI] [PubMed] [Google Scholar]
- 9.Kurttio P, Pukkala E, Kahelin H, Auvinen A, Pekkanen J. Arsenic concentrations in well water and risk of bladder and kidney cancer in Finland. Environ Health Perspect. 1999;107:705–710. doi: 10.1289/ehp.99107705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer- a mechanism for early oncogenic pathway addiction. Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 11.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 12.Marsit CJ, Karagas MR, Danaee H, et al. Carcinogen exposure and gene promoter hypermethylation in bladder cancer. Carcinogenesis. 2006;27:112–116. doi: 10.1093/carcin/bgi172. [DOI] [PubMed] [Google Scholar]
- 13.Marsit CJ, Houseman EA, Schned AR, Karagas MR, Kelsey KT. Promoter hypermethylation is associated with current smoking, age, gender and survival in bladder cancer. Carcinogenesis. 2007;28:1745–51. doi: 10.1093/carcin/bgm116. [DOI] [PubMed] [Google Scholar]
- 14.Wolff EM, Liang G, Jones PA. Mechanisms of Disease: genetic and epigenetic alterations that drive bladder cancer. Nat Clin Pract Urol. 2005;2:502–10. doi: 10.1038/ncpuro0318. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 16.Jiang L, Gonda TA, Gamble MV, et al. Global hypomethylation of genomic DNA in cancer-associated myofibroblasts. Cancer Res. 2008;68:9900–8. doi: 10.1158/0008-5472.CAN-08-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chalitchagorn K, Shuangshoti S, Hourpai N, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–6. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 18.Hsiung DT, Marsit CJ, Houseman EA, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–14. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 19.Issa J. Aging, DNA methylation and cancer. Crit Rev Oncol Hematol. 1999;32:31–43. doi: 10.1016/s1040-8428(99)00019-0. [DOI] [PubMed] [Google Scholar]
- 20.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–62. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 22.Smith SS, Crocitto L. DNA Methylation in Eukaryotic Chromosome Stability Revisited: DNA Methyltransferase in the Management of DNA Conformation Space. Mol Carcinog. 1999;26:1–9. doi: 10.1002/(sici)1098-2744(199909)26:1<1::aid-mc1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 23.Figueiredo JC, Grau MV, Wallace K, et al. Global DNA hypomethylation (LINE-1) in the normal colon and lifestyle characteristics and dietary and genetic factors. Cancer Epidemiol Biomarkers Prev. 2009;18:1041–9.32. doi: 10.1158/1055-9965.EPI-08-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect. 2008;116:1547–52. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarantini L, Bonzini M, Apostoli P, et al. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect. 2009;117:217–22. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bollati V, Baccarelli A, Hou L, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–80. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 27.Moore LE, Pfeiffer RM, Poscablo C, et al. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol. 2008;9:359–66. doi: 10.1016/S1470-2045(08)70038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graziano F, Kawakami K, Ruzzo A, et al. Methylenetetrahydrofolate reductase 677C/T gene polymorphism, gastric cancer susceptibility and genomic DNA hypomethylation in an at-risk Italian population. Int J Cancer. 2006;118:628–32. doi: 10.1002/ijc.21397. [DOI] [PubMed] [Google Scholar]
- 29.Karagas MR, Tosteson TD, Blum J, Morris JS, Baron JA, Klaue B. Environ Health Perspect. Vol. 106. 1998. Design of an epidemiologic study of drinking water arsenic exposure and skin and bladder cancer risk in a U.S. population; pp. 1047–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karagas MR, Stukel TA, Morris JS, et al. Skin cancer risk in relation to toenail arsenic concentrations in a US population-based case-control study. Am J Epidemiol. 2001;153:559–65. doi: 10.1093/aje/153.6.559. [DOI] [PubMed] [Google Scholar]
- 31.England RPM Pyro Q-CpG2: quantitative analysis of methylation in multiple CpG sites by PyrosequencingR. Nat Methods. 2005;2:1–2. [Google Scholar]
- 32.Karagas MR, Park S, Nelson HH, et al. Methylenetetrahydrofolate reductase (MTHFR) variants and bladder cancer: a population-based case-control study. Int J Hyg Environ Health. 2005;208:321–7. doi: 10.1016/j.ijheh.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Wallace K, Kelsey KT, Schned A, Morris JS, Andrew AS, Karagas MR. Selenium and risk of bladder cancer: a population-based case-control study. Cancer Prev Res (Phila Pa) 2009;2:70–3. doi: 10.1158/1940-6207.CAPR-08-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Maarri O, Becker T, Junen J, et al. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet. 2007;122:505–14. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- 35.Zhong CX, Mass MJ. Both hypomethylation and hypermethylation of DNA associated with arsenite exposure in cultures of human cells identified by methylation-sensitive arbitrarily-primed PCR. Toxicol Lett. 2001;122:223–34. doi: 10.1016/s0378-4274(01)00365-4. [DOI] [PubMed] [Google Scholar]
- 36.Zhao CQ, Young MR, Diwan BA, Coogan TP, Waalkes MP. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc Natl Acad Sci USA. 1997;94:10907–12. doi: 10.1073/pnas.94.20.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H, Li S, Liu J, Diwan BA, Barrett JC, Waalkes MP. Chronic inorganic arsenic exposure induces hepatic global and individual gene hypomethylation: implications for arsenic hepatocarcinogenesis. Carcinogenesis. 2004;25:1779–86. doi: 10.1093/carcin/bgh161. [DOI] [PubMed] [Google Scholar]
- 38.Pilsner JR, Liu X, Ahsan H, et al. Folate deficiency, hyperhomocysteinemia, low urinary creatinine, and hypomethylation of leukocyte DNA are risk factors for arsenic-induced skin lesions. Environ Health Perspect. 2009;117:254–60. doi: 10.1289/ehp.11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pilsner JR, Liu X, Ahsan H, et al. Genomic methylation of peripheral blood leukocyte DNA: influences of arsenic and folate in Bangladeshi adults. Am J Clin Nutr. 2007;86:1179–86. doi: 10.1093/ajcn/86.4.1179. [DOI] [PubMed] [Google Scholar]
- 40.Kaneda A, Tsukamoto T, Takamura-Enya T, et al. Frequent hypomethylation in multiple promoter CpG islands is associated with global hypomethylation, but not with frequent promoter hypermethylation. Cancer Sci. 2004;95:58–64. doi: 10.1111/j.1349-7006.2004.tb03171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–36. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sassaman DM, Dombroski BA, Moran JV, et al. Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]