Abstract
Caloric restriction (CR) without malnutrition slows the aging process and extends lifespan in diverse species by unknown mechanisms. The inverse linear relationship between calorie intake and lifespan suggests that regulators of energy metabolism are important in CR’s actions. Studies in several species reveal tissue-specific changes in energy metabolism with CR and suggest that metabolic reprogramming plays a critical role in its mechanism of aging retardation. We herein describe common signatures of CR and suggest how they may slow aging. We discuss recent advances in understanding the function of key metabolic regulators that likely coordinate the response to altered nutrient availability with CR, and how the pathways they regulate may retard the aging process.
Aging and CR impact energy metabolism
Caloric restriction (CR) extends average and maximum lifespan and delays the onset of age-associated phenotypes in diverse species [1,2,3]. In mammals, age-associated phenotypes include diseases such as cardiovascular disease, cancer, and diabetes as well as muscle atrophy. Our recent demonstration of the conserved effect of CR on “health-span” and survival in non-human primates [4] confirms the translatability of CR research as a means to explore aging and disease onset, and underscores the importance of elucidating CR’s underlying mechanisms.
Studies in diverse species suggest that dysregulation of mitochondrial energy metabolism is a hallmark of aging and disease [1]. In contrast, a coordinated increase in the expression of genes involved in energy metabolism is a prominent feature of CR in mouse tissues including heart, skeletal muscle, and white adipose tissue. These transcriptional alterations are indicative of metabolic reprogramming, a change in how energy is generated and how fuel is utilized. This altered metabolic state results in a shift from fat storage to fat utilization, impacting stress signaling pathways and reactive oxygen species production. Multiple cellular pathways, previously considered to be functionally distinct, have now been shown to influence energy metabolism. A new perspective is emerging where this complex interplay of factors coordinates the cellular metabolic response to changes in nutrient availability, energetic demand or redox status. The fact that many of these regulatory molecules have been associated with longevity suggests that maintenance of the integrity of metabolic regulation is a fundamental aspect of youth and a vulnerable element in the process of aging.
Common signatures of aging and CR
A primary challenge in the study of aging arises from the biological complexity of the process itself. In any given tissue, distinction of events that may be considered as causative in aging from those that are compensatory or symptomatic requires the resolution of a highly integrated signaling network. Gene expression profiling using microarrays has proven to be an informative approach for the analysis of complex processes. Large-scale analysis of age-associated transcriptional changes among 16 different mouse tissues demonstrates that most age-related changes in gene expression are of relatively low magnitude (< 2 fold) and that the number and identity of genes that are regulated with age are tissue specific [5]. Gene set enrichment analysis screens for coordinated regulation of functionally related genes and has the potential to reveal cellular pathways that are important in directing aging. A number of gene sets display a high degree of tissue specificity; for example, the effect of age in increasing expression of the cytosolic ribosome gene set in neural tissues is opposite that observed in other tissues examined. The apparent aging of each tissue evaluated based on the transcriptional profiles correlates among tissues indicating that, despite tissue-specific differences in the effect of age on transcriptional profiles, the rate of aging across tissues appears to be coordinated [5]. This observation is suggestive of a role for systemic factors in orchestrating the aging of the animal as a whole.
Mitochondrial energy metabolism
A comparison across humans, mice, flies and worms has identified downregulation of nuclear-encoded genes of the mitochondrial electron transport system (ETS) as the only age effect common to all four species [5]. A meta-analysis of age-related gene expression profiles from 27 datasets generated from human, rat and mouse tissues identified several signatures of aging, including increased expression of genes involved in inflammation and immune responses, and reduced expression of genes involved in mitochondrial energy metabolism [6]. At the level of individual tissues, CR prevents the majority of these age-associated changes in gene expression [7].
The decline in mitochondrial function with age is opposed by CR, where the transcription of nuclear encoded genes involved in the ETS is enhanced and mitochondrial activities are maintained [1]. The impact of CR on mitochondria may not be fully appreciated. In vitro measures of enzymatic activities in isolated mitochondria reflect the activities of recoverable mitochondria and often reveal maximal capacity rather than innate activity. Furthermore, differences in mitochondrial size and density can confound normalization of data. A comparison of mitochondria isolated from skeletal muscle from young (3 months old) or late-middle age (18 months old) mice revealed a greater number of mitochondria in the mature animals and an overt impairment in respiration when measured per mitochondrion [8]. These changes were associated with increased oxidative damage at the protein level. UCP2 and UCP3 uncoupling proteins have been previously associated with reduced reactive oxygen species (ROS) production, and UCP3 expression is increased in tissues from CR animals [9]. Despite elevated levels of UCP3, proton leak is lower in mitochondria from CR animals suggesting that its fatty acid transporter activity may be more important than its uncoupling capacity in this context.
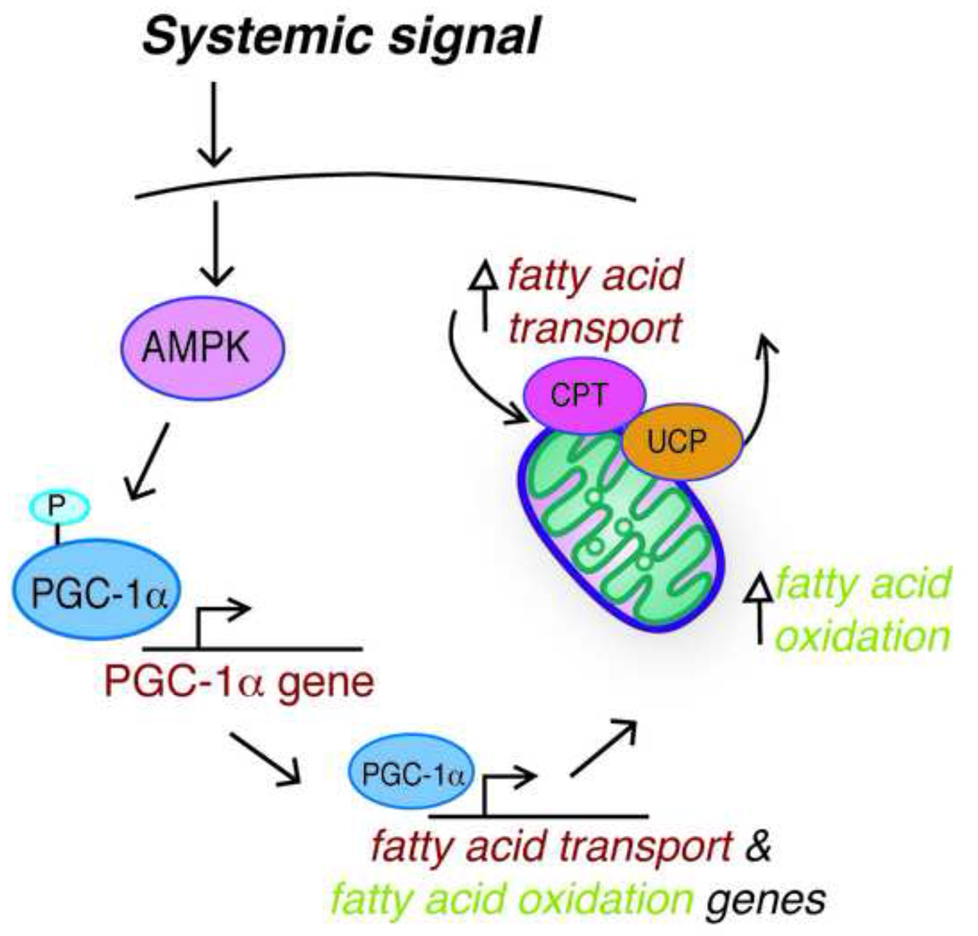
An interesting connection between systemic signaling and cellular metabolic function lies in the dependence of the gut hormone ghrelin on UCP2 for regulation of feeding behavior. Ghrelin levels increase in the unfed state; in the hypothalamus, ghrelin induces mitochondrial proliferation, increases respiration and reduces ROS [10]. Ghrelin signaling leads to increased fatty acid metabolism by an AMPK (AMP-activated protein kinase), fatty acid transporter CPT1 (carnitine palmitoyltransferrase 1), and UCP2 dependent mechanism [10]. It will be interesting to see whether a similar cellular fuel switch mechanism is important in CR’s actions (Figure 1).
Figure 1. Systemic signaling regulates the shift in mitochondrial fuel utilization in response to CR.
We propose a model where an AMPK and PGC-1α dependent signaling pathway permits a coordinate shift in fuel utilization in response to systemic input. Mitochondrial fatty acid oxidation is promoted through enhanced expression of genes encoding enzymes involved in β-oxidation and molecules involved in fatty acid transport.
A role for p53 in mitochondrial respiration has been described, where p53 deficiency is associated with a metabolic switch away from respiration toward glycoloysis [11]. In skeletal muscle of p53 deficient adult mice, mitochondrial content is diminished, respiration is lower and ROS production is elevated [12]. These mice display diminished physical performance and appear resistant to mitochondrial driven apoptosis. Aging is associated with an increase in the p53-mediated transcriptional program in mice [13]. In skeletal muscle, age induces a coordinated increase in genes activated by p53 with a concomitant reduction in expression of genes that inhibit p53. This signature of increased p53 activation is not limited to skeletal muscle but is also evident in multiple tissues and is substantially prevented by CR [13].
IGF/Insulin signaling
Insulin resistance is a well-recognized phenotype of mammalian aging, while improved insulin sensitivity is a hallmark of CR. Many long-lived mouse models display increased insulin sensitivity compared to wildtype littermates connecting this arm of endocrine signaling to longevity [14]. The extent to which insulin or IGF (insulin-like growth factor) signaling pathways are involved in CR’s actions has been investigated in several species, and any mechanistic overlap appears partial [15].
Reduced energy intake by CR or by increased energy expenditure through exercise improves glucose tolerance and insulin sensitivity in mice [16] and in humans [17]. A moderate 24 week CR diet (18% restriction) lowered circulating levels of IGF-1 in mice [16]; however, 1 year of a 20% restriction diet did not reduce IGF-1 levels in humans [18]. This difference may be due to dietary composition rather than a species-specific impact of CR. Therefore, it remains unclear whether reduced IGF-1 directly contributes to the beneficial effects of CR.
In mice, CR induces a marked increase in expression of genes involved in energy metabolism in white adipose tissue, including a striking and coordinated induction of nuclear genes encoding components of the mitochondrial ETS, and increased mitochondrial activity [19,20]. Fat-specific insulin receptor knockout mice (FIRKO) have lower fat mass, lower body weight, and a somewhat extended lifespan (but not as long as seen for mice on CR). Similar to CR mice, expression of genes involved in multiple metabolic pathways is increased in white adipose tissue from FIRKO mice [21], as well as increased expression of nuclear encoded mitochondrial genes (~1.5 – 2.0 fold range in adipose tissue from FIRKO mice). Gene set enrichment analysis suggests that CR preserves IGF and mTOR (target of rapamycin) signaling, pathways that are generally associated with promotion of growth [20]. It seems likely that insulin/IGF signaling is involved in CR’s actions in mammals, though perhaps through a shift in the balance of downstream cellular signaling rather than a simple decrease in receptor signaling.
Both insulin receptor substrates IRS1 and IRS2 have been implicated in longevity [22,23]. The influence of reduced IRS1 on longevity is gender specific; IRS1 null female mice live longer than their wildtype littermates, whereas IRS1 null males show no changes in longevity [22]. In addition to being very small (~17g at 22 months of age compared with ~35g for wildtype controls), IRS1 null females have much lower body fat (~50%) without differing in average food intake. The case for IRS2 is somewhat controversial: IRS2 heterozygote mice have been reported as long-lived in one study [23] and as having no impact on longevity in the above study [22] even though both genetic manipulations were in the same C57Bl/6 background. An overt difference in the study where IRS2 impacted lifespan lies in the diet and resulting large size of the mice (~55–60g at 22 months for both IRS2+/− and wildtype controls). The mice in these two studies would not be expected to be metabolically comparable due to overt differences in body size and composition. The disparity in longevity effects between the studies underscores the importance of dietary composition and knowing caloric intake in determining the impact of any manipulation on longevity.
A possible connection between mitochondrial function and insulin signaling stems from the observation that excess fat intake is associated with insulin resistance in skeletal muscle. A high fat diet induces increased mitochondrial hydrogen peroxide emission in permeabilized muscle fibers [24]. When increased hydrogen peroxide levels are blocked by either an antioxidant compound targeted to mitochondria or genetic introduction of mitochondrial targeted catalase, insulin sensitivity is preserved despite excess fat intake [24]. These data argue that changes in mitochondrial function contribute to alterations in insulin signaling, and suggest that the reduction in ROS production with CR may contribute to improved insulin sensitivity.
Adiponectin
One of the most overt phenotypes of CR is the change in body composition. The reduction in total body weight usually reflects the level of CR (i.e., 30% less food lowers body weight by ~30%), but weight loss is not distributed equally among tissues. Proportionately more fat mass is lost in mice, monkeys, and humans fed a CR diet (i.e., 30% CR leads to ~70% less fat). Until fairly recently, white adipose tissue was seen as a biologically inert lipid storage depot. The current view embraces the systemic role of white adipose tissue in influencing metabolism, inflammation, and immune responses that is implemented through a suite of adipose-derived signaling factors collectively known as adipokines [25,26].
Adiponectin is one such adipokine that may be an important mechanistic component of CR’s action. In rats, serum adiponectin levels correlate negatively with fat mass and are elevated with CR [27]. These increased adiponectin levels are associated with increased fatty acid oxidation in adipose tissue and reduced lipid accumulation in peripheral tissues [28]. In adipose tissue from CR mice, adiponectin is detected at increased levels in the smaller adipocytes that result from CR.
Adiponectin circulates in the blood as a multimeric protein of three distinct molecular weight forms, and its ability to multimerize is essential for proper function [29]. The interaction of adiponectin with growth factors is multimer-specific, and the activation of downstream targets of adiponectin signaling is multimer- and tissue-specific. Importantly, CR increases relative levels of the high molecular weight form [30], suggesting that adiponectin activity may be qualitatively different in CR animals in addition to being quantitatively greater.
Adiponectin has a profound effect on systemic factors associated with metabolism and inflammation. Transgenic overexpression of adiponectin in mice prevents several abnormalities induced by a high fat diet, including elevated serum glucose and insulin levels [31]. Despite the increased caloric intake in adiponectin-overexpressing mice fed a high fat diet, both adipocyte size and macrophage infiltration of adipose tissue are decreased, compared to wildtype controls. Furthermore, chronic adiponectin treatment of a genetic mouse model of diabetes (db/db) alleviates symptoms such as insulin resistance, hyperglycemia, and hypertriglycermia [32]. These data indicate that adiponectin is a critical regulator of metabolic homeostasis and suggest that the elevated adiponectin levels observed in CR animals may contribute to improvements in glucoregulatory function.
Adiponectin signaling regulates mitochondrial energy metabolism in skeletal muscle in mice, and adiponectin knockout mice display insulin resistance and reduced mitochondrial enzyme activity [33]. Adiponectin treatment of human myotubes causes an AMPK dependent increase in mitochondrial biogenesis and enzymatic activities and reduces ROS production. Activation of mitochondria is concomitant with an increase in the levels of mitochondrial regulator PGC-1α (peroxisome proliferator activated receptor gamma co-activator 1 alpha) [33], a direct target of AMPK phosphorylation that increases its own expression when activated by AMPK [34]. In human skeletal muscle, CR induces mitochondrial biogenesis, and PGC-1α levels are increased [35]. PGC-1α stability is regulated by GSK3β glycogen synthase kinase 3 beta) [36], and adiponectin also influences GSK3β activity though modulation of inhibitory phosphorylation of GSK3β [37]. GSK3β is negatively regulated by CR in adipose tissue [36], suggesting that this kinase may also contribute to CR-induced metabolic reprogramming.
The cardioprotective effect of CR is abrogated in transgenic adiponectin anti-sense mice and in mice treated with AMPK inhibitor [30], further supporting a role for the adiponectin-AMPK pathway in CR. CR accelerates the revascularization of ischemic muscle in an adiponectin-dependent manner. In this case, adiponectin acts through AMPK to activate eNOS (endothelial nitric oxide synthase) [38]. Adiponectin-mediated stimulation of eNOS also protects against cerebral ischemic injury [39]. eNOS has previously been associated with CR where it influences mitochondrial biogenesis and expression of the NAD dependent deacetylase SIRT1 in multiple tissues [40]. Interestingly, in cultured adipocytes, SIRT1 has been implicated in adiponectin secretion through its negative impact on factors that regulate secreted protein release from the endoplasmic reticulum [41]. It is not clear whether this is a direct effect on adiponectin processing, because SIRT1 activation prevents adipogenesis and adiponectin is released from mature adipocytes only.
Together these findings have three important implications: first, that adiponectin plays a pivotal role in CR’s mechanism at the systemic level; second, that changes in mitochondrial function are a critical downstream event in adiponectin signaling; and third, that white adipose tissue function may be a key factor in CR and in the aging process.
Candidate regulatory pathways in CR
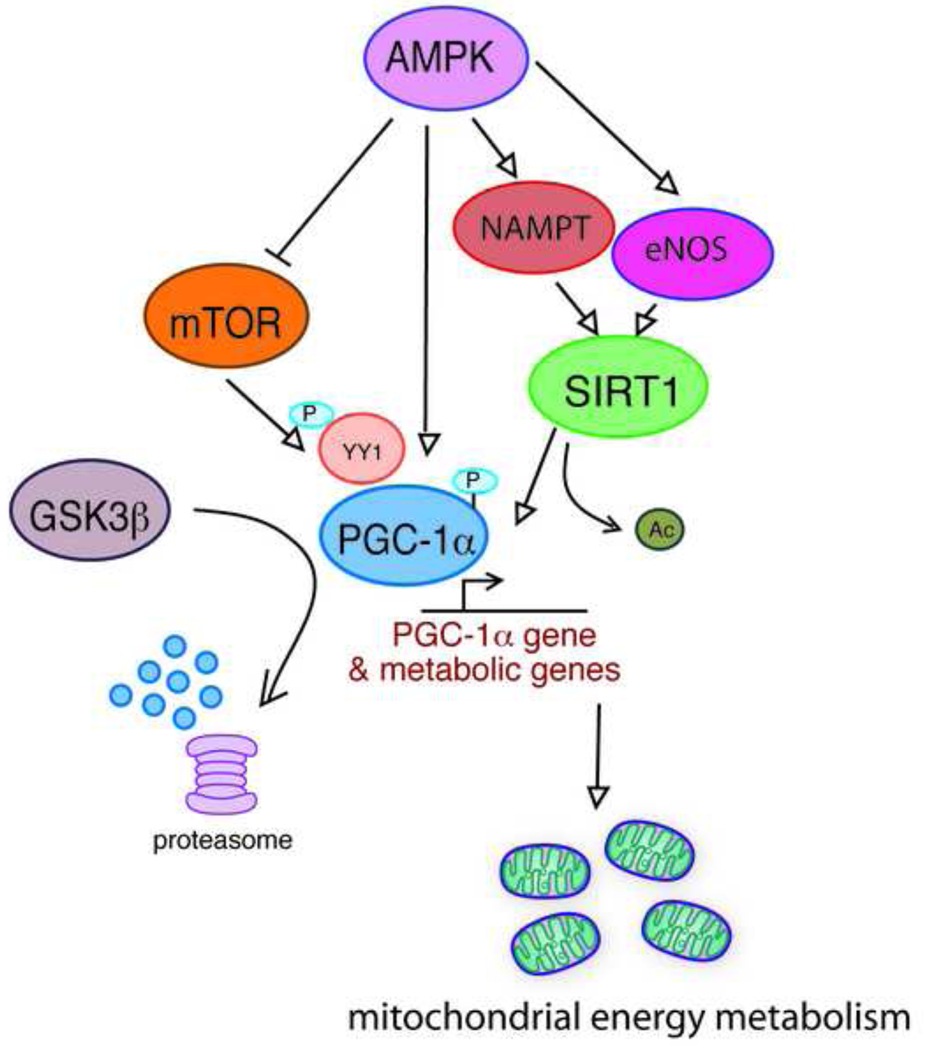
At the most simple level, the mechanisms of CR appear rooted in the response to reduced energy availability, resulting in the induction of an altered metabolic state that promotes longevity. The conserved role of mitochondrial energy metabolism in aging and in CR focuses attention on factors that influence mitochondrial number and function, and supports nutrient sensitive metabolic regulators as being strong candidates in the mechanisms of CR (Figure 2). We describe below some of the more prominent factors associated with the regulation of longevity, or metabolism, or both. There is a vast literature describing studies of the functionality and regulation of these factors, from which we sample just a few that we find particularly striking.
Figure 2. Nutrient sensitive metabolic regulators are strong candidates as effector molecules in the mechanisms of CR.
A complex network of interactions among these molecules has been described, although much remains to be learned about their regulation specifically in the context of CR. A unifying theme is the regulation of energy metabolism: alterations in energy metabolism are a hallmark of CR and likely are critical events in CR’s mechanisms of aging retardation.
NAD dependent deacetylase SIRT1
Sirtuins are a family of proteins with homology to yeast regulator of silencing Sir2. SIRT1 null mice have defects in liver metabolism, shortened lifespan and are not responsive to CR [42]. A number of reports indicate that CR regulates SIRT1, the closest mammalian homologue. CR increases SIRT1 in white adipose tissue [36,40,43,44], although there is a lack of agreement on the impact of CR on SIRT1 liver and muscle [43,44,45]. The importance of tissue-specificity in the SIRT1 response to CR can be inferred from studies of knockout and transgenic mouse models. The liver-specific knockout SIRT1 mouse is resistant to metabolic derangements arising from a high fat diet [43]; however, activation of SIRT1 with a synthetic activator or transgenic expression of SIRT1 protect against a high fat diet [46,47,48].
In mice fed a high fat diet, activation of SIRT1 enhances fat oxidation in peripheral tissues and enhances expression of genes involved in the ETS [47], metabolic alterations that are likely responsible for the protective effect of increased SIRT1. Interestingly, stimulation of NADH oxidation increases the NAD+/NADH ratio, protects against diet induced obesity, and is associated with enhanced mitochondrial fatty acid oxidation, increased PGC-1α and increased SIRT1 [49]. SIRT1 gain-of-function mice have improved glucose tolerance [46,48] and increased serum adiponectin levels despite no change in body weight compared to wildtype controls [46]. In contrast, mice with SIRT1 overexpression primarily in white adipose tissue have lower body weights and reduced levels of adiponectin [50]. These data underscore the importance of tissue-specific functions of SIRT1 and, relevant to the mechanisms of CR, implicate SIRT1 in regulation of adiponectin. A better understanding of the effect of altered SIRT1 levels and activity may be gleaned through further genetic approaches using conditional and tissue-specific manipulations. It will be particularly important to establish SIRT1 functions that are physiologically normal and those that manifest only with manipulated large-scale changes in SIRT1.
Regulator of mitochondrial metabolism PGC-1α
As the role of PGC-1α in aging and CR has been recently reviewed [51], herein we briefly discuss some potentially relevant new data relating to PGC-1α in oxygen sensing and ROS signaling.
The CR dependent increase in PGC-1α expression in multiple tissues is coincident with increased expression of eNOS [40]. Inhibition of eNOS during CR prevents the activation of SIRT1, an activator of PGC-1α, and abrogates the protective effect of CR. eNOS null mice fail to initiate mitochondrial reprogramming that is associated with CR, possibly due to reduced levels of SIRT1 [40]. CR enhances ischemic tolerance by a nitric oxide dependent mechanism [45]. In endothelial cells, PGC-1α levels are decreased by chronic inhibition of eNOS but show the opposite effect upon acute treatments [52], suggesting that the PGC-1α mediated response to nitric oxide is highly dynamic and temporally sensitive. PGC-1α also regulates mitochondrial adaptation as part of the cellular response to oxidative stress [36]. PGC-1α activity is regulated through changes in its cellular localization and protein stability allowing the rapidity and persistence of the PGC-1α response to be independently controlled. In skeletal muscle cells, extended exposure to oxidative stress activates AMPK leading to induction of PGC-1α expression and decreased ROS production [53].
Interestingly, high levels of PGC-1α overexpression increase respiration and induce HIF-1α (hypoxia inducible factor 1 alpha) signaling by a mechanism that involves stabilization of the HIF-1α protein [54]. This feed forward signal presumably allows cells to respond appropriately to changes in oxygen availability. The composition of the ETS can be regulated to optimize respiration efficiency through a HIF-1α dependent mechanism [55]. A switch between two distinct COX4 Complex IV subunit isoforms alters ATP production, oxygen consumption and ROS generation. HIF-1α activity correlates with its cellular protein levels; in contrast, HIF-2α activity is regulated by post-translational modifications including deacetylation by SIRT1 [56]. HIF-1α is downregulated in white adipose tissue from CR mice [57], and recently HIF-1α deficiency has been implicated in longevity and in CR in C. elegans [58]. These data place PGC-1α in a central role in the cellular response to oxygen availability, and suggest that alterations in mitochondrial energy metabolism with CR may be communicated through ROS to impact diverse cellular pathways.
Energy and nutrient sensitive regulators AMPK and mTOR
Molecular interactions studied outside of the realm of aging research lend support for AMPK and mTOR playing a role in the mechanism of CR. Importantly, activation of either AMPK or mTOR positively influences mitochondrial respiration, although the impact of tissue specificity in CR’s influence on these factors remains underappreciated. Activation of AMPK in mice has a pronounced effect on skeletal muscle metabolism, where indicators of mitochondrial density are increased and UCP3 is upregulated [59]. AMPK directly phosphorylates PGC-1α and enhances its ability to co-activate its own promoter [34], and therefore may be the principal means by which AMPK influences mitochondrial function.
The ability of AMPK to regulate mitochondrial biogenesis in skeletal muscle becomes attenuated with age, and the response to either chronic or acute AMPK stimulation is muted in older animals [60]. Although AMPK directly activates PGC-1α by phosphorylation, it also acts indirectly by enhancing the activity of SIRT1, the enzyme that deacetylates and thereby activates PGC-1α [61]. AMPK has the ability to increase the cellular NAD+/NADH ratio through its influence on respiration and fatty acid β-oxidation, and in this way indirectly leads to SIRT1 activation. Glucose restriction activates AMPK and also increases the cellular NAD+/NADH ratio [62], although in this case the NAD salvage pathway enzyme NAMPT (nicotinamide phosphoribosyltransferase) not only influences cellular NAD+/NADH but also reduces levels of the SIRT1 inhibitor nicotinamide. The elaborate crosstalk between these highly conserved pathways for energy sensing and metabolic cofactor production may be a fundamental aspect of the link between longevity and the regulation of metabolism.
In contrast to AMPK that responds to energy stress, mTOR is activated by increased nutrient availability. In mice, short-term CR (4 weeks) results in increased AMPK phosphorylation and activation, and decreased mTOR phosphorylation and downstream signaling in liver [63]. AMPK inhibits mTOR complex I by two distinct mechanisms: i) stimulating TSC2 (tuberous sclerosis complex 2), a negative regulator of mTOR, and ii) phosphorylating raptor, an essential binding partner for the induction of ribosome biogenesis and translation by mTOR Complex I [64]. In addition to stimulating AMPK activity by CR in liver, AKT kinase activity is reduced, alleviating the inhibitory phosphorylation of TSC2 [63]. These findings indicate that there is a coordinated inhibition of mTOR in liver with CR, with multiple facets of mTOR regulation at play.
In mammalian cells, in contrast to yeast (Box 1), mTOR Complex I activity correlates positively with oxidative capacity such that cells with increased association of mTOR and raptor respire more [65]. Treatment of muscle cells with rapamycin causes a decrease in the expression of mitochondrial genes and reduces oxygen consumption, an effect that is also observed in skeletal muscle of rapamycin-treated mice [66]. mTOR and raptor together regulate PGC-1α expression by influencing PGC-1α coactivation of its own promoter. The role of mTOR in CR is likely tissue-specific, in that CR reduces mTOR signaling in liver but CR increases mitochondrial ETS expression in skeletal muscle, a finding that is not consistent with reduced mTOR signaling. Gene set enrichment analysis indicates enhanced mTOR signaling in adipose tissue with CR [20]. Interestingly, adipose tissue-specific knockout of raptor (component of mTOR Complex I) protects against diet-induced obesity [67]. In contrast to what might be expected, the absence of raptor in adipose tissue is associated with reduced serum levels of adiponectin even though there was a reduction in adiposity in these mice. These findings suggest that mTOR might be important in regulating adiponectin secretion.
Box 1. Convergence of yeast longevity pathways on NAD and respiration
In yeast, there are two distinct copies of TOR that participate in separate complexes only one of which (Tor1:Complex I) is rapamycin-sensitive. Reduced Tor1 signaling extends replicative lifespan in yeast by a mechanism that requires the nicotinamidase PNC1, a gene in the NAD salvage pathway [70]. This gene is also required for lifespan extension by CR in yeast, and the mechanism of induction of PNC1 by either CR or rapamycin treatment involves the same transcription factors activated through increased nuclear localization. Pnc1 depletes nicotimamide, similar to the mammalian NAMPT, and activates Sir2, the homologue of mammalian SIRT1. Deletion of TOR1 also extends chronological lifespan in yeast and is associated with increased respiration and increased expression of genes encoding components of the mitochondrial ETS [71]. Yeast do not favor respiration when grown in glucose; however, deletion of TOR1 permits de-repression of respiration and increased expression mitochondrial encoded genes.
The beneficial effect of mTOR inhibition on lifespan in mice has been reported [68], where rapamycin treatment was initiated in relatively old mice. Unlike CR, rapamycin treatment shows sexual dimorphism in mice, being more efficacious in females. These data suggest that the role of mTOR in the mechanisms of CR is likely part of a complex network of effector molecules that in combination orchestrate the reprogramming of energy metabolism.
Conclusions
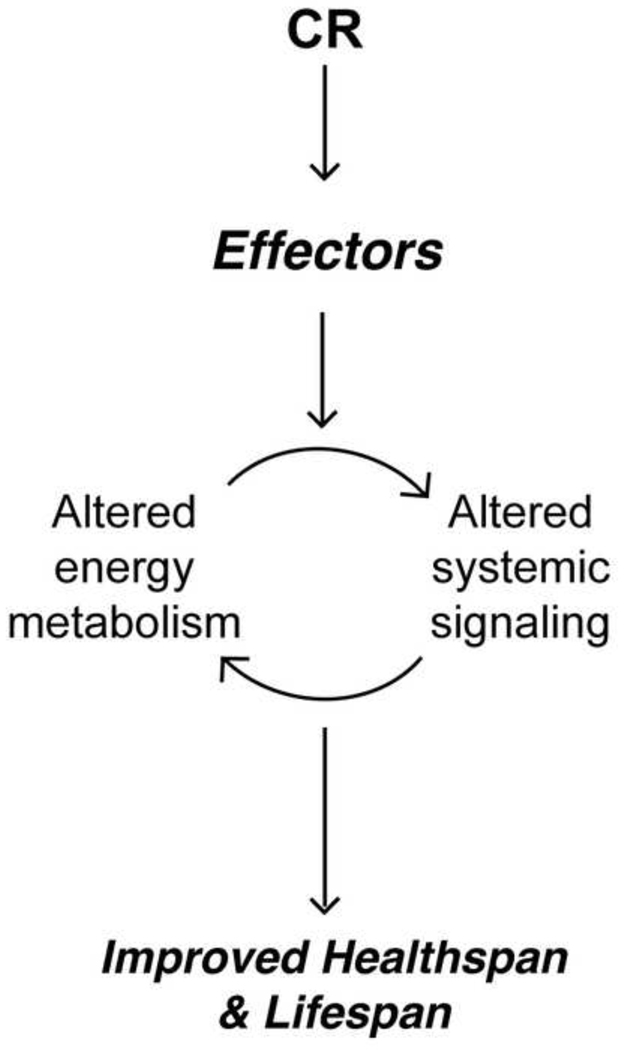
We have outlined some of the common signatures of CR including altered mitochondrial energy metabolism, enhanced sensitivity of insulin signaling, and increased circulating levels of the adipokine, adiponectin. We highlight elements of overlap between signatures and discuss how these hallmarks of CR may be mechanistically connected. We describe several candidate effectors in the mechanisms of CR based on the fact that they can be linked to the above signatures, are firmly established as regulators of metabolism, and have been associated directly or indirectly in the regulation of longevity. We suggest that metabolic reprogramming is an essential contributor to the mechanisms of CR, and that the factors described herein likely play an important role in establishing the CR-induced altered metabolic state that promotes health and longevity (Figure 3).
Figure 3. The improvement in healthspan and lifespan by CR is a coordinated active response.
Key effector molecules implement a reprogramming of energy metabolism in response to reduced energy availability. This altered metabolic state is maintained and perpetuated though systemic signaling.
A recent and very interesting Drosophila study further supports a critical role for mitochondrial energy metabolism in the mechanisms of CR [69]. In flies on CR, transcripts of mitochondrial ETS components are preferentially processed due to the structure of their upstream untranslated regions. This attribute ensures a relative enrichment in translation of these transcripts and ultimately results in increased activity of ETS complexes. The process is regulated by 4E-BP, a translational repressor specifically upregulated by CR in flies, and a component of the TOR signaling pathway. These data reveal the multiplicity of inputs in the regulation of mitochondrial function and introduce a novel element of translational regulation in the mechanisms of CR.
This is an exciting time in CR research. Critical insights into the function and regulation of key elements in aging and CR are being drawn from a broad range of studies in diverse models and species; these include short-lived and long-lived organism studies, cell culture and whole animal studies, and genetic and pharmacological manipulation studies. The contribution of each facet permits us to view an increasingly complex network by which CR appears to influence aging. Continued advances are anticipated, with the promise of translation of this knowledge to the eventual benefit of human health (Box 2).
Box 2. Outstanding questions
To what extent are mechanisms of CR identified in short-lived species conserved in primate species?
Which tissues contribute most significantly to aging retardation by CR?
Which of the prominent regulators of energy metabolism is/are most suitable as a target for the development of nutraceuticals or drugs to mimic the beneficial effects of CR?
Acknowledgement
This work was supported by grants from NIH/NIA (AG11915), NIH/NCI (CA103697) and NIH/NCRR/CTSA (UL1RR025011). We thank Mike Polewski and Josef Clarke for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson RM, Weindruch R. Metabolic reprogramming in dietary restriction. Interdiscip Top Gerontol. 2007;35:18–38. doi: 10.1159/000096554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennedy BK, et al. Ruminations on dietary restriction and aging. Cell Mol Life Sci. 2007;64:1323–1328. doi: 10.1007/s00018-007-6470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piper MD, Bartke A. Diet and aging. Cell Metab. 2008;8:99–104. doi: 10.1016/j.cmet.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Colman RJ, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zahn JM, et al. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Magalhaes JP, et al. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park SK, Prolla TA. Lessons learned from gene expression profile studies of aging and caloric restriction. Ageing Res Rev. 2005;4:55–65. doi: 10.1016/j.arr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Figueiredo PA, et al. Aging impairs skeletal muscle mitochondrial bioenergetic function. J Gerontol A Biol Sci Med Sci. 2009;64:21–33. doi: 10.1093/gerona/gln048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asami DK, et al. Effect of aging, caloric restriction, and uncoupling protein 3 (UCP3) on mitochondrial proton leak in mice. Exp Gerontol. 2008;43:1069–1076. doi: 10.1016/j.exger.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrews ZB, et al. UCP2 mediates ghrelin's action on NPY/AgRP neurons by lowering free radicals. Nature. 2008;454:846–851. doi: 10.1038/nature07181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matoba S, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 12.Saleem A, et al. Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiol Genomics. 2009;37:58–66. doi: 10.1152/physiolgenomics.90346.2008. [DOI] [PubMed] [Google Scholar]
- 13.Edwards MG, et al. Gene expression profiling of aging reveals activation of a p53-mediated transcriptional program. BMC Genomics. 2007;8:80. doi: 10.1186/1471-2164-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiba T, et al. Role of insulin and growth hormone/insulin-like growth factor-I signaling in lifespan extension: rodent longevity models for studying aging and calorie restriction. Curr Genomics. 2007;8:423–428. doi: 10.2174/138920207783591726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- 16.Huffman DM, et al. Effect of exercise and calorie restriction on biomarkers of aging in mice. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1618–R1627. doi: 10.1152/ajpregu.00890.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss EP, et al. Improvements in glucose tolerance and insulin action induced by increasing energy expenditure or decreasing energy intake: a randomized controlled trial. Am J Clin Nutr. 2006;84:1033–1042. doi: 10.1093/ajcn/84.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fontana L, et al. Long-term effects of calorie or protein restriction on serum IGF-1 and IGFBP-3 concentration in humans. Aging Cell. 2008;7:681–687. doi: 10.1111/j.1474-9726.2008.00417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higami Y, et al. Adipose tissue energy metabolism: altered gene expression profile of mice subjected to long-term caloric restriction. Faseb J. 2004;18:415–417. doi: 10.1096/fj.03-0678fje. [DOI] [PubMed] [Google Scholar]
- 20.Linford NJ, et al. Transcriptional response to aging and caloric restriction in heart and adipose tissue. Aging Cell. 2007;6:673–688. doi: 10.1111/j.1474-9726.2007.00319.x. [DOI] [PubMed] [Google Scholar]
- 21.Katic M, et al. Mitochondrial gene expression and increased oxidative metabolism: role in increased lifespan of fat-specific insulin receptor knock-out mice. Aging Cell. 2007;6:827–839. doi: 10.1111/j.1474-9726.2007.00346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selman C, et al. Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J. 2008;22:807–818. doi: 10.1096/fj.07-9261com. [DOI] [PubMed] [Google Scholar]
- 23.Taguchi A, et al. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science. 2007;317:369–372. doi: 10.1126/science.1142179. [DOI] [PubMed] [Google Scholar]
- 24.Anderson EJ, et al. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. J Clin Invest. 2009;119:573–581. doi: 10.1172/JCI37048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 26.Lago F, et al. The emerging role of adipokines as mediators of inflammation and immune responses. Cytokine Growth Factor Rev. 2007;18:313–325. doi: 10.1016/j.cytogfr.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Zhu M, et al. Circulating adiponectin levels increase in rats on caloric restriction: the potential for insulin sensitization. Exp Gerontol. 2004;39:1049–1059. doi: 10.1016/j.exger.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 28.Zhu M, et al. Adipogenic signaling in rat white adipose tissue: modulation by aging and calorie restriction. Exp Gerontol. 2007;42:733–744. doi: 10.1016/j.exger.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, et al. Post-translational modifications of adiponectin: mechanisms and functional implications. Biochem J. 2008;409:623–633. doi: 10.1042/BJ20071492. [DOI] [PubMed] [Google Scholar]
- 30.Shinmura K, et al. Cardioprotective effects of short-term caloric restriction are mediated by adiponectin via activation of AMP-activated protein kinase. Circulation. 2007;116:2809–2817. doi: 10.1161/CIRCULATIONAHA.107.725697. [DOI] [PubMed] [Google Scholar]
- 31.Otabe S, et al. Overexpression of human adiponectin in transgenic mice results in suppression of fat accumulation and prevention of premature death by high-calorie diet. Am J Physiol Endocrinol Metab. 2007;293:E210–E218. doi: 10.1152/ajpendo.00645.2006. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, et al. Post-translational modifications of the four conserved lysine residues within the collagenous domain of adiponectin are required for the formation of its high molecular weight oligomeric complex. J Biol Chem. 2006;281:16391–16400. doi: 10.1074/jbc.M513907200. [DOI] [PubMed] [Google Scholar]
- 33.Civitarese AE, et al. Role of adiponectin in human skeletal muscle bioenergetics. Cell Metab. 2006;4:75–87. doi: 10.1016/j.cmet.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jager S, et al. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Civitarese AE, et al. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson RM, et al. Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell. 2008;7:101–111. doi: 10.1111/j.1474-9726.2007.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, et al. Adiponectin modulates the glycogen synthase kinase-3beta/beta-catenin signaling pathway and attenuates mammary tumorigenesis of MDA-MB-231 cells in nude mice. Cancer Res. 2006;66:11462–11470. doi: 10.1158/0008-5472.CAN-06-1969. [DOI] [PubMed] [Google Scholar]
- 38.Kondo M, et al. Caloric restriction stimulates revascularization in response to ischemia via adiponectin-mediated activation of endothelial nitric-oxide synthase. J Biol Chem. 2009;284:1718–1724. doi: 10.1074/jbc.M805301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimura M, et al. Adiponectin prevents cerebral ischemic injury through endothelial nitric oxide synthase dependent mechanisms. Circulation. 2008;117:216–223. doi: 10.1161/CIRCULATIONAHA.107.725044. [DOI] [PubMed] [Google Scholar]
- 40.Nisoli E, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 41.Qiang L, et al. Adiponectin secretion is regulated by SIRT1 and the endoplasmic reticulum oxidoreductase Ero1-L alpha. Mol Cell Biol. 2007;27:4698–4707. doi: 10.1128/MCB.02279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boily G, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen D, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen HY, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 45.Shinmura K, et al. Impact of 6-mo caloric restriction on myocardial ischemic tolerance: possible involvement of nitric oxide-dependent increase in nuclear Sirt1. Am J Physiol Heart Circ Physiol. 2008;295:H2348–H2355. doi: 10.1152/ajpheart.00602.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banks AS, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feige JN, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 48.Pfluger PT, et al. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hwang JH, et al. Pharmacological stimulation of NADH oxidation ameliorates obesity and related phenotypes in mice. Diabetes. 2009;58:965–974. doi: 10.2337/db08-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bordone L, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 51.Anderson R, Prolla T. PGC-1alpha in aging and anti-aging interventions. Biochim Biophys Acta. 2009;1790:1059–1066. doi: 10.1016/j.bbagen.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borniquel S, et al. Nitric oxide regulates mitochondrial oxidative stress protection via the transcriptional coactivator PGC-1alpha. FASEB J. 2006;20:1889–1891. doi: 10.1096/fj.05-5189fje. [DOI] [PubMed] [Google Scholar]
- 53.Irrcher I, et al. Interactions between ROS and AMP kinase activity in the regulation of PGC-1alpha transcription in skeletal muscle cells. Am J Physiol Cell Physiol. 2009;296:C116–C123. doi: 10.1152/ajpcell.00267.2007. [DOI] [PubMed] [Google Scholar]
- 54.O'Hagan KA, et al. PGC-1alpha is coupled to HIF-1alpha-dependent gene expression by increasing mitochondrial oxygen consumption in skeletal muscle cells. Proc Natl Acad Sci U S A. 2009;106:2188–2193. doi: 10.1073/pnas.0808801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukuda R, et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 56.Dioum EM, et al. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- 57.Higami Y, et al. Energy restriction lowers the expression of genes linked to inflammation, the cytoskeleton, the extracellular matrix, and angiogenesis in mouse adipose tissue. J Nutr. 2006;136:343–352. doi: 10.1093/jn/136.2.343. [DOI] [PubMed] [Google Scholar]
- 58.Chen D, et al. HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000486. doi: 10.1371/journal.pgen.1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Putman CT, et al. AMPK activation increases uncoupling protein-3 expression and mitochondrial enzyme activities in rat muscle without fibre type transitions. J Physiol. 2003;551:169–178. doi: 10.1113/jphysiol.2003.040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reznick RM, et al. Aging-associated reductions in AMP-activated protein kinase activity and mitochondrial biogenesis. Cell Metab. 2007;5:151–156. doi: 10.1016/j.cmet.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Canto C, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fulco M, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang W, et al. Dietary energy restriction modulates the activity of AMPactivated protein kinase, Akt, and mammalian target of rapamycin in mammary carcinomas, mammary gland, and liver. Cancer Res. 2008;68:5492–5499. doi: 10.1158/0008-5472.CAN-07-6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schieke SM, et al. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- 66.Cunningham JT, et al. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 67.Polak P, et al. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 2008;8:399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 68.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zid BM, et al. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Medvedik O, et al. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bonawitz ND, et al. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]