Abstract
Various methods of protein footprinting use hydrogen peroxide as oxidant. Its removal by various solid-phase desalting methods, catalase treatment, or freeze-drying after the footprinting is critical to insure no uncontrolled oxidation. Although catalase treatment removes hydrogen peroxide with little loss of protein or additional protein oxidation, we discovered that freeze-drying or freezing of the protein in peroxide solution does lead to protein oxidation. Interestingly, the oxidation is not a result of freeze or thaw processes but depends on the temperature and length of time for incubation. After 2 h, apomyoglobin undergoes nearly complete single oxidation at −80 °C and double oxidation at −15 °C. The oxidation extents are in addition to the number of methionine residues. Minimal oxidation is observed at 4 °C and 22 °C compared to oxidation at −80 °C or −20 °C. The concentration of hydrogen peroxide is critical; 75 mM (0.2%) is required to oxidize > 50% of the protein at −15 °C and 100-mM (0.3%) at −80 °C. In addition to Met, ~ 5% of the tryptophan and tyrosine residues are oxidized as well as lower amounts of His and Phe. Oxidation of Val 68 and Val 17 (a buried residue) also occurs, with the oxidation of Val 17 likely occurring by electron transfer from one of two of the oxidized aromatic residues contacting Val 17. We describe here the need to remove the hydrogen peroxide prior to cold storage of proteins, and we also report some preliminary results pertaining to the mechanism of cold, solid-state oxidation.
Keywords: Protein Oxidation, Hydrogen Peroxide, Ice Lattice, Protein Footprinting, Mass Spectrometry
INTRODUCTION
Proteomics has become an essential science in part because identifying proteins that are differentially expressed in a cell is important for understanding disease processes1, 2. Many disease states arise from mis-regulation of proteins and the complexes in which they participate. Part of the goal for the field of systems biology is to map all protein-protein interactions through experimental methods such as yeast two-hybrid assays 3-5 or affinity-purification methods. Mass spectrometry plays a major role in the latter method by identifying the proteins that are present in the complex, and documenting changes in the composition of a protein complex, including post-translational modifications 6, 7.
The next challenge is to determine the structure of protein complexes. Recent developments demonstrate that labeling of solvent-accessible residues gives significant insight into the location of protein-ligand interactions including protein-small molecule footprinting combined with MS identifies residues with altered reactivity as a result of interface formation. Examples of chemical probes are deuterium (H/D amide exchange), amine-specific probes, and hydroxyl radicals, which target principally aromatic side chains and sulfur-containing groups on amino acid residues.
Chance and Brenowitz12 have made significant contributions to hydroxyl-radical chemical footprinting by extending the DNA footprinting approach, introduced by Tullius in 1986 for RNA folding13 and DNA-protein footprinting12, 14. Chance and coworkers10 further expanded the approach to protein allostery15 and to localization of protein-protein interactions. To provide a deeper understanding of radical reactions, they16 characterized the relative reactivity and oxidation products of different amino acid residues.
The synchrotron radiolysis method, used by Chance, is the standard for investigating protein-ligand interactions using the hydroxyl-radical probe. To make these experiments less dependent on access to the beam line on a synchrotron, other labs are focusing on reactions that form hydroxyl radicals by chemical or photochemical processes, including the Fenton reagent, which causes the iron-catalyzed reduction of hydrogen peroxide17, the photolytic cleavage of 5 M hydrogen peroxide with low flux 254-nm light18, and high-voltage electrospray oxidation19. Two pulsed-laser methodologies were described in 2005 20, 21. They both exploited nanosecond pulses of UV light to cleave homolytically hydrogen peroxide forming two hydroxyl radicals. There are important experimental differences in the two approaches, however. The method introduced by Aye uses 100 mM H2O2 (0.3 %) and no chemical scavenger, whereas that introduced by us uses 15 mM H2O2 (0.05 %) and a chemical scavenger to control the lifetime of the hydroxyl radicals, limiting the duration of reaction to less than one microsecond. Both groups are conscientious about the long term effect of H2O2 on proteins and remove it from the solution after the experiment. Aye and coworkers submitted the irradiated sample to flash freezing to quench the reaction and then freeze-dried the samples to remove the H2O2, whereas we added immobilized catalase to consume residual peroxide.
In the research reported in this paper, we investigated the freeze-drying approach to quenching and made the surprising discovery that incubation of apomyoglobin and H2O2 in the solid state results in apomyoglobin oxidation (only when the protein is frozen in the presence of H2O2). We also show that the extent of oxidation depends on the concentration of H2O2, incubation time, and temperature of the solid state; the oxidation also occurs by freeze-drying apomyoglobin in the presence of 100 mM H2O2.
As far as we are able to ascertain, there is no other description of this effect in the literature. Certainly, freezing of cells can cause oxidative damage, but this was traced to iron released during the freeze process, which after thawing allows oxidation of proteins via the Fenton reaction 22-25. There are a few reactions that are accelerated by freezing; interestingly, all involve the reaction of oxygen with small molecules; for example, nitrite 26-28, sulfite 28, ascorbic acid 29, and potassium iodide (at acidic pH) 30. There can be a five-fold acceleration of the rate of oxidation caused by freezing sulfide with H2O2 at basic pH (HS− must be present)31, 32 but in these cases, the authors showed that the operative effect is freeze concentration. As a solution is frozen, product formation increased until the solution was frozen completely (as verified by visual inspection). After this point, no additional product was formed. The research reported here addresses the question of whether oxidation of a protein occurs primarily when its solution is frozen or while frozen. The approach is to evaluate the extent of oxidation as a function of time spent in the frozen matrix and the temperature of the matrix.
EXPERIMENTAL PROCEDURES
Materials & Methods
To investigate oxidation of apomyoglobin (unless otherwise noted, all reagents were from Sigma-Aldrich, St. Louis, MO), a solution with a final concentration of 20 μM protein, 10 mM NaH2PO4 (pH = 7.8) and 150 mM NaCl was used for all experiments. Hydrogen peroxide was added from a 1 M or 150 mM H2O2 stock solution. The sample (20 μL in an Eppendorf vial) was flash frozen by immediately plunging the tube into liquid nitrogen. Following incubation at various temperatures, samples were thawed by handwarming, and the hydrogen peroxide was removed by adding 1 Λ of 1 μM catalase. Freeze-drying was carried out in a speed-vac for 30 min with a 10−3 Torr vacuum without heating.
A solution of 0.5 μL (containing 10 pmol total protein) of the sample was loaded onto a C18 Opti-guard column from Optimize Technologies (Oregon City, OR) that was pre-equilibrated with 150 μL of water. The sample was desalted with 350 μL of water. The protein was eluted from the column into the mass spectrometer by using 20 μL/min 50% CH3CN in 0.2% formic acid. Data acquisition was performed with the Waters Ultima Global Quadrupole time-of-flight mass spectrometer (Milford, MA), operating in the “V mode” at ~10,000 mass resolving power. The summed data were deconvoluted with MaxEnt 1 after selecting appropriate mass ranges, a resolution of 0.5, and a Gaussian damage model of 0.025 to model the true width of the protein isotope envelope.
The protein remaining after the mass spectrometric analysis was digested with trypsin at a 1:20 ratio of enzyme:protein. Following 24 h incubation at 37 °C, the resulting peptide solution was desalted using a Millipore C18 ZipTip (Billerica, MA). The resulting solution was freeze-dried and resuspended in 10 μL of solvent A (97% H2O, 3% CH3CN, 0.1% FA). The sample (2.5 μL) was loaded onto a silica capillary column with a PicoFrit™ tip (New Objective, Inc, Woburn, MA) custom-packed with C18 reverse-phase material (Delta-Pak, 0.075 × 100 mm, 5-μm, 300-Å, Waters Corp., Milord, MA). The gradient was pumped by using an Eksigent NanoLC-1D (Livermore, CA) and was from 0% solvent B (97% CH3CN, 3% H2O, 0.1% FA) to 60% solvent B over 60 min, then to 80% solvent B for 10 min at a flow of 260 nL/min followed by a 10 min re-equilibration step. The flow was directed into the entrance of the heated capillary of an LTQ-FT mass spectrometer (ThermoFisher, San Jose, CA). A mass spectrum of eluting peptides was obtained at high mass resolving power, allowing accurate mass assignments by using the FT mass spectrometer component while MS/MS experiments on ions representing the nine most abundant eluents were rapidly performed in the LTQ at a collision energy of 33%, selecting the precursor mass over a range of 0.5 m/z below to 4.0 m/z above and using wide-band activation. Various m/z ions that were submitted to activation (MS/MS) were placed in a dynamic exclusion list for 30 sec.
Data processing
The data were searched by using Mascot (Matrix Sciences, London, UK) against an E. coli database containing myoglobin to identify the unmodified peptides from myoglobin. The positively identified unmodified peptide product-ion spectra (formatted as SEQUEST dta files) were correlated against all dta files in the dataset by using custom-designed software. The correlation algorithm identified dta files that had both zero and non-zero mass shift correlations. In this manner, peptides modified, for example, by +16 m/z were identified if fragment ions with both no mass shift and some with +16 m/z modifications were seen. The correlation was made for mass shifts from −50 m/z to +50 m/z. Product-ion mass spectra of the identified peptides were manually interpreted by comparing them with the product-ion spectra of the unmodified tryptic peptides, and an assignment was made when there was certainty that a particular residue had been modified. All unmodified peptides were also manually searched for known oxidations that lead to mass modifications of +16, +32 and −22, adjusted according to the charge state of the precursor ion. Product-ion spectra of the identified peptides were manually interpreted by comparing them with the product-ion spectra of the unmodified tryptic peptides, and an assignment was made when there was certainty that a particular residue had been modified.
To calculate solvent exposure of all residues, the solvent exposure calculator available on-line at http://molbio.info.nih.gov/structbio/basic.html was used. This calculator, which was implemented by S. Chacko based on previously published work33, allowed us to compare readily the apo and holo forms of myoglobin as the calculator was able to account for the porphyrin group in the holo form.
Percent oxidized peptide was calculated by dividing the area under the curve for the modified peptide by the area under the curve for the unmodified peptide. The area under the curve (A.U.C.) was determined by the ICIS algorithm in the Qualbrowser package from ThermoFisher (San Jose, CA). We used a baseline window of 40, area noise factor of 5, and peak noise factor of 10. The noise method utilized the repetitive noise feature. Minimum peak width was 2 with a multiplet resolution of 10, area tail extension of 5, and area scan window of 0.
RESULTS AND DISCUSSION
What is an effective way to remove H2O2 post oxidation?
During our development of fast photochemical oxidation of proteins (FPOP)21, we noticed that proteins stored overnight with H2O2 at −80 °C could be oxidized. As this was surprising to us, we initiated further inquiries to illuminate more fully the phenomenon. We first sought to demonstrate that peroxide could be removed completely without oxidizing apomyoglobin. There are a variety of ways to accomplish this removal. One method is to desalt the protein using a C4 ZipTip prior to nanospray analysis17. Similarly, one can use the tC2 SepPak from Waters34, but this is more suitable for larger amounts of protein than used here. A third approach is to use catalase to remove peroxide21, 35, and a fourth is to freeze-dry the solution to evaporate the peroxide20. (We also tried Microcon 10 kDa MW cut-off membranes from Millipore, G-25 Sephadex beads in a micro spin column, and the tC2 SepPak, and found that these approaches were prone to loss of oxidized protein (data not shown)). One issue with oxidized proteins is that they can unfold, exposing hydrophobic residues that may bind tightly to various surfaces. Catalase treatment followed by on-line guard-column desalting showed no detectable loss of protein when compared with on-line guard-column desalting alone (data not shown).
In the footprinting method, where synchrotron radiolysis is used to prepare OH radicals, some small amount of H2O2 is generated, and this is removed by treatment with catalase prior to storage, improving quantification of methionine-containing peptides 35. We also previously showed that catalase is capable of removing a 15-mM concentration of H2O2 with no observable protein oxidation.
What is the effect of catalase on protein oxidation?
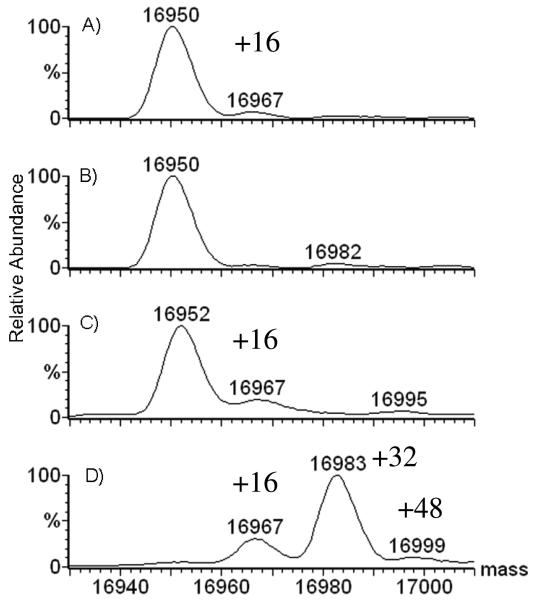
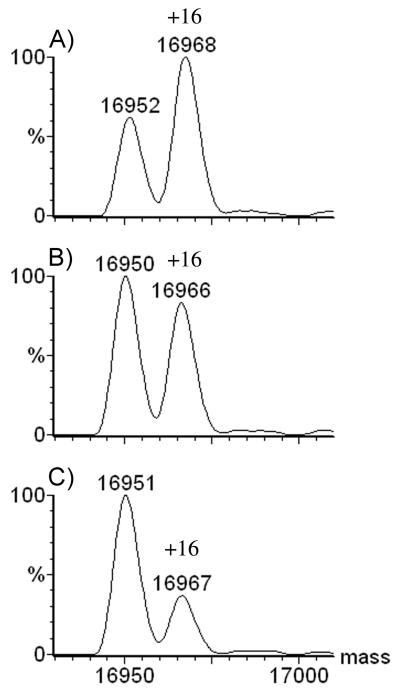
In the experiments described here, we tested significantly higher concentrations of H2O2 to insure that the oxidant can be removed from a sample without causing oxidation of the protein. A sample of apomyoglobin containing 500 mM H2O2 was flash frozen in liquid nitrogen, hand-thawed, incubated at room temperature (22 °C) for 2 h, and quenched by adding 1 μL of 1 μM catalase while causing only trace oxidation based on the low abundance of the A + 16 peak in the mass spectrum (Figure 1A). Did the small amount of oxidation occur in the freeze/thaw cycle, during the 2 h at room temperature, or in the catalase quenching step? No oxidation occurred by pretreating the sample with catalase to remove the H2O2 prior to flash freezing followed by storage at −200 °C for 2 h (Figure 1B). Similarly, if the sample containing H2O2 was flash frozen, thawed immediately, and treated with catalase, no detectable oxidation occurred (data not shown). These experiments demonstrate that protein oxidation does not occur during the catalase treatment or in the subsequent freeze/thaw cycle. The small amount of oxidation observed at 22 °C with 500 mM H2O2 is due to the 2 h incubation of protein with 500 mM H2O2 at room temperature.
Figure 1.
Mass spectra of 20 μM apomyoglobin (MWtheoretical = 16951) that was: A) frozen, thawed, then incubated at room temperature for 2 h in presence of 500 mM H2O2 prior to catalase treatment, B) incubated at −200 C for 2 h after removal of H2O2 by catalase, C) thawed, and catalase-treated after incubation with 500 mM H2O2 for 2 h at −200 C, D) freeze-dried for 0.5 h to remove the 500 mM H2O2, then resuspended in water for MS analysis.
Interestingly, when the protein was incubated with 500 mM H2O2 at −200 °C for 2 h (Figure 1C), slightly more oxidation occurred than for similarly long incubation at 22 °C, indicating that oxidation takes place in the ice lattice. As shown in Figure 1D, when the peroxide was removed by freeze-drying, significant protein oxidation occurred. This set of experiments confirms that the hydrogen peroxide can be removed from the solution by using catalase without causing oxidation, and that the freeze/thaw cycle in the presence of H2O2 does not cause the protein to be oxidized; hence this is not a freeze-concentration effect. Freeze-drying, however, is clearly unsuitable for removing peroxide from a solution containing protein. We now ask how various conditions during the incubation of the sample influence the amount of oxidation?
What is the effect of [H2O2] on protein oxidation?
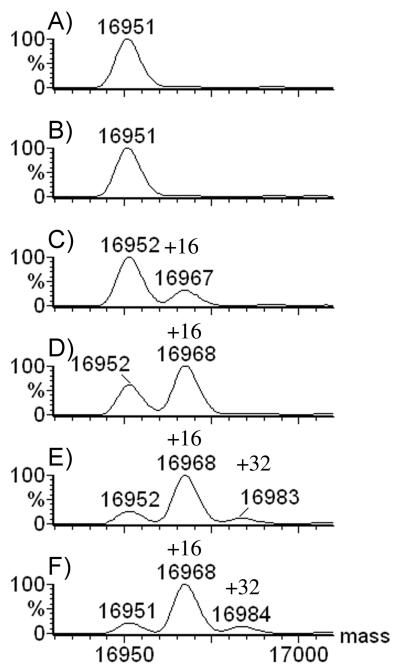
To determine the effect of H2O2 concentration, we prepared solutions of apomyoglobin that were identical in every respect, except for the varying concentration of hydrogen peroxide (0 to 500 mM). After pipet mixing of protein and peroxide, we flash-froze the samples immediately in liquid nitrogen, then stored them in a pre-chilled Eppendorf rack at −80 or −15 °C. The data in Figure 2 demonstrate that protein incubated for 2 h at −80 °C was not modified with 50 mM H2O2, but was partially oxidized at 75 mM. At 100-mM, over 50% of the protein was singly oxidized, and at 250 and 500 mM, nearly complete single oxidation occurred. Interestingly, the oxidation appears to level off at 80% at −80 °C. There were traces of a second oxidation when the concentrations of H2O2 are 250 and 500 mM, indicating that two oxidations per protein are possible, but rare at −80 °C. The deconvoluted mass spectra of myoglobin show clearly the unmodified protein at MW 16952 while singly and doubly oxidized protein appear at MW 16968 and 16984 (Figure 2).
Figure 2.
20 μM Apomyoglobin incubated for 2 h at −80 °C in varying concentrations of H2O2: A) 0 mM; B) 50 mM H2O2; C) 75 mM; D) 100 mM; E) 250 mM; F) 500 mM.
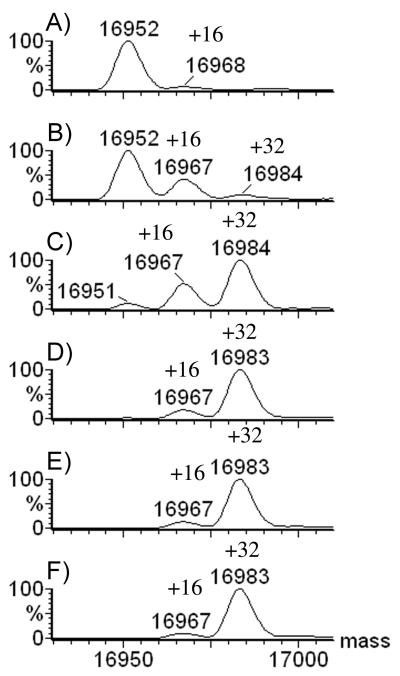
At −15 °C, the protein underwent a first oxidation when the concentration of H2O2 was 50 mM, but the extent of oxidation changed rapidly when [H2O2] was increased to 75 mM; at that concentration, most of the protein underwent double oxidation (Figure 3). By 100 mM H2O2, double oxidation was nearly complete, and minimal changes occurred with an increase to 500 mM H2O2. The temperature of the incubation has a major effect on the number of oxidations occurring in the protein.
Figure 3.
20 μM apomyoglobin incubated for 2 h at −15 °C with varying concentrations of H2O2: A) 0 mM; B) 50 mM H2O2; C) 75-mM; D) 100 mM; E) 250 mM; F) 500-mM.
What is the effect of time on protein oxidation?
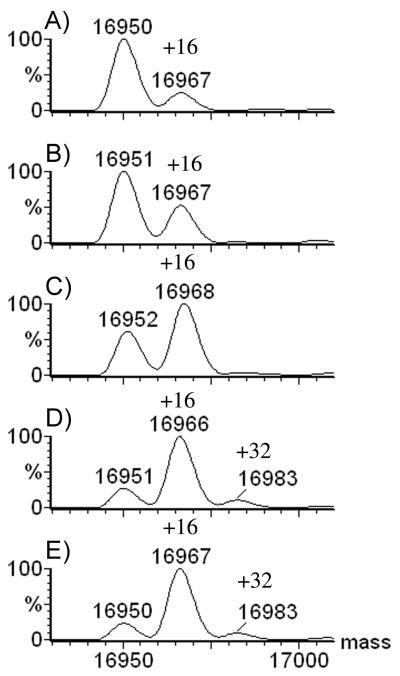
As 100 mM hydrogen peroxide afforded nearly full single oxidation at −80 °C, we fixed the concentration of H2O2 at that level and investigated the time dependence of oxidation. The results (Figure 4) demonstrate that there is minimal oxidation of apomyoglobin at zero time. Between 1 and 2 h of incubation, the extent of oxidation shifted to single oxidation. At longer times, a small amount of double oxidation occurred, but the decrease of the unmodified protein from 3 to 24 h was minimal. In fact, when apomyoglobin was incubated with 500 mM H2O2 for 24 h, there was no difference in the intensity of the signal for oxidized protein or in the number of modifications compared to 100 mM H2O2, suggesting that a small fraction of the protein cannot be oxidized. This contrasts with the 2 h incubation with 100 mM H2O2 at −15 °C, where nearly all the protein was doubly oxidized with only a small fraction of singly oxidized apomyoglobin that remains.
Figure 4.
Time course of a −80 °C incubation of 20-μM apomyoglobin with 100 mM H2O2 A) 30 min; B) 1 h; C) 2 h; D) 3 h; E) 24 h.
What is the effect of temperature on protein oxidation?
At −80 and −15 °C, significant oxidation occurred, with more oxidation at the higher temperature. The amount of oxidation, however, fell off precipitously when the protein was incubated at +4 °C. What is clear is that protein oxidation is strongly accelerated when the protein is in an ice lattice as opposed to being in liquid water at +4 °C. We will speculate in the following on the reasons for this.
Why are only one or two oxidations observed?
Apomyoglobin has two methionine residues that are readily oxidized. Oxidation should occur at much lower concentrations of peroxide than those used here because its concentration (100 mM) is in large excess (5000:1) relative to the protein concentration of 20 μM. The solution, however, is frozen, a situation that presumably stops diffusion. Thus, the number of H2O2 molecules around the protein is considerably less than 5000:1. To estimate the number of H2O2 molecules in contact with the protein in the solid state, we calculated the solvent-exposed surface of the holomyoglobin crystal structure. Although there is no high resolution structure of apomyoglobin, we used the structure of the holo form because it is very similar to that of the apo form36.
Using the solvent-exposed surface area calculator available at http://molbio.info.nih.gov/structbio/basic.html37, we estimated a surface area of 6,840 Å2 for holomyoglobin. Dividing this by the surface area of a water molecule (radius = 1.4 Å; area = 6.2 Å2) allows us to estimate that the protein is in direct contact with approximately 1,100 water molecules. Water, at a concentration of 55 M, is in large excess compared to 100 mM peroxide (550:1). Therefore, given that the polar characteristics of H2O and H2O2 are similar, we estimate that, on average, two [(1100 water / protein) /(550 water per H2O2) = 2 H2O2 / protein] peroxide molecules are in contact with the surface of the protein when the solution is flash frozen at −200 °C. We emphasize that this is only an estimate for argument purposes. At −15 °C, the peroxide molecules are presumably able to reach the methionine residues nearly quantitatively, explaining the maximum number of oxidations that we observed. As we will show later, the methionines are heavily oxidized, indicating that there is sufficient diffusion around the protein at −15 °C and that the approximately the two H2O2 molecules have some ability to diffuse in the water layer surrounding the protein in the frozen matrix but not into the bulk matrix. One may view this as motion in two dimensions given that the H2O2 molecules in contact with the protein surface and cannot move away from it. There is decreased diffusion at −80 °C, however, and in 2 h, only one peroxide molecule can diffuse into the proximity of the methionine residues and oxidize them. This suggests that the hydration shell around the protein is thinner in some regions or that certain areas of water around the protein present diffusion barriers owing to the protein surface composition Given that at 50 mM H2O2, the protein is in contact with one H2O2 molecule on average, the limited diffusion in the ice lattice prevents methionine oxidation rationalizing the near absence of oxidation, even at −15 °C (Figure 3 B).
This proposal also indicates why the protein is oxidized less at +4 °C or +22 °C. At these temperatures, the H2O2 molecules are constantly moving close to and away from the protein; they are moving in three dimensions compared to the nearly two-dimensional motion when the solution is frozen. Thus, the oxidation reaction proceeds slowly owing to the diffusion of the oxidant in the bulk solution. In this situation, the oxidant spends considerable time away from the protein, whereas when frozen, the few molecules of H2O2 that are in surface contact are trapped and cannot diffuse away from the protein.
If the two H2O2 molecules at the surface of the apomyoglobin, as per our estimate, react readily with two methonines in the sequence, then a smaller surface area protein with a single methionine should have primarily one oxidation under these conditions. In a flash photolysis experiment of ubiquitin (which has one methionine) following which the solution was freeze-dried in the presence of 100 mM H2O2, Aye et al.20 observed single oxidation of the protein. The solvent-exposed surface area for the ubiquitin crystal structure 1UBQ38 is 4,510 Å2. This equates to 730 water molecules, and at a 550:1 ratio of water:H2O2, there are on average 1.3 peroxide molecules contacting the protein in the frozen matrix, correlating well with the single oxidation that was reported. Aye et al. also found that the optimal concentration is 100 mM H2O2. These results fit well with our proposed explanation of the number of observed oxidations and the concentration requirements for the freeze-induced oxidation to occur. In the solid state, there are a limited number of H2O2 molecules at the protein surface. Oxidation is limited to those oxidant molecules because there is neglibile diffusion from frozen bulk solvent. This corroborates our explanation as to why two oxidations are observed for apomyoglobin.
Is oxidation caused by radical diffusion through ice?
If the oxidant were capable of diffusing through the solid state, we reasoned that a quencher for H2O2 should eliminate all oxidation when it is in large excess compared to the concentration of the protein. Prior experiments in solution21 show that 20 mM phenylalanine (Phe) effectively quenches the hydroxyl radical oxidation of 10 μM apomyoglobin; these were conditions under which both the radical and protein diffused freely. Therefore, we measured the extent of oxidation after adding 10 and 70 mM Phe to 100 mM H2O2 reaction solutions. In both cases, there was a large excess of quencher compared to a 20 μM apomyoglobin concentration. Indeed, protein oxidation was reduced at both concentrations of phenylalanine (Figure 5); however, the extent of oxidation with 10-mM Phe was only slightly reduced whereas that with 70 mM Phe was more significantly reduced. But neither concentration of the scavenger caused complete quenching of the oxidation as we observed with freely diffusing radicals in solution. These results are in accord with the hypothesis that the oxidant does not diffuse freely through the ice lattice. At 10 mM Phe, on average, there are 0.2 molecules of phenylalanine contacting the protein, and we observed a decrease of 27% in the relative signal for the singly oxidized protein. When [Phe] = 70 mM, the average number of molecules of phenylalanine contacting apomyoglobin increases to 1.6, and the extent of oxidation decreased by 55%.
Figure 5.
Mass spectra after A) a 2 h, −80 °C incubation of 20 μM apomyoglobin with 100 mM H2O2; B) same as A) but with phenylalanine added to 10 mM; C) same as A) but with phenylalanine added to 70 mM.
At the higher concentration of Phe, the number of Phe molecules in contact with the surface of the protein is comparable to those of H2O2 (1.6:2.0). In the “micro environment” of the protein surface, oxidation of the protein is competitive with that of Phe, and the result is diminished oxidation of the protein. One may picture this “micro environment” as one in which the ice lattice is disrupted at the protein surface, permitting limited diffusion of the oxidant in a liquid-like protein hydration shell existing near the surface. Diffusion from the bulk, however, is prevented by the solid-state ice lattice.
Why are there different extents of oxidation at different temperatures?
Although the estimates of H2O2 and scavenger molecules contacting the protein agree with the decrease in the extent of oxidation, it remains unclear why only singly oxidized protein is observed at −80 °C, whereas at −15 °C, double oxidation is favored. Further, one might expect that at 500 mM H2O2, more than two oxidations should occur as there are approximately 10 molecules of peroxide in contact with the protein surface.
One explanation is that the oxidant modifies principally reactive amino acid side chain (e.g., those of methionine or of aromatic amino acids) at −80 °C. At −15 °C, additional channels of reactivity open, resulting in more oxidations located at other residues in the protein. Another explanation is that the protein unfolds owing to strains imposed by the ice lattice. If unfolding were the source of the increased oxidation at −15 °C, we would expect to see modifications on residues that are buried in the native-state protein. These residues may be accessible at −15 °C because that temperature, with respect to −80 °C, permits greater disruption of the ice lattice, allowing the protein conformation to change. At −80 °C, however, the ice lattice would restrict protein motion more than at −15 °C, constraining the protein structure and slowing unfolding.
Other explanations can also be forwarded. Perhaps some diffusion of H2O2 can occur in the lattice. At −80 °C, the ice lattice would restrict the diffusion of the oxidant compared to −15 °C. If there is no reactive residue within the diffusion volume of the H2O2, the protein will not be oxidized. We would expect, however, that the same residues would be oxidized at −80 °C as at −15 °C, but the abundance of each oxidation would be different. Thus, the average number of oxidations per protein would decrease, but the residues that become modified would not change.
What amino-acid residues are oxidized?
To answer this question, we determined the modifications that occur to specific amino acids after incubation at both −80 and −15 °C (Table 1).
Table 1.
Percent oxidations after cold chemical oxidation of apomyoglobin. The %'s were calculated from the ratio of the signal for the modified peptide divided by that for the unmodified peptide; the apomyoglobin was incubated for 2 h with 100 mM H2O2 at −80 or −15 °C. The water exposure and % oxidized were calculated as described in the experimental section.
| Residue Type | % Ox (−80 °C) | %Ox (−15 °C) | Protein Water Exposure (Å2) |
|---|---|---|---|
| Trp-7 | 3% | 2% | 19 |
| Trp-14 | Ya | Ya | 7 |
| Tyr-103 | 0.2% | 0.2% | 28 |
| Tyr-146 | 6% | 6% | 18 |
| Met-55 | Y | Y | 16 |
| Met-131 | 56% | 82% | 0 |
| Phe-33 | ndb | ndb | 6 |
| Phe-106 | 0.01% | 0.01% | 36 |
| Phe-123 | Y | Y | 9 |
| Phe-138 | 2% | 2% | 30 |
| Phe-151 | Y | Y | 38 |
| His-24 | 1% | 0.8% | 12 |
| His-36 | 0.9% | 0.8% | 29 |
| His-64 | Y | Y | 28 |
| His-81 | xc | xc | 64 |
| His-82 | x | x | 9 |
| His-93 | x | x | 37 |
| His-97 | x | x | 38 |
| His-113 | Y | Y | 48 |
| His-116 | 0.03% | 0.1% | 47 |
| His-119 | 0.2% | 0.7% | 24 |
| Val-10 | nd | nd | 0 |
| Val-13 | nd | nd | 8 |
| Val-17 | 0.5% | 0.2% | 0 |
| Val-28 | nd | nd | 0 |
| Val-67 | nd | nd | 112 |
| Val-68 | 0.2% | 0.3% | 44 |
| Val-114 | nd | nd | 10 |
Y annotation indicates that the oxidized peptide was detected but could not be quantified owing to interference and/or poor chromatographic resolution.
The nd annotation indicates that the modified peptide was not detectable although the unmodified peptide was. The detection limits were in all cases better than 1% and for 80% of the detections, the detection limits were 0.2% or better. Detection limits are variable owing to differences in the ionization efficiencies of the tryptic peptides.
The x annotation indicates that the unmodified peptide carrying the denoted residue was not observed (was not retained on the reverse phase column); therefore, no conclusion regarding the reactive residue can be drawn.
As for the results for apomyoglobin by FPOP 21 where all methionine, tryptophan and tyrosine residues were modified, oxidation in the solid state also modified these reactive residues. Indeed, the oxidation occurred more readily on the most reactive residues at both temperatures (see Table 1), and the majority of oxidation occurs at Met 131, which is not solvent-exposed. Although an accurate relative abundance measurement cannot be made for Met 55 (it binds poorly in the HPLC separation), we did observe considerable signal for oxidized peptide in the solvent band. It would not be surprising that this residue has significant levels of oxidation based on the significant oxidation of Met 131. The facile oxidation of Met is also in accord with the double oxidation of the protein, even at concentrations of H2O2 as high as 500 mM. Oxidation of Met may occur by mechanisms in which electrons are transferred from the surface to a buried Met by a relay process, hence other residues can be modified, but they pass the radical on to the methionine. This process has been documented as a method to protect proteins in biological systems from radical inactivation39.
Tryptophan, tyrosine, and phenylalanine appear to be the next most abundantly oxidized residues, particularly Trp 7, Tyr 146, and Phe 138, which have relatively high solvent exposures. Other aromatic amino acid residues (e.g., Tyr 103, Phe 106 and 151) also have relatively high solvent accessibility but undergo less oxidation. This is difficult to explain. One explanation is that quantification by ESI has pitfalls. Another is that some oxidized residues at the surface may return to a reduced state by an electron-relay processes. Alternatively, the protein surface may favor limited diffusion for entropic reasons.
Most histidines were modified, but at relatively low levels (< 1%). For example, His 24, 36 and 119 have relatively high solvent exposure and underwent readily detectable oxidations. His 64, 113, 116, on the other hand, are also solvent accessible but showed less oxidation. For peptides containing multiple histidine modifications, it was not possible to identify unambiguously the site of the oxidation. His oxidation may play a role in the oxidation of Phe 123, which is unexpected because it has low exposure and does not undergo oxidation in the solution FPOP or with synchrotron-generated OH radicals. It may be that there is cold-induced unfolding or electron transfer from His 116 or 119, the former of which is clearly oxidized. These matters are subjects for future study.
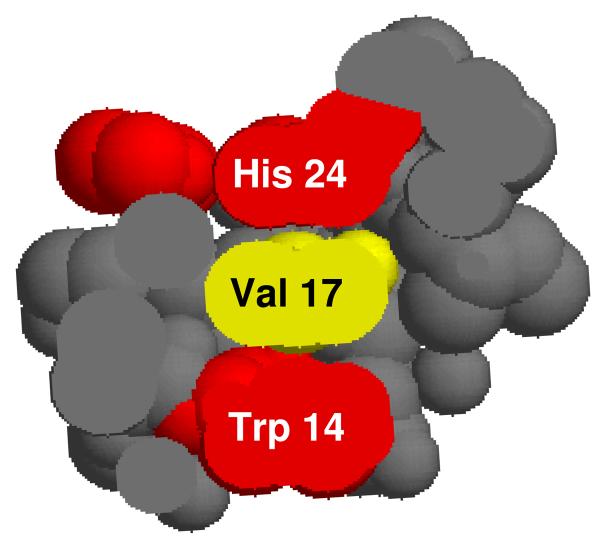
More conclusive evidence for electron transfer to the surface is found for the oxidation of Val 17. This residue has no solvent-exposed surface, but the Cβ and both Cγ carbons are buried between the aromatic side chains of Trp 14 and His 24, both of which are oxidized in this work. Given that only two of seven valines are oxidized, and these both adjoin oxidized aromatics, we favor an explanation whereby electron transfer occurs between the interior and the surface of a protein as opposed to invoking unfolding to produce solvent exposure of the valine residues.
In support of the electron-transfer hypothesis are results from De Felippis and Faragi 40, 41 who demonstrated electron transfer in proteins by a distance-dependent mechanism. The tryptophan-to-tyrosine electron transfer rate constant is 7.4 × 104 s−1, as was determined by monitoring the formation of an indolyl radical that is located at a distance of 5 Å. Although tryptophan radical is a good electron donor, the rate is probably reduced in our experiment because valine is not a good electron acceptor. It may be that a hydrogen atom is abstracted from valine, forming a radical that is then further oxidized. The same argument applies to the oxidation of Val 68. This residue has significantly less solvent exposure than Val 67. Val 68 is within van der Waals contact of His 64, which was oxidized, but Val 67 does not contact any aromatic residue. The oxidation on both Val 17 and Val 68 supports, but does not prove, the electron-transfer hypothesis.
Given that the same residues were oxidized at both −80 and −15 °C, no new channels of reactivity (to modify other residues) are opened at the higher temperature, and it is unlikely that apomyoglobin is more unfolded at −15 °C than at −80 °C. These data are in accord with the hypothesis that the H2O2 is less constrained by the ice lattice at −15 °C than at −80 °C, resulting in oxidation of the same residues, but producing higher abundance products at the higher temperature. In particular, the oxidant can diffuse and modify both methionine residues at −15 °C, explaining why a maximum of two oxidations were observed, whereas at −80 °C, diffusion is more limited, resulting in a maximum of one oxidation of the protein.
CONCLUSIONS
Freeze-drying is not suitable for the removal of H2O2 in protein footprinting studies. Instead, its removal can be carried out successfully by using catalase. More interestingly, oxidation of a protein occurs in the solid state when H2O2 is present. It occurs only when the protein is frozen with H2O2 and it generates nearly quantitative single oxidation after 2 h of incubation. Although the mechanism of “cold oxidation” is not yet established, we have evidence that the oxidant diffuses over some portion of the protein surface in the solid state, and oxidizes the most reactive amino acid residues. As a chemical footprinting method, the results of cold oxidation, however, do not correlate well with solvent exposure as calculated from the crystal structure, because the mechanism of oxidation is likely to be different than that in solution when H2O2 is photochemically converted into OH radicals. Secondly, the availability of oxidant is considerably reduced in the solid state, and only the H2O2 that is situated at the surface of the protein appears to react. Furthermore, buried residues are also oxidized, suggesting that electron transfer by a relay process between the interior of the protein and solvent-exposed residues can occur in the solid state. Although the cold chemical oxidation phenomenon may provide insight into the question of protein structure in frozen media, it is less likely that it will provide insight on protein structure in solution. In any case, cold oxidation must be avoided in protein footprinting studies. .
Figure 6.
Space-filling model of Val 17 with His 24 and Trp 14 showing no more than 5 Å separation and permitting radical transfer from aromatic H24 or W14 to V17.
ACKNOWLEDGMENT
This research was supported by the National Centers for Research Resources of the NIH (Grant P41RR000954) and by a supplemental grant from NCRR. We thank Drs. H. Rohrs, I. Vidavsky, J. Walters, D. Rempel, and C. Chung for their help in data collection and analysis.
REFERENCES
- 1.Godovac-Zlmmermann J, Brown LR. Mass Spectrometry Reviews. 2001;20:1–57. doi: 10.1002/1098-2787(2001)20:1<1::AID-MAS1001>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 2.Bradshaw RA, Burlingame AL. IUBMB Life. 2005;57:267–272. doi: 10.1080/15216540500091536. [DOI] [PubMed] [Google Scholar]
- 3.Aebersold R, Mann M. Nature (London, United Kingdom) 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 4.Gingras AC, Aebersold R, Raught B. J Physiol. 2005;563:11–21. doi: 10.1113/jphysiol.2004.080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aderem A. Cell. 2005;121:511–513. doi: 10.1016/j.cell.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 6.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 7.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 8.Zhu MM, Rempel DL, Du Z, Gross ML. J. Am. Chem. Soc. 2003;125:5252–5253. doi: 10.1021/ja029460d. [DOI] [PubMed] [Google Scholar]
- 9.Hager-Braun C, Tomer KB. Expert Rev Proteomics. 2005;2:745–756. doi: 10.1586/14789450.2.5.745. [DOI] [PubMed] [Google Scholar]
- 10.Guan J-Q, Takamoto K, Almo SC, Reisler E, Chance MR. Biochemistry. 2005;44:3166–3175. doi: 10.1021/bi048021j. [DOI] [PubMed] [Google Scholar]
- 11.Guan J-Q, Almo SC, Chance MR. Accounts of Chemical Research. 2004;37:221–229. doi: 10.1021/ar0302235. [DOI] [PubMed] [Google Scholar]
- 12.Tullius TD, Dombroski BA. Proc Natl Acad Sci U S A. 1986;83:5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sclavi B, Woodson S, Sullivan M, Chance MR, Brenowitz M. J Mol Biol. 1997;266:144–159. doi: 10.1006/jmbi.1996.0775. [DOI] [PubMed] [Google Scholar]
- 14.Tullius TD, Dombroski BA. Science. 1985;230:679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- 15.Xu G, Liu R, Zak O, Aisen P, Chance MR. Mol Cell Proteomics. 2005 doi: 10.1074/mcp.M500095-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Xu G, Chance MR. Anal Chem. 2005;77:4549–4555. doi: 10.1021/ac050299+. [DOI] [PubMed] [Google Scholar]
- 17.Sharp JS, Becker JM, Hettich RL. Analytical Biochemistry. 2003;313:216–225. doi: 10.1016/s0003-2697(02)00612-7. [DOI] [PubMed] [Google Scholar]
- 18.Sharp JS, Guo J.-t., Uchiki T, Xu Y, Dealwis C, Hettich RL. Analytical Biochemistry. 2005;340:201–212. doi: 10.1016/j.ab.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Wong JWH, Maleknia SD, Downard KM. Journal of the American Society for Mass Spectrometry. 2005;16:225–233. doi: 10.1016/j.jasms.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Aye TT, Low TY, Sze SK. Anal Chem. 2005;77:5814–5822. doi: 10.1021/ac050353m. [DOI] [PubMed] [Google Scholar]
- 21.Hambly DM, Gross ML. J Am Soc Mass Spectrom. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Rauen U, Elling B, Gizewski ER, Korth HG, Sustmann R, de Groot H. Free Radic Biol Med. 1997;22:17–24. doi: 10.1016/s0891-5849(96)00273-0. [DOI] [PubMed] [Google Scholar]
- 23.Kerkweg U, Li T, de Groot H, Rauen U. Hepatology. 2002;35:560–567. doi: 10.1053/jhep.2002.31869. [DOI] [PubMed] [Google Scholar]
- 24.Rauen U, Petrat F, Li T, De Groot H. Faseb J. 2000;14:1953–1964. doi: 10.1096/fj.00-0071com. [DOI] [PubMed] [Google Scholar]
- 25.Bhaumik G, Srivastava KK, Selvamurthy W, Purkayastha SS. Int J Biometeorol. 1995;38:171–175. doi: 10.1007/BF01245384. [DOI] [PubMed] [Google Scholar]
- 26.Takenaka N, Ueda A, Daimon T, Bandow H, Dohmaru T, Maeda Y. J. Phys. Chem. 1996;100:13874–13884. [Google Scholar]
- 27.Betterton EA, Anderson DJ. Journal of Atmospheric Chemistry. 2001;40:171–189. [Google Scholar]
- 28.Takenaka N, Ueda A, Maeda Y. 1993:24–32. [Google Scholar]
- 29.Hatley RHM, Franks F, Day H. Biophysical Chemistry. 1986;24:187–192. doi: 10.1016/0301-4622(86)80013-8. [DOI] [PubMed] [Google Scholar]
- 30.Honda K, Yamasaki N, Emoto M, Mori Y, Fujieda S. 2001:622. [Google Scholar]
- 31.Takenaka N, Furuya S, Sato K, Bandow H, Maeda Y, Furukawa Y. International Journal of Chemical Kinetics. 2003;35:198–205. [Google Scholar]
- 32.Sato K, Furuya S, Takenaka N, Bandow H, Maeda Y, Furukawa Y. Bull. of the Chem. Soc. of Japan. 2003;76:1139–1144. [Google Scholar]
- 33.Tsai J, Gerstein M. Bioinformatics. 2002;18:985–995. doi: 10.1093/bioinformatics/18.7.985. [DOI] [PubMed] [Google Scholar]
- 34.Sharp JS, Becker JM, Hettich RL. Analytical Chemistry. 2004;76:672–683. doi: 10.1021/ac0302004. [DOI] [PubMed] [Google Scholar]
- 35.Xu G, Kiselar J, He Q, Chance MR. Anal Chem. 2005;77:3029–3037. doi: 10.1021/ac048282z. [DOI] [PubMed] [Google Scholar]
- 36.Eliezer D, Wright PE. J Mol Biol. 1996;263:531–538. doi: 10.1006/jmbi.1996.0596. [DOI] [PubMed] [Google Scholar]
- 37.Sanner MF, Olson AJ, Spehner JC. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 38.Vijay-Kumar S, Bugg CE, Cook WJ. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 39.Levine RL, Moskovitz J, Stadtman ER. IUBMB Life. 2000;50:301–307. doi: 10.1080/713803735. [DOI] [PubMed] [Google Scholar]
- 40.DeFelippis MR, Faraggi M, Klapper MH. Journal of the American Chemical Society. 1990;112:5640–5642. [Google Scholar]
- 41.Faraggi M, DeFelippis MR, Klapper MH. Journal of the American Chemical Society. 1989;111:5141–5145. [Google Scholar]