Abstract
Results of behavioral genetic and molecular genetic studies have converged to suggest that genes substantially contribute to the development of attention deficit/hyperactivity disorder (ADHD), a common disorder that onsets in childhood. Yet, despite numerous linkage and candidate gene studies, strongly consistent and replicable association has eluded detection. To search for ADHD susceptibility genes, we genotyped approximately 600,000 SNPs in 958 ADHD affected family trios. After cleaning the data, we analyzed 438,784 SNPs in 2803 individuals comprising 909 complete trios using ADHD diagnosis as phenotype. We present the initial TDT findings as well as considerations for cleaning family-based TDT data. None of the SNP association tests achieved genome-wide significance, indicating that larger samples may be required to identify risk loci for ADHD. We additionally identify a systemic bias in family-based association, and suggest that variable missing genotype rates may be the source of this bias.
Introduction
With a prevalence of eight to twelve percent worldwide (Faraone and others 2003), attention deficit/hyperactivity disorder (ADHD) is among the most common childhood psychiatric disorders. ADHD is characterized by developmentally inappropriate levels of hyperactive, impulsive and inattentive behaviors that give rise to significant clinical and psychosocial impairments. The disorder is clinically heterogeneous with considerable variation in the profile of the core symptoms of hyperactivity, impulsivity and inattention and the pattern of associated comorbidities (Faraone 2005). In spite of this heterogeneity and some shift in diagnostic criteria (American Psychiatric 1987), ADHD is among the best-validated childhood diagnoses from clinical, longitudinal and neurobiological perspectives (Faraone and Biederman 1998) (Faraone and Biederman 2000) (Faraone 2006) (Faraone and Biederman 2004). This feature, along with observations that family members of children with ADHD are at elevated risk for ADHD (Morrison and Stewart 1971) (Chen and others 2008) and that the heritability of ADHD is in the region of 60–90% with an average across studies of 76%, makes this condition a promising target for molecular genetic studies (Faraone and others 2005) (Faraone 2004).
In the search for ADHD susceptibility genes, several groups have conducted genome-wide linkage scans. A study of 126 American affected sib-pairs found three regions showing some evidence of linkage (LOD scores >1.5): 5p12, 10q26, 12q23, and 16p13 (Fisher and others 2002). An expanded sample of 203 families found stronger evidence for the 16p13 region with a maximum LOD score of 4 (Smalley and others 2002). A study of 164 Dutch affected sib-pairs found a peak LOD score of 3.5 at 15q15, with suggestive linkage signals also on chromosomes 4p16, 7p13, 9q33 and 13q33 (Bakker and others 2003). A genome-wide scan of families from Columbia implicated 8q12, 11q23, 4q13, 17p11, 12q23, and 8p23 (Arcos-Burgos and others 2004). A study of 155 sib-pairs from Germany reported a maximum LOD score of 2.59 for chromosome 5p at 17cM, and also reported nominal evidence for linkage to chromosomes 6q, 7p, 9q, 11q, 12q and 17p, which had been identified in previous scans (Hebebrand and others 2006). Linkage analysis in a sample of mixed ethnicity revealed some evidence for risk loci on chromosome 8 and 15 in an affected sib pair design in 217 families (Faraone and others 2007). A study of 142 combined type ADHD affected sibling pairs from the International Multicentre ADHD Genetics (IMAGE) study, including some of the probands included in this study, implicated regions on 16q23 (LOD=3.1) and 9q22 (LOD=2.1) reported in some of the previous scans (Asherson and others 2008). Finally, using quantitative trait measures of ADHD in the IMAGE sample, a locus at 1p36 that overlaps with the dyslexia locus DYX8, was identified with an empirical genome-wide significance of 0.05 (Zhou and others in press). None of these linkage findings has yielded a replicable association, but suggest there may be some molecular
Although there is some overlap in nominally significant linkage peaks among studies, there is no evidence for the replication of a genome-wide significant finding using strict criteria (Lander and Kruglyak 1995). Given the fact that linkage studies are predominantly suited to pick up genetic factors of strong effect (>10% of the variance), the most likely conclusion we can draw from these studies is that the average effects of specific genes influencing ADHD-risk, as with most complex traits, cannot be very large. Furthermore, the observed heritability may be the consequence of additive and interaction effects involving multiple genetic and environmental risk factors (Kuntsi and others 2006). This, in turn, suggests that the discovery of ADHD genes will need to rely on the method of association applied to either targeted candidate genes (Brookes and others 2006) (Wang and others 2006) or a genome-wide association scan (GWAS). Although the testing of candidate genes has produced some consistent results in meta-analyses (Faraone and others 2005), with two variants within or close to the dopamine D4 and D5 receptor genes even reaching genome-wide levels of significance (Li and others 2006), this approach is limited by the need to specify candidate genes. Thus, to discover novel biological pathways and common variants for ADHD, we have applied the method of genome-wide association analysis to a large series of ADHD parent-child trios.
Methods
Subjects
Families were collected by the International Multicenter ADHD Genetics (IMAGE) project. Families were identified through ADHD probands aged 5 to 17 attending outpatient clinics at the data collection sites in Europe and Israel. A total of 958 affected proband-parent trios were initially selected for the GWAS scan. Family members were primarily of Western European origin hailing from eight countries including Belgium, Germany, Ireland, Israel, the Netherlands, Spain, Switzerland, and the United Kingdom. Of these, 893 probands were initially ascertained as having DSM-IV combined type ADHD. Sixty-five probands who did not meet combined subtype ADHD diagnosis were included because they either met the criteria for the inattentive or hyperactive subtypes, or they missed the DSM-IV combined type diagnosis by a single item. Exclusion criteria were autism, epilepsy, IQ < 70, brain disorders and any genetic or medical disorder associated with externalizing behaviors that might mimic ADHD.
Clinical Measures
Prior to entry into the study, all probands underwent clinical evaluations by a pediatrician or child psychiatrist and both existing and new patients were included in the study. Patients had to meet clinical criteria for ADHD-combined type before being enrolled in the study. Wherever possible, families withdrew stimulant medication for one week prior to research assessments to allow for more accurate ascertainment of the current level of ADHD symptoms and behaviors. Alternatively we ensured as far as possible that ratings were based on medication free periods. Probands were excluded from the study if the last medication free period was more than 2 years ago.
Parental Account of Childhood Symptom (PACS)
PACS is a semi-structured, standardized, investigator-based interview developed as an instrument to provide an objective measure of child behavior (Taylor and others 1986). A trained interviewer administers PACS with parents, who are asked for detailed descriptions of the child’s typical behavior in a range of specified situations. Such situations are defined either by external events (e.g. watching television, reading a book or comic, playing alone, playing with friends, going to bed, traveling) or by behaviors shown (e.g. crying, worried talk, tempers, fighting with siblings). Interviewers then make their own ratings on the basis of a formal training and written definitions of the behaviors to be rated, on a 4-point scale of severity and frequency in the previous week and previous year. Inter-rater reliability is high with product-moment correlations for pairs of interviewers ranging from 0.79 to 0.96. The Hyperactivity subscale is made up of attention span (time spent on a single activity, rated separately for four different kinds of activity), restlessness (moving about during the same activities), fidgetiness (movements of parts of the body during the same activities), and activity level (rated for structured situations such as mealtimes and car journeys), with other subscales covering defiant, emotional and other comorbid disorders including autistic spectrum disorders.
Rating Scales
Rating scales used to quantify ADHD symptoms included the Long Version of Conners’ Parent Rating Scale (CPRS-R:L), Long Version of Conners’ Teacher Rating Scale (CTRS-R:L), parent version of the Strengths and Difficulties Questionnaires (SDQ) and teacher version of SDQ (Conners 1996). In order to exclude autism spectrum disorders that might confound the analysis of ADHD, both probands and siblings were screened using the Social Communication Questionnaire in conjunction with the pro-social scale from the SDQ. Individuals falling above these thresholds were further evaluated using the autism spectrum disorder section of the PACS interview.
DSM-IV Diagnoses
A standardized algorithm is applied to PACS to derive each of the 18 DSM-IV ADHD items, providing operational definitions for each behavioral symptom. These are combined with items that scored 2 or 3 from the teacher rated Conners’ ADHD subscale, to generate the total number of items from the DSM-IV symptom checklist. Situational pervasiveness was defined as some symptoms occurring within two or more different situations from the PACS interview, as well as the presence of one or more symptoms scoring 2 or more from the ADHD subscale of the teacher rated Conners’. Several probands in the initial sample (prior to QC) only met criteria for DSM-IV inattentive subtype (N=13), hyperactive subtype (N=33) or missed one of the ADHD diagnoses by a single item on the structured interview (N=19). We retained these latter families because, upon review of the medical record and structured interview data, the ADHD diagnosis was confirmed.
Genotyping
Genotyping was conducted at Perlegen Sciences using their 600K genotyping platform, which comprises approximately 600,000 tagging SNPs designed to be in high linkage disequilibrium with untyped SNPs for the three HapMap populations. This study is part of the Genetic Association Information Network (GAIN), a public-private partnership of the Foundation for the National Institutes of Health, Inc. (FNIH) that currently involves the National Institutes of Health (NIH), Pfizer, Affymetrix, Perlegen Sciences, Abbott, and the Eli and Edythe Broad Institute (of MIT and Harvard University) (http://www.fnih.org/GAIN2/home_new.shtml). The DNA was extracted from blood for all samples and stored at the Rutgers depository. Genotype data were cleaned by The National Center for Biotechnology Information (NCBI). Quality Control analyses were processed using the GAIN QA/QC Software Package (version 0.7.4) developed by Gonçalo Abecasis and Shyam Gopalakrishnan at the University of Michigan. A copy of the software is available by e-mailing gopalakr@umich.edu or goncalo@umich.edu.
The quality control procedure can be broadly split into two categories: individual/family removal and SNP removal. Sample exclusion criteria and SNP exclusion criteria are found in Table 1 and 2, respectively.
Table 1.
Sample filtering and exclusion criteria from the full dataset
| Quality control metric | Individuals excluded |
|---|---|
| Call rate < 87% | 10 |
| Gender discrepancy | 1 |
| Sample heterozygosity < 32% | 16 |
| Per-family Mendelian errors of >2% | 6 |
| Total | 33 |
This table annotates the exclusion of individuals from the initial GWAS. Call rate is based on all SNPs at the outset. The 87% threshold was chosen based on the distribution of missinginess, as was sample heterozygosity. Mendelian errors are impossible configurations of transmission from parent to offspring. A rate higher than 2%, we took to imply either sample mix-up or non-paternity. These exclusions were applied to all individuals in the study.
Table 2.
SNP filtering and exclusion criteria
| Quality control metric | SNPs excluded | SNPs remaining |
|---|---|---|
| Call rate conditional on minor allele frequency | 144,511 | 454,634 |
| Mendel errors > 4 | 15,387 | 439,247 |
| Duplicate sample discordance (>1/15) | 185 | 439,062 |
| Hardy-Weinberg Disequilibrium (P < 0.000001) | 278 | 438,784 |
| Total | 160,361 | 438,784 |
The call rate conditional on minor allele frequency aims to identify distributional properties of the association test statistics. Mendel errors are impossible parent child transmission patterns. 15 samples were duplicated across the plates to assess the quality of the genotyping. If more than one pair of duplicates did not agree, then the SNP was excluded. Hardy-Weinberg Disequilibrium was tested in all individuals, with a strict threshold to protect against false positives on the HWD test being artificially excluded.
Our key data quality control consideration presented in table 2 is the call rate conditional on minor allele frequency. Missing (Hirschhorn and Daly 2005) or inaccurate (Cutler and others 2001) (Gordon and others 2002) genotype data has been shown to introduce artefactual association in family-based settings, with low minor allele frequency SNPs being particularly sensitive to these effects. Thus our primary quality control consideration was to carefully evaluate the joint effect of call rate and minor allele frequency (MAF) on our association tests. We binned SNPs into categories defined by MAF (0–1%, 1–2%, 2–3%, 3–4%, 4–5%, 5–10%, and 10–50%) and call rate (90–95%, 95–96%, 96–97%, 97–98%, 98–99%, and 99–100%). Then the distribution of association test statistics was inspected for each of these cells, with a particular focus on the λ(defined as the observed median χ2 divided by the expected median χ2). Based on these distributions, we selected SNPs under the following three conditions: 0.01 ≤ MAF < 0.05 and call rate ≥ 99%; 0.05 ≤ MAF < 0.10 and call rate ≥ 97%; and 0.10 ≥ MAF and call rate ≥ 95%. The SNPs from these three conditions comprise the primary set for analysis. As a way of generating a secondary and “super-clean” set of SNPs, we can remove the SNPs that failed the quality control metrics for the other two GAIN Perlegen studies (for Major Depression Disorder (MDD) and Psoriasis). Such a selection approach is premised that the individual assay may not generally perform well, and these SNPs were included in our study, because of random variation in the quality control metrics. From the MDD filtering, an additional 49,707 SNPs are lost (~48K because of low call rates) yielding 389,077. From the Psoriasis filtering, an additional 3,942 SNPs are lost yielding 385,134.
Statistical Analyses
We used the transmission disequilibrium test (TDT) (Spielman and others 1993) as implemented in PLINK. To determine the quality of the genotype calling in the top of the distribution, we also inspected the cluster plots of the intensity for the genotypes(Anney and others submitted). In addition to performing the basic TDT, we attempted to identify any sources of potential bias, by attempting to predict the TDT χ2 test statistic as a function of major versus minor allele over-transmission at the locus. We also explored the existing candidate genes from the ADHD literature, to place the potential effects in context. We examined two sets of candidates. The first set comprises genes that have showed significant association with ADHD in meta-analyses performed by Faraone and colleagues (Faraone and others 2005). These are SNAP25, DRD4, SLC6A3, HTR1B, SLC6A4, and DBH. The second set consists of genes that had been nominated by the study investigators as good candidates for ADHD, based on a previous in depth analysis of 51 genes using an overlapping set of samples with this study (Brookes and others 2006), and own research. These are: NR4A2, PER2, SLC6A1, DRD3, SLC9A9, HES1, ADRA2C, ADRB2, ADRA1B, DRD1, HTR1E, DDC, STX1A, ADRA1A, NFIL3, ADRA2A, ADRB1, SLC18A2, TPH1, BDNF, FADS1, FADS2, ADRBK1, ARRB1, DRD2, HTR3B, TPH2, SYT1, HTR2A, SLC6A2, ARRB2, PER1, PNMT, CHRNA4, COMT, ADRBK2, CSNK1E, MAOA, MAOB, and HTR2C. A meta-analysis of COMT strongly suggest no role for ADHD (Cheuk and Wong 2006).
Multiple Testing
Our approach for dealing with multiple testing is to require genome-wide association significance to assert that a putative finding is a true positive. Recently, two papers have detailed an estimate for the effective number of independent tests in the genome (Dudbridge and Gusnanto 2008; Pe’er and others 2008). Both of the papers identified approximately 5×10−8 as the threshold for significance. We recognize that this may be a conservative approach, but this dataset is the first step in identifying risk variation for ADHD, not the final word.
Power
To assess the effect sizes detectable for our sample size, we performed power calculations assuming four different models and a number of variants associated with ADHD ranging from one to one hundred. The results are shown in Figure 1. Given our sample size, we should have been able to detect at least 1 association at a P-value of 5×10−8, for an odds ratio of 1.3 and a minor allele frequency of 0.20.
Figure 1. Power to detect at least 1 genome-wide significant association for 909 trios.
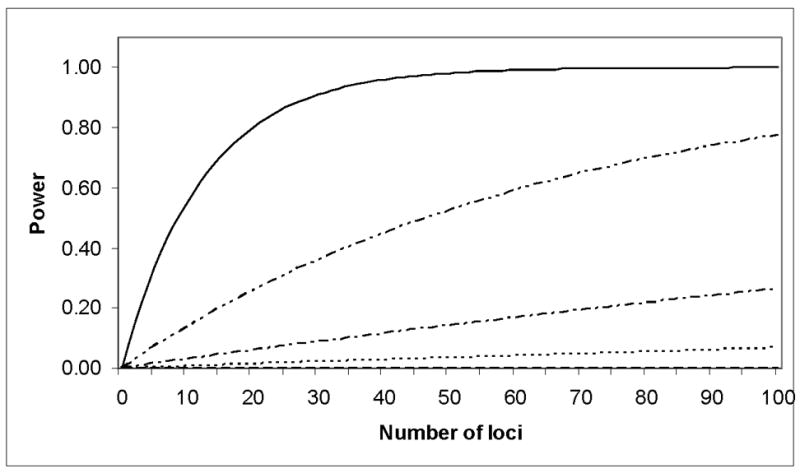
From top to bottom the series are based on models with an OR of 1.3 and MAF of 0.40, OR of 1.3 and MAF of 0.20, OR of 1.2 and MAF of 0.40, OR of 1.2 and MAF of 0.20. The power to detect at least one locus is equal to 1 − (1 − Q)N where Q is the power to detect an association at significance level of 5x10-8 of a given model and N is the number of loci with said effect.
Results
After the quality control procedures, 438,784 markers were adequate for use in the statistical analyses. A summary of the sample tested in the full TDT on the Perlegen arrays is listed in table 3. We restricted our TDT analysis to a subset of 909 complete family trios to prevent bias in the association statistic (Curtis and Sham 1995).
Table 3.
Descriptive Statistics on offspring in 909 trios used in the GWAS
| Sub-type | Counts | Country of Origin | Families |
|---|---|---|---|
| Hyperactive-Impulsive | 33 | Belgium | 36 |
| Inattentive | 13 | Germany | 99 |
| Combined | 845 | Holland | 303 |
| Sub-threshold | 18 | Ireland | 84 |
| Age at onset (s.d.) | Israel | 166 | |
| Hyperactive-impulsive (Hyp/Imp) | 2.77 (1.85) | Spain | 67 |
| Inattention (Ina) | 4.31 (1.96) | Switzerland | 25 |
| Average count of Ina symptoms | 7.98 (1.37) | United Kingdom | 129 |
| Average count of Hyp/Imp symptoms | 8.11 (1.10) | Male | 790 |
| Average total number of symptoms | 16.10 (1.92) | Female | 119 |
Included in this table is a breakdown of the different diagnoses for the TDT probands based on the IMAGE data. Additionally, average number of symptoms, the age of onset, and breakdown of the country of origin for the different families has been included for the 909 complete trios.
The Q-Q plot presented in Figure 2 illustrates the distribution of p-values from the SNPs in the TDT test. We observed a mild inflation across the distribution of association test statistics with a lambda of approximately 1.03. Splitting the dataset into groups of SNPs for which either the major or minor allele was over-transmitted yielded 222,089 and 208,836 SNPs respectively (McNemar χ2 = 407, P-value 10−90). The justification for the use of the paired McNemar χ2 test is Mendel’s law of segregation, as there is no expectation for a major or minor allele at a locus being overtransmitted. Furthermore, the median χ2 for the major and minor allele over-transmission was dramatically different (0.4899 and 0.4488 respectively, yielding λ’s of 1.077 and 0.9864). By removing additional SNPs that had failed QC in the MDD GAIN study, the median χ2 for major and minor allele transmission improved to 0.4836 and 0.4496, respectively. Similar improvement was observed when excluding the Psoriasis SNPs as well, with median chi squares of 0.485 and 0.4496.
Figure 2.
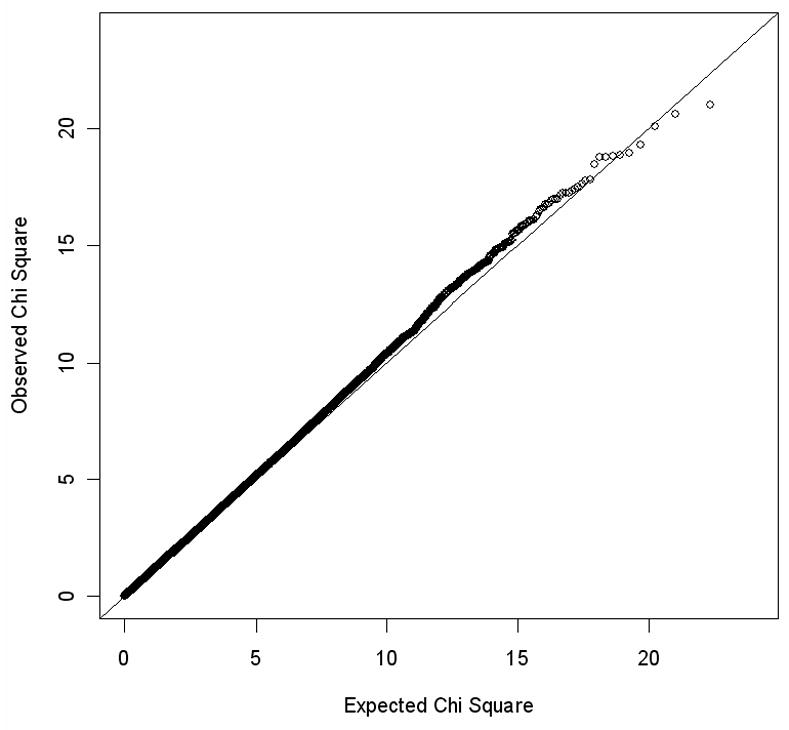
The QQ Plot of the TDT χ2 from the genome-wide data
Additionally, we endeavored to model the size of the χ2 test as a function of MAF and call rate. MAF shows no effect on the χ2, but call rate did show significant predictive power on this measure (p<10−10). However, the effect size was not particularly large, with the lowest call rate class, 95%, having a χ2 which was 0.164 larger than that of a non-missing SNP. As an additional check, we visually examined the cluster plots for our highest ranking SNPs to ensure the quality of these data. Such endeavors removed 468 SNPs from the top 2,000.
As missingness accounted for the vast majority of SNPs being excluded from the MDD and Psoriasis datasets and showed the most predictive power on the χ2, we explored the effect of non-random missingness on the TDT (see Supplementary Methods). Based on the observation that a small discrepancy in missing rates for each genotype class can yield the pattern of overtransmission of the major allele, we decided to keep all of the SNPs based on our original cleaning and apply the lambda correction. Upon application of the lambda we corrected for the overrepresentation of major allele overtransmission in the top end of the distribution. The top 25 hits from the uncorrected and lambda applied TDT results can be found in tables 4 and 5 respectively. These results are independent at an r2 > 0.8. As none of these results are genome-wide association significant, exploring the biological relevance to ADHD would be premature.
Table 4.
Top 25 Uncorrected TDT results
| Chr | SNP | A1:A2 | T:U | OR | χ2 | P-value |
|---|---|---|---|---|---|---|
| 6 | rs9389835 | C:T | 350:479 | 0.7307 | 20.07 | 7.45E-06 |
| 19 | rs9676447 | C:T | 106:51 | 2.078 | 19.27 | 1.14E-05 |
| 13 | rs1539549 | T:C | 328:449 | 0.7305 | 18.84 | 1.42E-05 |
| 6 | rs964647 | T:A | 170:99 | 1.717 | 18.74 | 1.50E-05 |
| 14 | rs1427324 | T:C | 301:416 | 0.7236 | 18.44 | 1.75E-05 |
| 6 | rs6919857 | C:T | 370:494 | 0.749 | 17.8 | 2.46E-05 |
| 4 | rs876477 | T:C | 118:62 | 1.903 | 17.42 | 2.99E-05 |
| 2 | rs4241112 | T:C | 277:384 | 0.7214 | 17.32 | 3.16E-05 |
| 22 | rs9608617 | C:G | 399:290 | 1.376 | 17.24 | 3.29E-05 |
| 4 | rs7657608 | T:C | 434:320 | 1.356 | 17.24 | 3.30E-05 |
| 21 | rs957795 | A:G | 117:189 | 0.619 | 16.94 | 3.86E-05 |
| 8 | rs2939678 | A:G | 273:378 | 0.7222 | 16.94 | 3.87E-05 |
| 4 | rs17689952 | G:A | 270:374 | 0.7219 | 16.8 | 4.16E-05 |
| 5 | rs17673653 | T:A | 160:242 | 0.6612 | 16.73 | 4.32E-05 |
| 15 | rs2439832 | T:C | 153:233 | 0.6567 | 16.58 | 4.66E-05 |
| 11 | rs17754282 | G:T | 294:204 | 1.441 | 16.27 | 5.51E-05 |
| 14 | rs2747100 | G:C | 391:512 | 0.7637 | 16.21 | 5.66E-05 |
| 12 | rs3782309 | T:G | 223:146 | 1.527 | 16.07 | 6.11E-05 |
| 4 | rs2323262 | A:G | 207:297 | 0.697 | 16.07 | 6.10E-05 |
| 15 | rs922781 | C:G | 493:375 | 1.315 | 16.04 | 6.20E-05 |
| 4 | rs12505502 | G:T | 525:403 | 1.303 | 16.04 | 6.21E-05 |
| 6 | rs10807124 | A:G | 313:421 | 0.7435 | 15.89 | 6.71E-05 |
| 1 | rs7528615 | C:T | 353:467 | 0.7559 | 15.85 | 6.86E-05 |
| 5 | rs4913069 | A:G | 136:210 | 0.6476 | 15.83 | 6.94E-05 |
| 18 | rs9973180 | T:C | 328:438 | 0.7489 | 15.8 | 7.05E-05 |
Chr is the chromosome, SNP is the SNP rs number, A1:A2 are the minor and major alleles respectively at the locus, T:U are the transmissions and untransmitted counts for the minor allele, and OR is the TDT odds ratio.
Table 5.
Top 25 TDT results after correction for TDT bias
| Chr | SNP | A1:A2 | T:U | OR | χ2 | P-value |
|---|---|---|---|---|---|---|
| 14 | rs2295426 | C:T | 304:428 | 0.7103 | 19.51 | 1.00E-05 |
| 19 | rs9676447 | C:T | 106:51:00 | 2.078 | 19.49 | 1.01E-05 |
| 18 | rs2311120 | G:A | 229:337 | 0.6795 | 19.14 | 1.22E-05 |
| 6 | rs964647 | T:A | 170:99 | 1.717 | 18.96 | 1.34E-05 |
| 6 | rs9389835 | C:T | 350:479 | 0.7307 | 18.64 | 1.58E-05 |
| 4 | rs876477 | T:C | 118:62 | 1.903 | 17.62 | 2.69E-05 |
| 6 | rs6570426 | T:A | 344:468 | 0.735 | 17.59 | 2.74E-05 |
| 13 | rs1539549 | T:C | 328:449 | 0.7305 | 17.50 | 2.88E-05 |
| 6 | rs9484448 | T:C | 353:478 | 0.7385 | 17.46 | 2.94E-05 |
| 4 | rs7657608 | T:C | 434:320 | 1.356 | 17.44 | 2.96E-05 |
| 22 | rs9608617 | C:G | 399:290 | 1.376 | 17.44 | 2.96E-05 |
| 1 | rs6657749 | T:C | 184:277 | 0.6643 | 17.42 | 2.99E-05 |
| 13 | rs17722514 | T:C | 178:108 | 1.648 | 17.33 | 3.14E-05 |
| 14 | rs1427324 | T:C | 301:416 | 0.7236 | 17.12 | 3.50E-05 |
| 11 | rs11221064 | C:G | 305:212 | 1.439 | 16.92 | 3.89E-05 |
| 6 | rs6919857 | C:T | 370:494 | 0.749 | 16.53 | 4.79E-05 |
| 10 | rs12772737 | G:C | 292:403 | 0.7246 | 16.4646 | 4.96E-05 |
| 11 | rs17754282 | G:T | 294:204 | 1.441 | 16.4591 | 4.97E-05 |
| 16 | rs7187223 | G:A | 37:83 | 0.4458 | 16.3718 | 5.21E-05 |
| 12 | rs3782309 | T:G | 223:146 | 1.527 | 16.2568 | 5.53E-05 |
| 5 | rs1541665 | T:C | 135:213 | 0.6338 | 16.2325 | 5.60E-05 |
| 4 | rs12505502 | G:T | 525:403 | 1.303 | 16.2264 | 5.62E-05 |
| 15 | rs922781 | C:G | 493:375 | 1.315 | 16.2264 | 5.62E-05 |
| 2 | rs4241112 | T:C | 277:384 | 0.7214 | 16.0839 | 6.06E-05 |
| 18 | rs2678787 | C:T | 279:386 | 0.7228 | 15.991 | 6.36E-05 |
Figure 3 presents the QQ plot for all GAIN SNPs in or near the candidate genes described in the methods section. Mild inflation is observed in the P-value distribution, suggesting that there may be weak evidence for association with ADHD for these candidate genes in the current study. However, none of SNPs in these genes show convincing evidence of association, i.e., none achieved genome-wide significance.
Figure 3.
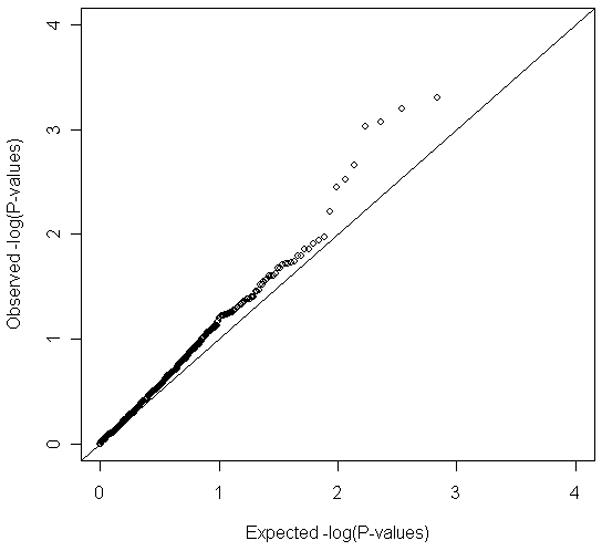
Distribution of P values for selected candidate genes
Discussion
We have completed the first genome-wide association scan of ADHD on a sample of 909 complete proband-parent triads with a child with the combined subtype of ADHD from the IMAGE project. We found no genome-wide significance for any SNP in this initial scan. These findings suggest that risk variants for ADHD must have very small average effects at the population level, conferring odds ratios of 1.3–1.4 at best and in many cases much lower. If this inference is correct, much larger samples will be needed to reliably detect ADHD susceptibility genes.
These negative results indicating the existence of very small genetic effects when individual variants are considered alone is not surprising given the recent results of initial examinations of other psychiatric disorders (e.g. bipolar disorder and schizophrenia) (Sklar and others 2008). For some complex diseases such as Type II Diabetes and Crohn’s disease mapping of one or a small number of disease-associated variants was successful in studies with similar sample sizes to the present study, but the vast majority of findings have emerged with the incorporation of multiple scans involving sample sizes many times larger than the one presented here (Rioux and others 2007) (Saxena and others 2007) and in most cases consisted of genetic loci conferring odds ratio in the regions of 1.1 – 1.4. While no single SNP achieved genome-wide association significance, there is likely to be some association signal in our data. The identification of this signal, however, requires additional data to improve the significance.
The general expectation from GWAS of complex disorders is for multiple genes of very small effect (Altshuler and Daly 2007). Backward power calculations on some of the initial true results from these diseases, however, indicate that many of the identified candidates were extremely unlikely to be detected from the initial study (Altshuler and Daly 2007). Thus, these initial studies were either fortunate or many such effects (potentially one hundred or more) with a similar effect size should be postulated. In this study we have not been fortunate, insofar as we did not identify a variant above genome-wide significance, which we define as 5×10−8 (Dudbridge and Gusnanto 2008; Pe’er and others 2008). Concerning the existing candidate genes for ADHD, the genome-wide association data do not provide support for any of the previously theorized genes (the analysis of these genes is discussed in more detail in Brookes et al., (2006)). That is not to say that these genes are to be unequivocally rejected from consideration, but rather that the effect sizes for each of these variants must be small if they are real effects. This is consistent with the results of existing meta-analyses (Faraone and others 2005; Li and others 2007).
The reasons for such small effects can arise for several potential reasons. First it may be correct that genetic risks for ADHD are due to numerous small additive effects of common risk variants. However, it is also possible that multiple rare variants of small to moderately large effect size could account for these findings. Alternative explanations include sample heterogeneity, the possible interaction of genetic variants either within or between genes; and their interaction with environmental risk factors. Such factors may also include copy number variants which may not be captured through the use of SNP association. Deeper coverage of the genome, different study designs, or greater homogeneity in the sample may yield further insight into the genetic causes of ADHD.
Additionally, we have shown that family-based association studies face a systemic bias in the transmission of the major versus the minor allele. As demonstrated in the supplementary methods, differential rates of missing genotypes may be the source of the problem. Given the clustering algorithms used for calling current genotyping technologies, the common homozygote is typically the class most accurately called, as cluster membership is high. Consequently, such bias in the TDT will be commonly observed. Clayton and colleagues demonstrated that differential missing rates in cases and controls can yield a bias in the association test statistic distribution. Differences in the probability of missing each genotype class, however, will not yield a bias in case control data, as long as they are equally applied to both groups.
Although the heritability of ADHD is high, this does not give an indication of the underlying genetic architecture, although it does imply that genetic influences are important for the aetiology of ADHD. Recent modeling of complex behavioural and biological traits in the mouse suggests that as heritability increases the number of genetic variants involved increases, though effect sizes of individual variants remain small (Flint and Munafo 2007). For ADHD, our expectation is that novel genes for ADHD will be identified from GWAS once sufficient whole genome association data has been accumulated from the analysis of 10,000 – 20,000 cases.
As the current study is part of the GAIN initiative, all data are available online at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap. Access to the dataset requires gaining approval from the Data Access Committee, but the results disclosed here will be made available on the dbGap server for browsing. Additionally the top results from the TDT analysis described here are available upon request.
Supplementary Material
Acknowledgments
The IMAGE project is a multi-site, international effort supported by NIH grant R01MH62873 to S.V. Faraone. Site Principal Investigators are Philip Asherson, Tobias Banaschewski, Jan Buitelaar, Richard P. Ebstein, Stephen V. Faraone, Michael Gill, Ana Miranda, Fernando Mulas, Robert D. Oades, Herbert Roeyers, Aribert Rothenberger, Joseph Sergeant, Edmund Sonuga-Barke, and Hans-Christoph Steinhausen. Senior coinvestigators are Margaret Thompson, Pak Sham, Peter McGuffin, Robert Plomin, Ian Craig and Eric Taylor. Chief Investigators at each site are Rafaela Marco, Nanda Rommelse, Wai Chen, Henrik Uebel, Hanna Christiansen, U.Mueller, Cathelijne Buschgens, Barbara Franke, Lamprini Psychogiou. We thank all the families who kindly participated in this research.
References
- Altshuler D, Daly M. Guilt beyond a reasonable doubt. Nat Genet. 2007;39(7):813–5. doi: 10.1038/ng0707-813. [DOI] [PubMed] [Google Scholar]
- American Psychiatric A. Diagnostic and statistical manual of mental disorders: DSM-III-R. Washington, D.C: American Psychiatric Association; 1987. p. 567. %7 3rd-Revised p. [Google Scholar]
- Anney R, Kenny E, O’Dushlaine C, Lasky-Su J, Franke B, Asherson P, Morris DW, Neale B, Asherson P, Faraone SV, et al. Non-Random Error in Genotype Calling Procedures: implications for family-based and case-control genome-wide association studies. doi: 10.1002/ajmg.b.30836. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcos-Burgos M, Castellanos FX, Pineda D, Lopera F, David Palacio J, Guillermo Palacio L, Rapoport JL, Berg K, Bailey-Wilson JE, Muenke M. Attention-deficit/hyperactivity disorder in a population isolate: linkage to Loci at 4q13.2, 5q33.3, 11q22, and 17p11. Am J Hum Genet. 2004;75(6):998–1014. doi: 10.1086/426154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asherson P, Zhou K, Anney RJ, Franke B, Buitelaar J, Ebstein R, Gill M, Altink M, Arnold R, Boer F, et al. A high-density SNP linkage scan with 142 combined subtype ADHD sib pairs identifies linkage regions on chromosomes 9 and 16. Mol Psychiatry. 2008;13(5):514–21. doi: 10.1038/sj.mp.4002140. [DOI] [PubMed] [Google Scholar]
- Bakker SC, van der Meulen EM, Buitelaar JK, Sandkuijl LA, Pauls DL, Monsuur AJ, van ‘t Slot R, Minderaa RB, Gunning WB, Pearson PL, et al. A whole-genome scan in 164 Dutch sib pairs with attention-deficit/hyperactivity disorder: suggestive evidence for linkage on chromosomes 7p and 15q. Am J Hum Genet. 2003;72(5):1251–1260. doi: 10.1086/375143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes K, Xu X, Chen W, Zhou K, Neale B, Lowe N, Aneey R, Franke B, Gill M, Ebstein R, et al. The analysis of 51 genes in DSM-IV combined type attention deficit hyperactivity disorder: association signals in DRD4, DAT1 and 16 other genes. Mol Psychiatry. 2006 doi: 10.1038/sj.mp.4001869. [DOI] [PubMed] [Google Scholar]
- Chen W, Zhou K, Sham P, Franke B, Kuntsi J, Campbell D, Fleischman K, Knight J, Andreou P, Arnold R, et al. DSM-IV combined type ADHD shows familial association with sibling trait scores: A sampling strategy for QTL linkage. Am J Med Genet B Neuropsychiatr Genet. 2008 doi: 10.1002/ajmg.b.30672. [DOI] [PubMed] [Google Scholar]
- Cheuk DK, Wong V. Meta-analysis of Association Between a Catechol-O-Methyltransferase Gene Polymorphism and Attention Deficit Hyperactivity Disorder. Behav Genet. 2006 doi: 10.1007/s10519-006-9076-5. [DOI] [PubMed] [Google Scholar]
- Conners K. Rating Scales in ADHD 1996 [Google Scholar]
- Curtis D, Sham PC. A note on the application of the transmission disequilibrium test when a parent is missing. Am J Hum Genet. 1995;56(3):811–2. [PMC free article] [PubMed] [Google Scholar]
- Cutler DJ, Zwick ME, Carrasquillo MM, Yohn CT, Tobin KP, Kashuk C, Mathews DJ, Shah NA, Eichler EE, Warrington JA, et al. High-throughput variation detection and genotyping using microarrays. Genome Res. 2001;11(11):1913–25. doi: 10.1101/gr.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32(3):227–34. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone SV. Genetics of adult attention-deficit/hyperactivity disorder. Psychiatr Clin North Am. 2004;27(2):303–21. doi: 10.1016/S0193-953X(03)00090-X. [DOI] [PubMed] [Google Scholar]
- Faraone SV. The scientific foundation for understanding attention-deficit/hyperactivity disorder as a valid psychiatric disorder. Eur Child Adolesc Psychiatry. 2005;14:1–10. doi: 10.1007/s00787-005-0429-z. [DOI] [PubMed] [Google Scholar]
- Faraone SV. Advances in the genetics and neurobiology of attention deficit hyperactivity disorder. Biological Psychiatry. 2006;60(10):1025–7. doi: 10.1016/s0006-3223(98)00240-6. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Biederman J. Neurobiology of attention-deficit hyperactivity disorder. Biological Psychiatry. 1998;44(10):951–958. doi: 10.1016/s0006-3223(98)00240-6. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Biederman J. Nature, nurture, and attention deficit hyperactivity disorder. Developmental Review. 2000;20:568–581. [Google Scholar]
- Faraone SV, Biederman J. Neurobiology of Attention Deficit Hyperactivity Disorder. In: Charney DS, Nestler EJ, editors. Neurobiology of Mental Illness. 2. New York, NY: Oxford University Press; 2004. [Google Scholar]
- Faraone SV, Doyle AE, Lasky-Su J, Sklar PB, D’Angelo E, Gonzalez-Heydrich J, Kratochvil C, Mick E, Klein K, Rezac AJ, et al. Linkage analysis of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2007 doi: 10.1002/ajmg.b.30631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick J, Holmgren MA, Sklar P. Molecular genetics of attention deficit hyperactivity disorder. Biological Psychiatry. 2005;57(11):1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Sergeant J, Gillberg C, Biederman J. The Worldwide Prevalence of ADHD: Is it an American Condition? World Psychiatry. 2003;2(2):104–113. [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, Francks C, McCracken JT, McGough JJ, Marlow AJ, MacPhie IL, Newbury DF, Crawford LR, Palmer CG, Woodward JA, et al. A genomewide scan for loci involved in attention-deficit/hyperactivity disorder. Am J Hum Genet. 2002;70(5):1183–96. doi: 10.1086/340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Munafo MR. The endophenotype concept in psychiatric genetics. Psychol Med. 2007;37(2):163–80. doi: 10.1017/S0033291706008750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Finch SJ, Nothnagel M, Ott J. Power and sample size calculations for case-control genetic association tests when errors are present: application to single nucleotide polymorphisms. Hum Hered. 2002;54(1):22–33. doi: 10.1159/000066696. [DOI] [PubMed] [Google Scholar]
- Hebebrand J, Dempfle A, Saar K, Thiele H, Herpertz-Dahlmann B, Linder M, Kiefl H, Remschmidt H, Hemminger U, Warnke A, et al. A genome-wide scan for attention-deficit/hyperactivity disorder in 155 German sib-pairs. Mol Psychiatry. 2006;11(2):196–205. doi: 10.1038/sj.mp.4001761. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6(2):95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Kuntsi J, Neale BM, Chen W, Faraone SV, Asherson P. The IMAGE project: Methodological issues for the molecular genetic analysis of ADHD. Behav Brain Funct. 2006;2(1):27. doi: 10.1186/1744-9081-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nature Genetics. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Li D, Sham PC, Owen MJ, He L. Meta-analysis shows significant association between dopamine system genes and attention deficit hyperactivity disorder (ADHD) Hum Mol Genet. 2006 doi: 10.1093/hmg/ddl152. [DOI] [PubMed] [Google Scholar]
- Li J, Wang Y, Hu S, Zhou R, Yu X, Wang B, Guan L, Yang L, Zhang F, Faraone SV. The monoamine oxidase B gene exhibits significant association to ADHD. Am J Med Genet B Neuropsychiatr Genet. 2007 doi: 10.1002/ajmg.b.30606. [DOI] [PubMed] [Google Scholar]
- Morrison JR, Stewart MA. A family study of the hyperactive child syndrome. Biological Psychiatry. 1971;3:189–195. [PubMed] [Google Scholar]
- Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–5. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39(5):596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K, Nimgaonkar VL, McQueen MB, Faraone SV, Kirby A, et al. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008 doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley SL, Kustanovich V, Minassian SL, Stone JL, Ogdie MN, McGough JJ, McCracken JT, MacPhie IL, Francks C, Fisher SE, et al. Genetic Linkage of Attention-Deficit/Hyperactivity Disorder on Chromosome 16p13, in a Region Implicated in Autism. Am J Hum Genet. 2002;71(4):959–63. doi: 10.1086/342732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM) American Journal of Human Genetics. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Taylor E, Schachar R, Thorley G, Wieselberg M. Conduct disorder and hyperactivity: I. Separation of hyperactivity and antisocial conduct in British child psychiatric patients. Br J Psychiatry. 1986;149:760–7. doi: 10.1192/bjp.149.6.760. [DOI] [PubMed] [Google Scholar]
- Wang B, Wang Y, Zhou R, Li J, Qian Q, Yang L, Guan L, Faraone SV. Possible association of the alpha-2A adrenergic receptor gene (ADRA2A) with symptoms of attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(2):130–4. doi: 10.1002/ajmg.b.30258. [DOI] [PubMed] [Google Scholar]
- Zhou K, Asherson P, Sham P, Franke B, Anney RJL, Buitelaar J, Ebstein R, Gill M, Brookes K, Buschgens C, et al. Linkage to Chromosome 1p36 for Attention-Deficit/Hyperactivity Disorder Traits in School and Home Settings. Bio Psy. doi: 10.1016/j.biopsych.2008.02.024. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


