Abstract
Although the protein synthesis inhibitor cycloheximide (CHX) has been known for decades, its precise mechanism of action remains incompletely understood. The glutarimide portion of CHX is seen in a family of structurally related natural products including migrastatin, isomigrastatin and lactimidomycin (LTM). LTM, isomigrastatin and analogs were found to have a potent antiproliferative effect on tumor cell lines and selectively inhibit protein translation. A systematic comparative study of the effects of CHX and LTM on protein translation revealed both similarities and differences between the two inhibitors. Both LTM and CHX were found to block the translocation step in elongation. Footprinting experiments revealed protection of a single cytidine nucleotide (C3993) in the E-site of the 60S ribosomal subunit, defining a common binding pocket for both inhibitors in the ribosome. These results shed new light on the molecular mechanism of inhibition of translation elongation by both CHX and LTM.
Keywords: Eukaryotic Translation, Elongation, Translocation, Lactimidomycin, Migrastatin, Isomigrastatin, Cycloheximide, Exit Site
Small molecule inhibitors of bacterial protein synthesis have served as powerful tools in the elucidation of the function of the prokaryotic ribosome. Even before the availability of high-resolution structures, the main functional aspects of the bacterial ribosome had been characterized largely with the help of antibiotics inhibiting various steps of the prokaryotic translational process1. In contrast to prokaryotes, far fewer compounds have been identified that inhibit eukaryotic translation. Given the essential role of translation in the proliferation and survival of eukaryotic cells, particularly fast-growing tumor cells, it seems likely that translation inhibitors may serve as leads in the development of new cancer therapeutics.
Among the known inhibitors of eukaryotic translation is cycloheximide (CHX, 1), the most common laboratory reagent used to inhibit protein synthesis (Fig. 1). CHX has been shown to block the elongation phase of eukaryotic translation. It binds the ribosome and inhibits eEF2-mediated translocation2. Surprisingly, CHX allows one complete translocation cycle to proceed before halting any further elongation3. It has been speculated that CHX requires an E-site bound deacylated tRNA for activity. Despite these mechanistic insights, however, significant gaps remain. For example, the exact binding site for CHX remains unknown. It is also unclear whether it directly interacts with eEF2, or whether translocation inhibition results from an indirect effect.
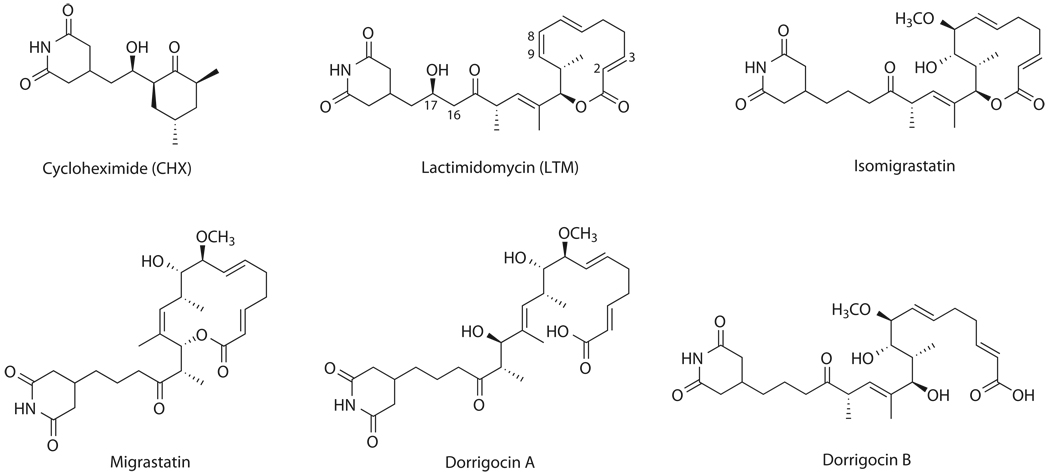
Figure 1. Chemical structures of glutarimide-containing natural products.
CHX, originally isolated from Streptomyces griseus, contains a glutarimide moiety. Recently, a new family of glutarimide-containing natural products were isolated, including migrastatin (2) from Streptomyces sp MK929-43F1, isomigrastatin (3) and dorrigocins from Streptomyces platensis, and lactimidomycin (LTM, 4) from Streptomyces, respectively (Fig. 1) 4,5. Migrastatin was found to inhibit tumor cell migration and has served as an anti-metastatic drug lead6,7. The dorrigocins (5,6) appear to inhibit a carboxyl methyltransferase involved in the processing of Ras-related proteins8,9. We have recently established that isomigrastatin is the nascent natural product and migrastatin and dorrigocins are shunt metabolites of isomigrastatin10. Upon exposure to water, Isomigrastatin undergoes ring-expansion or ring-opening rearrangements to migrastatin and dorrigocins, respectively. By optimizing isomigrastatin fermentation in S. platensis and LTM fermentation in S. amphibiosporus, as well as engineering the isomigrastatin and LTM biosynthetic machinery, we have subsequently produced a focused library of the glaturimide-containing polyketides featuring the isomigrastatin, migrastatin, dorrigocin, and LTM scaffolds (Fig. 1)4,11.
We screened the library for activity against the proliferation of several tumor cell lines. Remarkably, the 12-membered glutarimide-containing polyketide macrolides, exemplified by LTM and isomigrastatin, were found to possess potent anti-proliferative activity, while the 14-membered macrolides, represented by migrastatin, or the linear members, represented by the dorrigocins, showed little cytotoxicity. Further characterization revealed that LTM inhibited the elongation step of eukaryotic translation, in a similar but not identical fashion to CHX. Despite their structural similarity to migrastatin and their previous classification as cell migration inhibitors, isomigrastatin and LTM acted by a completely different mechanism. A systematic mechanistic study of LTM side-by-side with CHX allowed for the formulation of a comprehensive and coherent model for the mechanism of inhibition of eukaryotic translational elongation by LTM and CHX, including the binding site of these inhibitors on the 60S ribosome. Moreover, we also demonstrated that LTM possesses antitumor activity in vivo, suggesting that inhibitors of eukaryotic translation elongation may have potential of becoming novel anticancer agents.
RESULTS
Activity of the glutarimide-containing natural products
We have previously reported the construction of a focused library of the glutarimide-containing polyketides featuring the isomigrastatin, migrastatin, dorrigocin, and LTM scaffolds (Fig. 1)4. We screened the library of 35 compounds for inhibitory activity against the proliferation of three different tumor cell lines, HeLa, MDA-MB231 and Jurkat T cells (Supplementary Fig. 1). In the initial screens, each of the three tumor lines was incubated with every compound (2.5 µM) for 24 h before application of [3H]-thymidine to monitor cellular DNA synthesis. While most of the analogs based on the migrastatin and dorrigocin scaffolds did not drastically affect cell growth, LTM, isomigrastatin and selected analogs (7 and 8) proved highly active in the assay (Supplementary Fig. 1). An interesting structure-and-activity relationship (SAR) emerged from these studies.
First, all active analogs contain a 12-membered macrocycle as seen in LTM and isomigrastatin. None of the migrastatin-based 14-membered macrolides, nor any of the dorrigocin-based linear isomers showed any activity. Second, an intact glutarimide moiety is necessary but not sufficient for activity. Thus, alkylation of the glutarimide group led to an inactive analog (9). But neither migrastatin nor dorrigocin is active despite of the presence of an intact glutarimide moiety. Third, the 12-membered macrolide can tolerate some modifications (i.e., isomigrastatin vs. LTM) but not others (i.e., 10). Lastly, the hydroxyl group on C-17 in the linker region is dispensable. Interestingly, attachment of an ethoxycarbonylmethyl unit to the OH group (36) did not significantly decrease the activity of LTM (Supplementary Fig. 2).
It was striking that only isomigrastatin, LTM, and closely related analogs inhibited cell proliferation, while all migrastatin and dorrigocin analogs had no effect, in agreement with previous reports that neither compound had cytotoxic effects on mammalian cells8,9,12,13. Among all analogs tested, LTM stood out as the most potent inhibitor of cell proliferation (Supplementary Fig. 1), making it an ideal probe to investigate the mode of action of this family of cell proliferation inhibitors.
LTM and isomigrastatin inhibit eukaryotic translation
To investigate which biochemical process the active compounds might affect, we measured de novo protein synthesis through metabolic labeling with radioactive amino acids in the presence of each compound. HeLa cells were incubated with [35S]cysteine and methionine for two hours to allow for their incorporation into newly synthesized proteins. All compounds that inhibited cell proliferation also drastically decreased protein synthesis (Supplementary Fig. 1b, c).
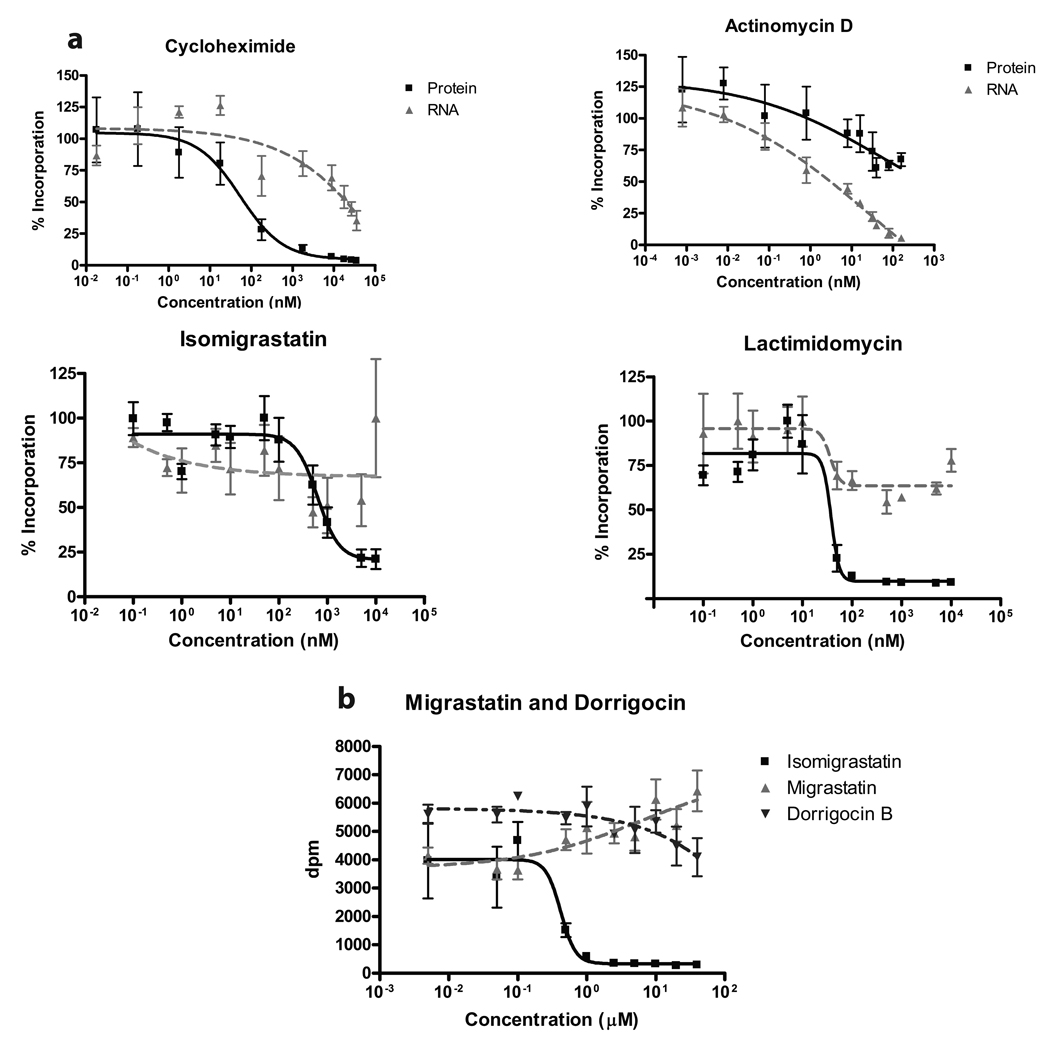
To verify that the observed effect of LTM and analogs on protein synthesis is specific, we determined their impact on both translation and transcription by metabolic labeling across a wide dose range. Transcriptional activity was monitored by incubation with [3H]uridine for two hours. Actinomycin D (ActD) and CHX served as controls as bona fide transcription and translation inhibitors, respectively. As expected, CHX strongly inhibited translation but only affected transcription at very high doses, while ActD concomitantly blocked transcription and translation as expected since protein synthesis requires a supply of mRNA (Fig. 2a). Similar to CHX, both LTM and isomigrastatin exclusively inhibited protein synthesis without a significant impact on transcription. Once again, LTM emerged as the most powerful inhibitor of translation, being about 10-fold more potent than CHX (Fig. 2a and Supplementary Table 1).
Figure 2. Inhibition of protein translation by LTM and isomigrastatin.
a. Dose-dependent inhibition of translation by LTM, isomigrastatin and analogs. HeLa cells were incubated with varying concentrations of each compound in presence of either [3H]uridine or [35S]cysteine/methionine for 2 h. Protein synthesis was measured by scintillation counting of TCA precipitated proteins on a PVDF membrane. Transcription was monitored by scintillation counting of nucleic acids bound to a GF/C glass fiber filter. b. Effects of isomigrastatin, migrastatin and dorrigocin on translation as measured in a. Each experiment was performed in triplicate and s.d. was shown.
Since all molecules in our collection share structural similarity with CHX, we confirmed our structure-activity findings by employing the global translation assay with migrastatin and dorrigocin B in comparison to isomigrastatin. Despite the molecules being isomers of one another, even high doses of migrastatin or dorrigocin B had no inhibitory effect on protein synthesis (Figure 2b), which corroborated our initial findings that neither compound affected cell proliferation (Supplementary Fig. 1b, d).
Cross-resistance of yeast strains against LTM and CHX
That LTM, like CHX, inhibited translation together with their structural similarity, raised the possibility that they might act upon the same target. CHX is known to inhibit translation in several strains of yeast and a few resistance mutations are known in Saccharomyces cerevisiae14. The most common mutation involves a change from glutamate to glutamine in ribosomal protein L28 known as cyh215 (Yeast L28 corresponds L27a in mammals; all proteins in this publication are numbered according to Planta and Mager16). We compared the sensitivity of four pairs of isogenic S. cerevisiae strains, each pair only differing by the presence or absence of cyh2. Each pair was exposed to varying concentrations of CHX or LTM and the growth was measured by optical density (Supplementary Table 2). While the IC50 concentrations differed between strains, likely due to distinct genetic backgrounds, all CHX resistant strains were also resistant to LTM, albeit to a lesser extent.
In addition to L27a, a neighboring ribosomal protein L41 (L36a in mammals) has been implicated in CHX resistance. A proline-to-glutamine transition in different genera of fungi, such as Candida or Kluyveromyces, renders them resistant to CHX17,18. Interestingly, Candida has also been reported to be resistant to LTM5. The cross-resistance of different mutants against both CHX and LTM suggested that the two inhibitors might share a similar mechanism of action by interacting with the same target, making LTM a useful molecular probe to gain insight into the mechanism of action of CHX.
Polysome profiles and toe-print between LTM and CHX
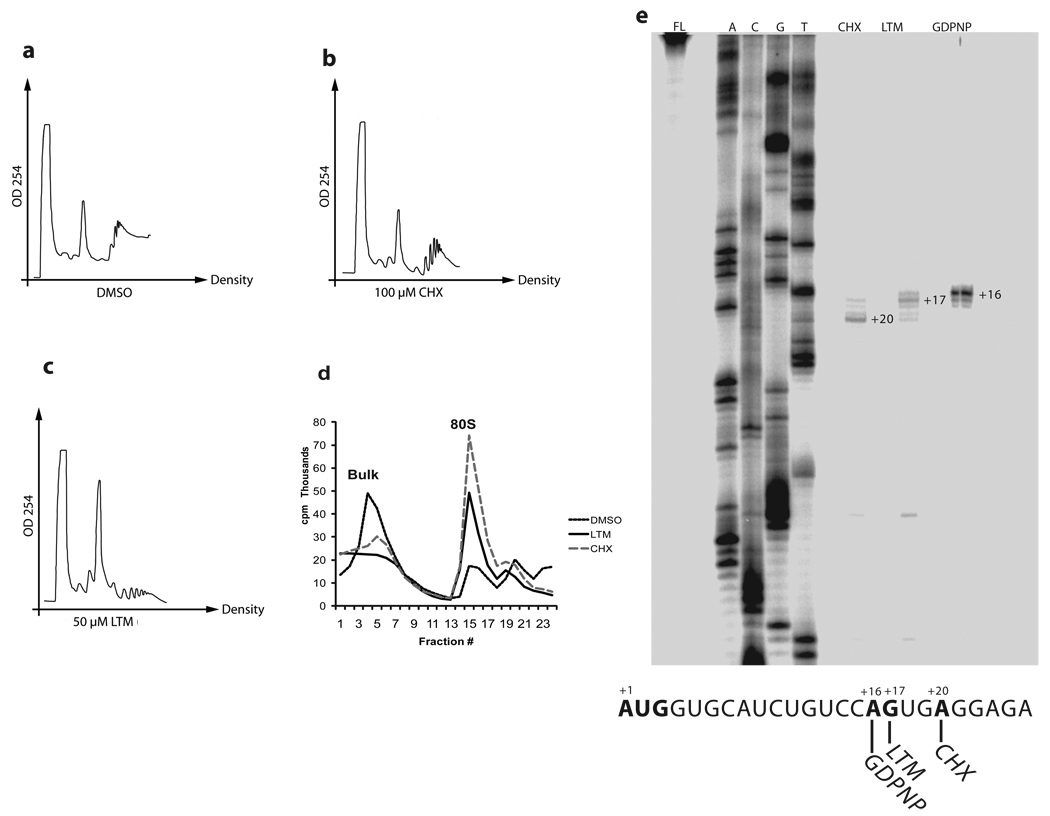
We compared the cellular distribution of RNA species after drug treatment through polyribosome profiling. HEK 293T cells were incubated with LTM or CHX for 30 min before lysis and cell lysates were applied to a sucrose density gradient. There was little difference between CHX and solvent control (Fig. 3a vs. b), though CHX seemed to slightly stabilize the RNA species, as has been observed before19. The profile displayed a modest 80S peak and distinct polysomes (Fig. 3b). In contrast, treatment with LTM led to a large increase in 80S ribosomes accompanied by depletion of polysomes (Fig. 3c). The LTM profile looked similar to the published profile of erythromycin in bacteria, suggesting a mechanistic difference between LTM and CHX20. Furthermore, even when 50 µM CHX was added 15 min before the addition of 1 µM LTM, the polysome profile still looked like the profile of LTM in absence of CHX (Supplementary Fig. 3). In vitro, both LTM and CHX had a similar effect, causing accumulation of 80S ribosomes in a cell free system (Fig. 3d). The accumulation of 80S suggested a blockade of either a late step in translation initiation or an early step in elongation.
Figure 3. Effects of LTM and cycloheximide on translation elongation in vitro and in vivo.
a–c. Polysome profiles of compounds in 293T cells. Cells were treated with each compound at the indicated concentrations before lysis and cell lysates were subjected to centrifugation through a 15–45% sucrose gradient. d. Polysome profiles in vitro. Capped [32P]-labeled rabbit β-globin RNA was incubated in rabbit reticulocyte lysate and indicated compound for 15 min before centrifugation through a 10-35% sucrose gradient. e. LTM prevents the ribosome from leaving the start codon. Toeprints of 2 mM Cycloheximide and 200 µM LTM compared to 1 mM GDPNP on rabbit β-globin mRNA (see METHODS SUMMARY for details). Each experiment was repeated at least once to ensure reproducibility.
To determine at which position this blockade occurred, we next proceeded to map where LTM arrested the ribosome on the mRNA by toeprinting (Fig. 3e). In this assay, a radio-labeled primer was hybridized to rabbit β-globin mRNA 3’ of the AUG start codon21. After incubation of the labeled mRNA in rabbit reticulocyte lysate (RRL) in the presence of various inhibiting agents, the reaction mixture was centrifuged through a sucrose gradient. The 80S fractions were removed to isolate stalled ribosomes on their mRNA template. The primer was extended with the β-globin mRNA as a template by avian myoblastoma virus (AMV) reverse transcriptase toward the stalled ribosome. When the reverse transcriptase reaches the stalled ribosome, it will fall off the template, yielding a defined transcript. Transcripts were resolved on polyacrylamide gels, which allowed mapping of the position at which the ribosome was stalled. The GTP analogue GDPNP prevents the initiation factor GTPases from functioning and stalls translation before ribosomal subunit joining. In accordance with published results, GDPNP yielded a transcript that mapped to A1621. CHX is known to allow one translocation process before preventing further elongation, resulting in a shortened transcript that mapped to A203. This represents a completely assembled 80S ribosome stalled on the second codon. LTM yielded a toeprint distinct from that of CHX and mapped to T17, exactly 3 nucleotides upstream of A20, suggesting that in presence of LTM the 80S ribosome formed but was stalled on the start codon without completing the first elongation cycle. These results revealed a subtle but potentially important difference in the effects of LTM and CHX on elongation.
Taking LTM’s higher potency into account, we repeated the toeprinting assay at three concentrations of CHX and LTM, respectively. At an excessive concentration of 10 mM, CHX did cause about half the ribosomal population to stop at the very first codon, yet even this high dose of CHX did not prevent a large amount of ribosomes to progress to the second codon. In contrast, even at 1/10,000th of the CHX concentration, LTM arrested the ribosome on the start codon (Supplementary Fig. 4).
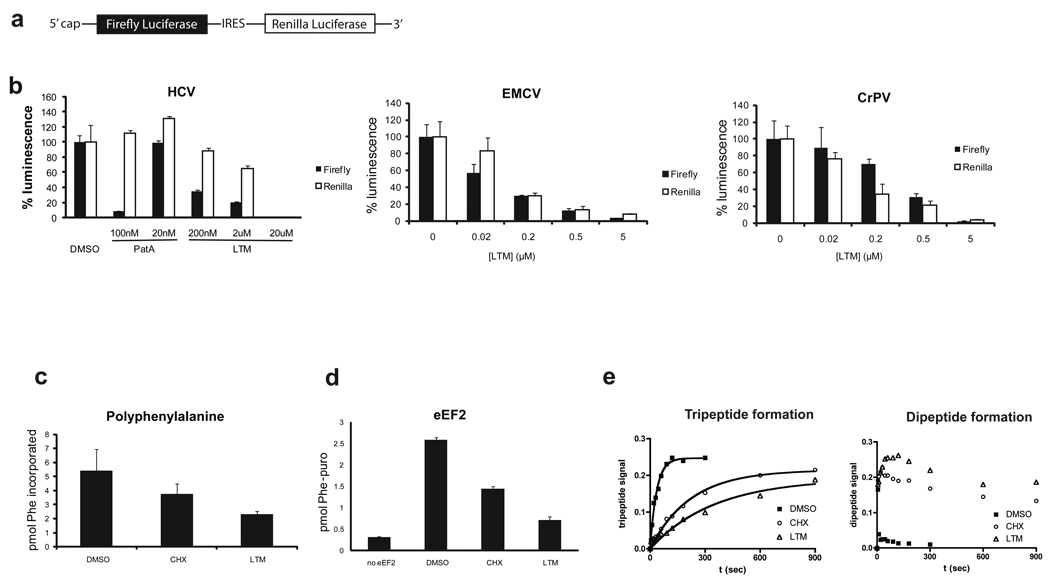
While CHX clearly inhibits elongation, the possibility remained that LTM inhibited the end of the initiation phase. To rule out this possibility, we employed bicistronic in vitro reporters with a conventional capped firefly luciferase followed by renilla luciferase under the translational control of the indicated IRES element (Fig. 4a)3,22,23 Normal cap-dependent initiation requires the concerted action of at least twelve initiation factors, which tightly regulate assembly of mRNA and initiator tRNA on the small ribosomal subunit, before allowing the joining of 40S and 60S subunits24,25. In contrast, the EMCV IRES circumvents the cap-binding protein eIF4E and allows translation of un-capped transcripts, while HCV obviates the need for the entire eIF4F complex, thereby eliminating the need not only for eIF4E, but also the helicase eIF4A and the scaffolding protein eIF4G. Furthermore, unlike either the EMCV or HCV IRES elements, the CrPV IRES does not require any initiation factors to enable translation of its transcript 22. These IRES elements allow for translation initiation independent of select initiation factors, making them resistant to inhibitors of translation initiation. Pateamine A (PatA), a marine natural product that binds to and interferes with the function of eIF4A, and hence blocking eukaryotic translation initiation, was included as a positive control. While PatA allowed translation off the HCV IRES 26, LTM inhibited translation from all three constructs to a similar degree (Fig. 4b), suggesting that LTM blocked translation elongation.
Figure 4. Effects of LTM and cycloheximide on different steps of translation elongation.
a. Configuration of the IRES reporters. Expression of firefly luciferase remains cap-dependent, while translation of renilla luciferase is under control of an IRES element. b. LTM inhibits IRES-mediated translation to a similar extent as cap-dependent translation. Pateamine A (PatA), which inhibits eIF4A-dependent translation initiation was chosen as a positive control. Error bars denote standard deviation. c. LTM inhibits poly-phenylalanine synthesis on a poly-uridine template. Phe-tRNA charged with [14C]phenylalanine was incubated with eEF1A, eEF2, ribosomes, poly(U) and GTP at 25°C for 2 min. Cycloheximide and LTM concentrations were both 200 µM. d. LTM inhibits eEF2-mediated translocation. Assay was performed as eEF1A assay, except for the use of GTP. After a 10-min preincubation, puromycin, indicated inhibitor, eEF2 and GTP were added. Formation of phenylalanyl puromycin was measured by scintillation counting of ethyl acetate extractable material. e. LTM and CHX decrease rate of tripeptide formation. The ability of pre-assembled initiation complexes to synthesize a tripeptide (Met-Phe-Phe) was measured over time. LTM and CHX treatments resulted in accumulation of didpeptides (right panel) and greatly reduced the rate of tripeptide formation (left panel). The measurements indicate the fraction of total input radioactivity. Bars in b–d represent s.d. from at least three repeats of each experiment.
LTM and CHX inhibited eEF2-mediated tRNA translocation
One cycle of translation elongation can be subdivided into at least three distinct steps: (i) binding of aminoacyl-tRNA to the ribosomal acceptor (A) site, which is dependent on eEF1A25,27,28; (ii) peptide bond formation and (iii) translocation of deacylated tRNA from the peptidyl (P) site to the E site and of the peptidyl-tRNA from A to P site, which requires eEF2. Judging from the toeprint, any of these three steps could have been interrupted by LTM. LTM inhibited polyphenylalanine synthesis from a poly(U) template using purified tRNA, ribosomes, eEF1A and eEF2 (Fig. 4c), ruling out involvement of another factor or binding partner and narrowing the potential targets to eEF1A, eEF2 and the ribosome itself.
The eEF1A-mediated binding of aminoacyl-tRNA was measured by filter binding using [14C]phenylalanine; it was not inhibited by either LTM or CHX, excluding aminoacyl-tRNA binding as the target for LTM (Supplementary Fig. 5a). Next, we turned to peptide bond formation. For measuring peptidyl transfer, [35S]methionyl tRNAMet was assembled onto initiation complexes on a short template29. Peptide bond formation was measured by monitoring [35S]methionyl-puromycin formation using thin layer chromatography (TLC). Again LTM did not affect peptide bond formation, even at millimolar concentrations. Sparsomycin was used as a positive control and prevented methyionyl-puromycin synthesis as expected (Supplementary Fig. 5b). Finally, we determined whether LTM affected eEF2-mediated translocation. Reactions were set up similar to the eEF1A-mediated binding assay, except for the presence of GTP instead of GDPNP that was used to stall the ternary complex in the aminoacyl-tRNA binding assay. After allowing tRNA binding to take place, inhibitors, eEF2 and puromycin were added. Phenylalanyl puromycin forms efficiently only if the acceptor tRNA becomes translocated into the P-site. Like CHX, LTM inhibited phenylalanyl puromycin formation, but with even higher potency (Fig. 4d).
Since LTM appears to arrest translation at the first translocation step without affecting tRNA binding or peptide bond formation, we predicted that ribosomes should only be able to produce dipeptides in presence of LTM. To test this prediction, initiation complexes with [35S]Methionyl-tRNA were assembled on a short template coding for three amino acids (Met-Phe-Phe-Stop)30. Reactions were initiated in presence of solvent alone, CHX or LTM. Aliquots were taken throughout the course of the reaction and resolved on an electrophoretic TLC system to distinguish between the di- and tripeptides (Fig. 4e). Indeed LTM greatly slowed down tripeptide formation, leading to the accumulation of dipeptides. Unexpectedly, CHX had a similar, albeit less pronounced effect.
A common binding site on the 60S ribosome for 4 and 2
The results described thus far provided a good description of LTM’s effect on translation. However, they could not fully explain the mechanism of action of either CHX or LTM. It remained unclear why the polysome profiles of LTM and CHX differed greatly while cycloheximide-resistant yeast also proved resistant to LTM. In particular, it remained puzzling why CHX primarily stalled ribosomes at the second codon, while LTM primarily prevented them from leaving the start site.
LTM’s higher potency compared to CHX made it seem likely that LTM would bind its target more tightly. Since the known resistance mutations are on ribosomal proteins, it seemed probable that LTM directly interacts with the ribosome. To assess this possibility, we applied chemical footprinting analysis to identify the potential binding site for LTM. Primers were designed based on previous studies with particular emphasis on rRNA in the vicinity of the cyh2 mutation in yeast31. For this purpose, primer sequences were overlaid with a previous model32. Unfortunately the rabbit ribosome has not yet been sequenced, but we found that primers designed on the basis of the mouse sequence generally worked well with only few exceptions (Supplementary Fig. 6). Mouse secondary structure information was obtained from the Comparative RNA Website and Database33. Hence all numbering refers to the murine 28S rRNA sequence.
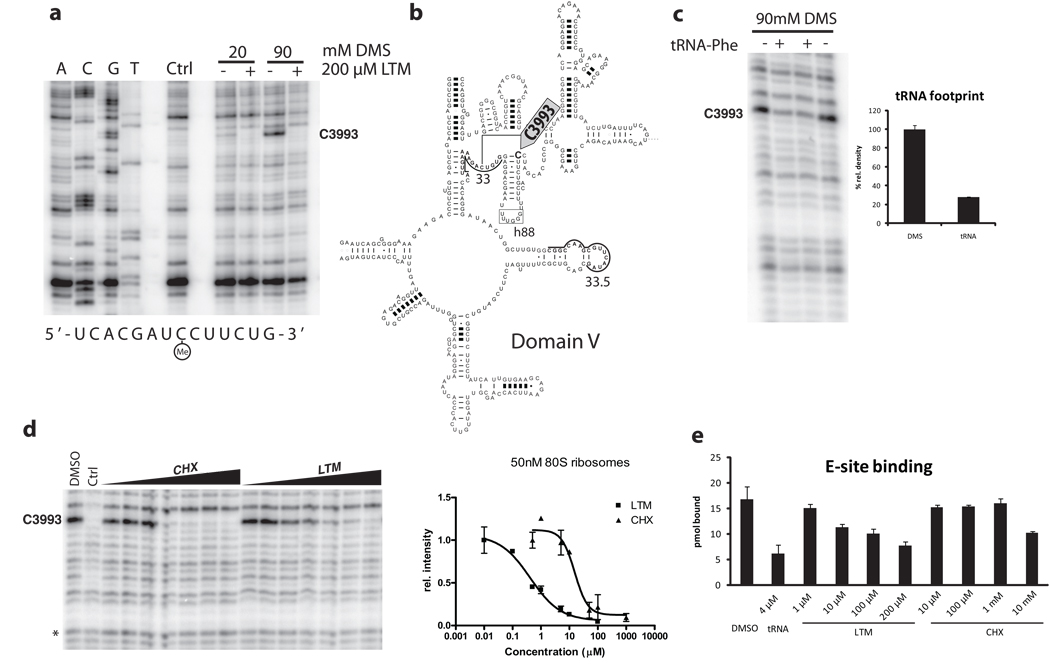
The 80S ribosomes were pre-incubated with individual compound and methylated with 20 or 90 mM dimethyl sulfate (DMS). Of all sites covered, we observed a single strong footprint on C3993 (Fig. 5a). The protected site lies at the base of hairpin 88 in domain V of the 28S rRNA (Fig. 5b). It was the only detectable footprint of LTM and attempts with kethoxal and CMCT did not reveal further sites of protection. In bacteria, the cytidine equivalent to C3993 had been identified as the interaction site between the 3’ end of tRNA and the E-site 34. We thus determined whether C3993 is involved in binding of tRNA to the eukaryotic ribosome. We were able to repeat the same result on the rabbit ribosome and observed about 70% protection in presence of deacylated tRNAPhe, the same value previously recorded in the bacterial system (Fig. 5c).
Figure 5. Footprinting analysis revealed the common binding sites of LTM and cycloheximide at the E-site of the larger ribosome subnit.
a. LTM binds to the 60S ribosomal exit site. 80S ribosomes were incubated with 200 µM LTM and methylated using 20 and 90 mM dimethyl sulfate. Extracted rRNA was hybridized to primer 33 or 33.5 (underlined) and reverse transcribed before electrophoresis. Ctrl denotes unmethylated rRNA. b. The binding site in domain V of the 28S rRNA at the base of hairpin 88 (arrow). c. The putative glutarimide-binding site coincides with the binding site of the 3’ end of deacylated tRNA at the E-site of the large ribosomal subunit. Deacylated Phe-tRNA was incubated with 80S ribosomes before DMS methylation and extraction. d. Both LTM and cycloheximide bind to the same site on the 60S ribosomal subunit in a dose-dependent manner. The KD values were estimated to be 500 nM for LTM and 15 µM for cycloheximide. e. LTM but not cycloheximide decrease binding of deacylated tRNA to the E-site. Ribosomes were incubated with [32P]-labeled deacylated Phe-tRNA in presence of LTM or cycloheximide at the indicated concentration. Excess cold tRNA was used as a positive control. Error bars denote standard deviation. Bars in c, d and e represent s.d..
The same footprint was also obtained with CHX. The protection was dose-dependent and allowed for estimation of a dissociation constant for each compound (Fig. 5d). Ribosome concentrations of 50 and 100 nM were repeatedly probed with increasing concentrations of each compound. LTM bound with a KD of about 500 nM, while CHX bound at 15 µM. Thus, LTM and CHX appear to share the same binding site on the ribosome but differ in their binding affinity. The common footprint uncovered here for LTM and CHX, along with the locations of resistant yeast mutants reported previously, defines a shared binding pocket for both inhibitors in the E-site of the 60S ribosome.
Protection of the same nucleotide by either tRNA or both compounds suggested that LTM and CHX might interfere with tRNA binding. Different doses of both CHX and LTM were incubated with ribosomes before addition of radioactively labeled deacylated tRNAPhe35. Buffer conditions were identical to the tRNA footprint and an excess amount of unlabeled tRNAPhe served as control. The association of tRNA was assessed by binding to nitrocellulose filters. While LTM decreased tRNA binding to the ribosome in a dose-dependent manner, only excessive amounts of CHX could interfere with tRNA-ribosome association (Fig. 5e). It had been observed that CHX interfered with deacylated tRNA release from the ribosome 2. This could mean that CHX binds together with a deacylated tRNA to block translation, while LTM occludes tRNA access to the E-site. The difference in association with deacylated tRNA thus provides a mechanistic explanation for the similar, yet distinct effects of LTM and CHX.
LTM inhibits breast cancer growth in vitro and in vivo
Although it was reported that LTM extended the survival of mice with P388 lymphoma5, it remains unclear whether it exhibits selective inhibition of tumor cells over non-transformed cells. We investigated its specificity for transformed cell lines and tested its effect on the proliferation of an array of breast cancer cell lines. LTM inhibited cell growth with IC50 concentrations in the low nanomolar range, but higher doses were necessary to inhibit growth of the non-tumorigenic breast cell line MCF10A (Supplementary Fig. 7a). This encouraging result prompted us to determine the effect of LTM on a solid tumor model in vivo. Two million MDA MB 231 cells were injected subcutaneously into female nude mice. Once tumors became palpable, mice received 0.6 mg/kg of LTM or solvent alone every day for one month. LTM had an appreciable effect on tumor growth in vivo, suggesting that LTM and other inhibitors of translation elongation may have potential as leads for developing anticancer agents (Supplementary Fig. 7b).
DISCUSSION
In this study, we identified a subset of the migrastatin family of glutarimide-containing natural products, including LTM and isomigrastatin, as potent inhibitors of eukaryotic translation elongation. Despite their structural similarity to the cell migration inhibitor migrastatin, LTM and isomigrastatin act by a completely different mechanism and their ability to inhibit cell migration very likely is only secondary to their effect on translation elongation. It is quite interesting to compare the structure and activity of migrastatin, isomigrastatin, dorrigocin and LTM. Although migrastatin, isomigrastatin and dorrigocin B share the same constituents and the glutarimide moiety, isomigrastatin features a 12-membered macrocycle, which can be readily converted to either the 14-membered migrastatin or the linear dorrigocins. Yet, the three natural products have completely distinct biological properties. While migrastatin inhibits cell migration, isomigrastatin inhibits translation and the dorrigocins possess neither activity. The shared inhibitory capacity on eukaryotic protein translation between isomigrastatin and LTM highlights the importance of the 12-membered macrolides for this activity. The lack of activity in the linear analogs including 8 and 11 is somewhat surprising. Streptimidone, which has a structure similar to that of CHX, essentially consisting of only the glutarimide and a linker without the macrolide present in LTM, also inhibits translation 2. However, extension of the linker region with a flexible chain in dorrigocin B (6) abolished any inhibitory activity.
The structural similarity between LTM and CHX and their common effect on eukaryotic translation elongation offered an opportunity to deconvolute their mechanisms of action. A systematic examination of their effects on different steps of translation elongation revealed that both inhibitors share a similar mechanism of action by blocking translation elongation through binding to the same position in the E site of the large ribosomal subunit. In addition, these experiments also revealed some subtle but definitive differences between LTM and CHX at their physiologically active concentrations. First, LTM is over ten-fold more potent than CHX for the inhibition of protein synthesis both in vitro and in vivo. Second, while LTM caused a significant depletion of polysomes in cell culture, CHX had little effect on the polysome profile. Third, LTM stalled the ribosome on the initiating AUG codon, but CHX appeared to allow one round of translocation such that the ribosome stalled on the second codon of the template mRNA. Both similarities and differences between the two structurally related inhibitors can now be reconciled based on the new observations made in this study.
The footprint at C3993 generated by both LTM and CHX, together with the cross-resistance to the yeast L28 (mammalian L27a) mutant towards both inhibitors, provides for the first time three key points defining a common binding pocket for both inhibitors in the ribosome. Taking the reports on L36a into account, this binding site lies between C3993 at the base of hairpin 88 of the 28S rRNA, the 38th amino acid of L27a and proline 54 of L36a. All three points of reference lie in vicinity of one another and in proximity to the two 3’-terminal nucleotides of the E-site tRNA (Supplementary Fig. 8). The location of the footprint corresponds to the same nucleotide that was protected by the 3’-end of the E-site tRNA in bacterial ribosomes34. Under the same conditions as used for the E. coli ribosome, we observed a footprint of Phe-tRNA on rabbit ribosomes as well, confirming the conserved function of the location.
It has been previously proposed that CHX likely acts via the E-site3. In this study, we offer direct experimental evidence corroborating the proposed model. The lower affinity of CHX compared to LTM alone, however, could not account for the differences in polysome profile and toeprinting pattern. The underlying cause of these differences likely stems from the larger size of LTM due to its unique 12-membered macrolide, which CHX lacks. When de-acylated tRNA was 3’ end-labeled with [32P], we observed a decrease in tRNA binding at increasing concentrations of LTM but not CHX. Only at a concentration of 10 mM did CHX cause a detectable decrease in the amount of bound tRNA. It is unclear, though, whether the effect of CHX at 10 mM is solely due to its binding to the E site or to some non-specific interactions with other sites of the ribosome or the de-acylated tRNA. It had been previously observed that CHX treatment of ribosomes decreases their ability to release deacylated tRNA2. With its 12-membered macrocycle, LTM is significantly larger in size and thus takes up considerably more space than the smaller CHX. Both molecules bind to the same pocket of the ribosome and given their structural similarity around the glutarimide moiety, it is tempting to speculate that the observed footprint is the binding site of the glutarimide portion of each inhibitor. In bacteria, interaction between the tRNA 3’-OH and the E-site is necessary to stimulate translocation36. This is consistent with our observation that both LTM and CHX inhibited eEF2-mediated translocation from the A to the P site and reduced the rate of tripeptide formation.
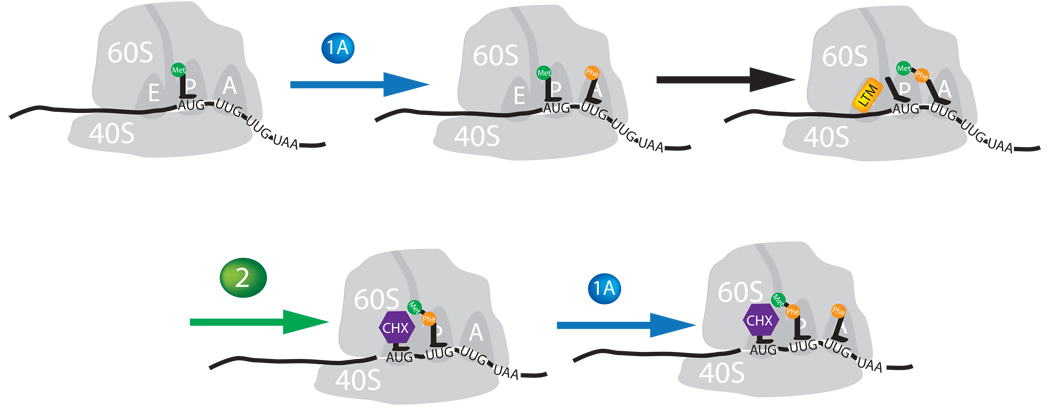
Taking all existing experimental results into consideration, we propose a mechanistic model for the action of both CHX and LTM (Fig. 6). In this model, both CHX and LTM share a largely similar mechanism through binding to the E site of the 60S ribosome. The binding of CHX and LTM to the E site blocks eEF2-mediated tRNA translocation. This model explains the similar effects of CHX and LTM on translational elongation and tripeptide formation (Fig. 4e). A difference between CHX and LTM lies in the ability of CHX to bind the E site together with the E-site tRNA while the larger LTM occludes deacylated tRNA from the E site. Thus, binding of CHX to the E site alone does not affect translocation while occupation of the E site by both CHX and deacylated tRNA stalls translocation, leading to an arrest of the ribosome on the second codon (Fig. 3e). This is in agreement with the observation by others that CHX allows for 2 rounds of translocation on a CrPV IRES template, because the cricket paralysis virus element initiates translation without initiator tRNA and begins translation from the A-site3. Consequently it takes two translocation events before deacylated tRNA reaches the E site. While LTM binds to the same site, its sheer size occludes deacylated tRNA. Thus LTM blocks the very first round of elongation and prevents the ribosome from leaving the start site. Unlike LTM, the smaller CHX presumably can also bind to actively translocating ribosomes, providing a plausible explanation for the distinct polysome profiles for LTM and CHX. In contrast to CHX, the mutually exclusive binding of LTM and deacylated tRNA to the E site leads to preferential binding of LTM to empty E site immediately after initiation, allowing dipeptide formation but blocking the translocation of the newly formed deacylated initiating tRNA from translocating from the P to E site, stalling the ribosome at the AUG start codon. Once elongation has been initiated and the E site is occupied by deacylated tRNA, it will be more difficult for LTM to gain access to the E site, leading to polysome depletion (Fig. 3c versus a). Unlike LTM, CHX can interrupt translation elongation at any time, as its binding to the E site is independent of the occupancy of E site by de-acylated tRNA. Thus, its polysome profile does not significantly differ from that of an untreated cell (Fig. 3b versus a). We note that this model does not seem to account for the effect of CHX on eEF-2 mediated translocation assay indirectly measured by phenylalanyl puromycin formation at first sight. However, there were plenty of deacylated tRNA present in the translocation assay, whose co-occupation of the E site may explain the inhibition seen by CHX.
Figure 6. Mechanistic models for inhibition of translation elongation by CHX and LTM.
Proposed Mechanisms of action of LTM and cycloheximide. LTM binds the ribosomal E-site and prevents translocation of the P-site tRNA into the E-site after eEF1A has delivered an aminoacyl-tRNA into the A-site and peptidyl transfer has occurred. Cycloheximide binds in the same location but stalls translocation by skewing the binding of deacylated tRNA to the E-site and hence allowing one complete round of translocation to proceed before inhibiting further elongation.
Increasing evidence points to a connection between protein synthesis and cancer cell growth. Didemnin B and homoharringtonine, two small molecule inhibitors of translation, have advanced to clinical trials37,38. Inhibitors of translation elongation in conjunction with an established chemotherapeutic agent such as doxorubicin have been shown to sensitize the tumor to therapy19. Furthermore, the development of drug resistance necessitates expression of anti-apoptotic proteins or drug transporters. Inhibition of translation should therefore greatly suppress the occurrence of resistance. Given LTM’s specificity for transformed cell lines and effect on in vivo tumor growths, our findings seem to corroborate a potential therapeutic use of translation elongation inhibitors in cancer treatment.
LTM furthermore extends the molecular toolbox for inhibiting a specific step in eukaryotic translation. Together with CHX, LTM makes it possible to dissect translation at the first and at the second translocation step. A comparison between LTM and CHX reveals how the CHX core structure is further elaborated through addition of a 12-member macrocycle to enhance its affinity for the E site of the ribosome and increase its potency against tumor cell lines. It remains to be determined which structural element of the E site of the ribosome, be it ribosomal RNA or protein, interacts with the macrocycle portion of LTM to confer higher potency. Deeper insights into the interaction between LTM and the E site of the ribosome may further our mechanistic understanding of translocation and guide the design of future small molecule inhibitors of eukaryotic translation. It is possible that chemical modifications of the macrolide portion of LTM and isomigrastatin will further enhance the potency and specificity of this family of natural products.
Methods
Isolation of LTM, isomigrastatin, migrastatin, dorrigocin and congeners
Production, isolation, and characterization of isomigrastatin, migrastatin, dorrigocins, LTM, and their analogs used in this studies are carried our as described previously4.
Phe-tRNA preparation
Phenylalanyl specific tRNA was charged with [14C]Phenylalanine using a yeast S-100 fraction. 10 µM tRNAPhe, 2 mM ATP, 15 µM [14C]Phenylalanine and 10% S-100 were incubated in 30 mM HEPES, pH 7.4, 15 mM MgCl2, 25 mM KCl and 4 mM DTT for 90 min. The product was extracted 3× with buffered phenol and 1× with chloroform before ethanol precipitation. Charging efficiency was around 20%.
eEF1A-dependent RNA binding
eEF1A-mediated tRNA binding was measured as previously described39. Briefly 89 pmol of ribosomes were incubated in 20 mM HEPES, pH 7.4, 100 mM KCl, 10 mM MgCl2 and 1 mM DTT in presence of 200 ng polyuridine RNA, 10 pmol of [14C]Phe-tRNAPhe, 2.2 µg eEF1A and 150 µM GDPNP in presence of 200 µM compound with the enzyme before tRNA was added. After a 10-min incubation at 37°C, the reactions were resuspended in 1 ml of buffer and immediately washed through a nitrocellulose filter (Millipore HA 45 µm) and rinsed with 3× 1ml of buffer. Filters were dried and scintillation counted. One picomole of charged tRNA emitted 1100 dpm.
eEF2-dependent translocation
Reactions were set up in the same manner as the eEF1A-dependent RNA binding assay, except for the use of GTP instead of GDPNP. After 5 min of incubation at 37°C, compound was added to a final concentration of 200 µM and samples were incubated for another 5min at room temperature before addition of 45 µl containing 4.5 µl of 10× buffer, 0.5 µg eEF2, 10 µl of 10 mg/ml puromycin and 6 µl 15 mM GTP. After incubation at 37°C for 10 min, 1.4 ml of cold ethyl acetate was added and samples were immediately vortexed. After centrifugation at top speed in a microcentrifuge at 4°C for 5min, 1 ml aliquots of the organic phase were collected, mixed with 4 ml of scintillation fluid and counted.
Peptidyl transfer
The peptidyl transfer assay was performed according to Lorsch and Herschlag29. Charged [35S]Met-tRNAiMet and mRNA were prepared as per Acker and Lorsch40. 25 µl reactions were set up at 26 °C in 32 mM HEPES, pH 7.4, 140 mM KOAc, 3.3 mM MgOAc2, 2.8 mM DTT and 4% glycerol containing 2 nmol [35S]Met tRNAiMet, 0.5 mM GTP, 1 µM mRNA and 60 nM ribosomes and as much HSW as the volume permitted. Addition of 400 nM puromycin initiated the reaction. 2 µl aliquots were removed at the indicated time intervals and quenched with 0.5 µl of 3 M NaOAc (pH 5.0). A 1 µl aliquot was spotted on Polygram IONEX-25 SA-Na cation exchange thin layer chromatography plates. The chromatography was carried out in 2 M NH4Cl with 10% acetonitrile. Plates were dried and exposed to a phosphoimager screen overnight.
Chemical footprints
RNA footprinting was performed using the procedures of Noller and Nygard with slight modifications41,42. An aliquot of 60 pmol of 80S ribosomes, which had been purified by centrifugation through two high-salt sucrose cushions, was diluted into 80 µl volumes containing 10 µl 10× Buffer A with 0.25M sucrose and 200 µM LTM in DMSO or solvent alone. After a 5-min preincubation at room temperature, 20 µl 450 mM or 100 mM dimethyl sulfate (DMS) in water were added to a final concentration of 20 and 90 mM, respectively, and reactions were incubated at 37°C for an additional 5 min. The reaction was quenched and RNA was extracted using the RNAqueous isolation kit (Ambion) according to the manufacturer’s instructions. Recovered rRNA was diluted to 0.4 mg/ml and stored at −80°C.
A sample of 2.5 µl rRNA was mixed with 1 µl 10 µM primer (for sequences see Supplementary Fig. 5) in 1 µl of 4.5x hybridization buffer (225 mM HEPES, pH 7.0, 450 mM KCl) and hybridized by heating to 90°C for 2 min and cooling at 1 degree/s to 47°C in a Peltier Thermo Cycler. To the hybrid, 2 µl of elongation mixture (87 mM Tris-HCl, pH 8.5, 6.67 mM MgCl2, 6.67 mM DTT, 6 µCi [α-32P]-TTP, 1.8 µM TTP, 33 µM dATP, dCTP and dGTP, as well as 2U of AMV reverse transcriptase) were added and incubated at 42°C for 55 min. The elongated product was precipitated in 120 µl precipitation buffer (83 mM NaOAc in 67% ethanol) at −20°C for 30min and then centrifuged in a microcentrifuge at 13,200 rpm at 4°C. The supernatant was removed and the precipitated pellets dried before 10 µl of gel loading buffer (90% formamide, 10% 10× TBE buffer, bromophenol blue and xylene cyanol) was added. The products were resolved on a polyacrylamide sequencing gel.
For KD determination, the same protocol was used except that the total volume was increased to 300 µl with an overall ribosome concentration of 50 or 100 nM. The tRNA binding conditions were adapted from Moazed and Noller34 . An aliquot of 60 pmol deacylated tRNA was incubated with the indicated inhibitor at the indicated concentrations with 20 pmol 80S ribosomes in 30 mM HEPES, pH 7.4, 100 mM KOAc, 20 mM MgCl2, 2 mM DTT and 0.25 M sucrose at room temperature for 5min before DMS was added to a final concentration of 90mM.
Deacylated tRNA binding
Deacylated tRNA was 3’ labeled with [32P] as described by Ledoux and Uhlenbeck35. Labeled tRNA was diluted to 80,000 cpm/pmol and 60 pmol were added to 20 pmol of ribosomes in the same buffer used for DMS methylation. Reaction mixtures were incubated with the inhibitor at the indicated concentrations or 4 µM cold deacylated tRNA. An aliquot of 1 ml of 1× buffer was added and the reactions were passed through a nitrocellulose filter disk and washed with 5× 2 ml of 1× buffer. Filters were dried and scintillation counted. Background radiation proved negligible.
For remaining experimental procedures, see Supplementary Methods.
Supplementary Material
Acknowledgements
We are indebted to Drs. Jef Boeke and Jonathan Warner for the CHX-resistant strains of S. cerevisiae and to Dr. Jerry Pelletier for providing us with the HCV and EMCV IRES reporter constructs, as well as Dr. Peter Sarnow for providing the CrPV vector. We would like to thank the laboratories of Drs. Jerry Hart, Paul Englund, Jon Lorsch, Saraswati Sukumar and Rajini Rao for use of specialized equipment and constructive advice. This work is supported in part by grants from NCI and FAMRI (J.O.L), CA106150 and CA113297 (B.S.).
Footnotes
Author Contributions
T.S-P. and J.O.L. designed the experiments; T.S-P., D.E.E., Y.D., and S.B. performed the experiments; J.J., W.C.M., R. G., and B.S. contributed reagents; T.S-P., D.E.E., Y.D., R. G., B. S. and J.O.L. analyzed data; and T.S-P. and J.O.L. wrote the manuscript.
Competing Financial Interests: None.
References
- 1.Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 2.Obrig TG, Culp WJ, McKeehan WL, Hardesty B. The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J Biol Chem. 1971;246:174–181. [PubMed] [Google Scholar]
- 3.Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ju J, Lim SK, Jiang H, Seo JW, Shen B. Iso-migrastatin congeners from Streptomyces platensis and generation of a glutarimide polyketide library featuring the dorrigocin, lactimidomycin, migrastatin, and NK30424 scaffolds. J Am Chem Soc. 2005;127:11930–11931. doi: 10.1021/ja053118u. [DOI] [PubMed] [Google Scholar]
- 5.Sugawara K, et al. Lactimidomycin, a new glutarimide group antibiotic. Production, isolation, structure and biological activity. J Antibiot (Tokyo) 1992;45:1433–1441. doi: 10.7164/antibiotics.45.1433. [DOI] [PubMed] [Google Scholar]
- 6.Gaul C, et al. The migrastatin family: discovery of potent cell migration inhibitors by chemical synthesis. J Am Chem Soc. 2004;126:11326–11337. doi: 10.1021/ja048779q. [DOI] [PubMed] [Google Scholar]
- 7.Shan D, et al. Synthetic analogues of migrastatin that inhibit mammary tumor metastasis in mice. Proc Natl Acad Sci U S A. 2005;102:3772–3776. doi: 10.1073/pnas.0500658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadam S, McAlpine JB. Dorrigocins: novel antifungal antibiotics that change the morphology of ras-transformed NIH/3T3 cells to that of normal cells. III. Biological properties and mechanism of action. J Antibiot (Tokyo) 1994;47:875–880. doi: 10.7164/antibiotics.47.875. [DOI] [PubMed] [Google Scholar]
- 9.Karwowski JP, et al. Dorrigocins: novel antifungal antibiotics that change the morphology of ras-transformed NIH/3T3 cells to that of normal cells. I. Taxonomy of the producing organism, fermentation and biological activity. J Antibiot (Tokyo) 1994;47:862–869. doi: 10.7164/antibiotics.47.862. [DOI] [PubMed] [Google Scholar]
- 10.Ju J, Lim SK, Jiang H, Shen B. Migrastatin and dorrigocins are shunt metabolites of iso-migrastatin. J Am Chem Soc. 2005;127:1622–1623. doi: 10.1021/ja043808i. [DOI] [PubMed] [Google Scholar]
- 11.Feng Z, et al. Engineered production of iso-migrastatin in heterologous Streptomyces hosts. Bioorg Med Chem. 2009;17:2147–2153. doi: 10.1016/j.bmc.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakae K, et al. Migrastatin, a new inhibitor of tumor cell migration from Streptomyces sp. MK929-43F1. Taxonomy, fermentation, isolation and biological activities. J Antibiot (Tokyo) 2000a;53:1130–1136. doi: 10.7164/antibiotics.53.1130. [DOI] [PubMed] [Google Scholar]
- 13.Nakae K, et al. Migrastatin, a novel 14-membered lactone from Streptomyces sp. MK929-43F1. J Antibiot (Tokyo) 2000;53:1228–1230. doi: 10.7164/antibiotics.53.1228. [DOI] [PubMed] [Google Scholar]
- 14.Fried HM, Warner JR. Molecular cloning and analysis of yeast gene for cycloheximide resistance and ribosomal protein L29. Nucleic Acids Res. 1982;10:3133–3148. doi: 10.1093/nar/10.10.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaufer NF, Fried HM, Schwindinger WF, Jasin M, Warner JR. Cycloheximide resistance in yeast: the gene and its protein. Nucleic Acids Res. 1983;11:3123–3135. doi: 10.1093/nar/11.10.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Planta RJ, Mager WH. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast. 1998;14:471–477. doi: 10.1002/(SICI)1097-0061(19980330)14:5<471::AID-YEA241>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 17.Stevens DR, Atteia A, Franzen LG, Purton S. Cycloheximide resistance conferred by novel mutations in ribosomal protein L41 of Chlamydomonas reinhardtii. Mol Gen Genet. 2001;264:790–795. doi: 10.1007/s004380000368. [DOI] [PubMed] [Google Scholar]
- 18.Kawai S, et al. Drastic alteration of cycloheximide sensitivity by substitution of one amino acid in the L41 ribosomal protein of yeasts. J Bacteriol. 1992;174:254–262. doi: 10.1128/jb.174.1.254-262.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert F, et al. Altering chemosensitivity by modulating translation elongation. PLoS ONE. 2009;4:e5428. doi: 10.1371/journal.pone.0005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tai PC, Wallace BJ, Davis BD. Selective action of erythromycin on initiating ribosomes. Biochemistry. 1974;13:4653–4659. doi: 10.1021/bi00719a029. [DOI] [PubMed] [Google Scholar]
- 21.Anthony DD, Merrick WC. Analysis of 40 S and 80 S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J Biol Chem. 1992;267:1554–1562. [PubMed] [Google Scholar]
- 22.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 23.Novac O, Guenier AS, Pelletier J. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 2004;32:902–915. doi: 10.1093/nar/gkh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Algire MA, Lorsch JR. Where to begin? The mechanism of translation initiation codon selection in eukaryotes. Curr Opin Chem Biol. 2006;10:480–486. doi: 10.1016/j.cbpa.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 26.Bordeleau ME, et al. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc Natl Acad Sci U S A. 2005;102:10460–10465. doi: 10.1073/pnas.0504249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodnina MV, et al. [Mechanism of tRNA translocation on the ribosome] Mol Biol (Mosk) 2001;35:655–665. [PubMed] [Google Scholar]
- 28.Merrick WC. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorsch JR, Herschlag D. Kinetic dissection of fundamental processes of eukaryotic translation initiation in vitro. EMBO J. 1999;18:6705–6717. doi: 10.1093/emboj/18.23.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmberg L, Melander Y, Nygard O. Probing the structure of mouse Ehrlich ascites cell 5.8S, 18S and 28S ribosomal RNA in situ. Nucleic Acids Res. 1994;22:1374–1382. doi: 10.1093/nar/22.8.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandramouli P, et al. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cannone JJ, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 35.Ledoux S, Uhlenbeck OC. [3'-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods. 2008;44:74–80. doi: 10.1016/j.ymeth.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lill R, Robertson JM, Wintermeyer W. Binding of the 3' terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO J. 1989;8:3933–3938. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 38.Rinehart KL. Antitumor compounds from tunicates. Med Res Rev. 2000;20:1–27. doi: 10.1002/(sici)1098-1128(200001)20:1<1::aid-med1>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 39.SirDeshpande BV, Toogood PL. Mechanism of protein synthesis inhibition by didemnin B in vitro. Biochemistry. 1995;34:9177–9184. doi: 10.1021/bi00028a030. [DOI] [PubMed] [Google Scholar]
- 40.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–145. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- 41.Smith CWJ. RNA-protein interactions : a practical approach, xxv. Oxford ; New York: Oxford University Press; 1998. p. 341. [Google Scholar]
- 42.Holmberg L, Melander Y, Nygard O. Probing the conformational changes in 5.8S, 18S and 28S rRNA upon association of derived subunits into complete 80S ribosomes. Nucleic Acids Res. 1994;22:2776–2783. doi: 10.1093/nar/22.14.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.