Abstract
This work was designed to clarify details in repair pathways acting to remove DNA double strand breaks (DSB) induced by temozolomide (TMZ). Cultured mouse embryonic fibroblasts were used which were deficient in their DNA DSB repair ability. Cell sensitivity to drug treatments was assessed using colony forming assays. The most effective molecular target which was correlated with TMZ cell sensitivity was DNA Ligase IV (Lig4). In addition, it was found that siRNA for Lig4 efficiently enhanced cell lethality induced by TMZ in human glioblastoma A172 cells. These findings suggest that down regulation of Lig4 might provide a useful tool for cell sensitization during TMZ chemotherapy.
Keywords: TMZ, DSB, NHEJ, HR, Lig4
1. Introduction
Alkylating drugs are the oldest class of anti-cancer drugs which are still commonly in use, and they remain important in the treatment of several types of cancers [1,2]. Temozolomide (TMZ) is a methylating agent which prolongs survival when administered during and after radiotherapy used as a first-line treatment for glioblastoma [3], and TMZ also has significant activity against recurrent glioma [4,5]. However, the therapeutic efficacy of TMZ is limited because of tumor drug resistance.
TMZ causes the methylation of guanine (G) on its N7and O6 atoms, and the methylation of adenine (A) on its N3 atom [6]. O6-methylG is repaired through the action of O6-methylG-DNA methyltransferase (MGMT) [7]. Early studies showed that MGMT deficient cells were unable to repair O6-methylG damage and were therefore more sensitive to the effects of methylating agents than cells with normal levels of MGMT [8,9]. This observation has been utilized experimentally and in many clinical trials, because MGMT can be inhibited by the O6-methylG analogue O6-benzylG and by other agents [10–12]. Elevated levels of MGMT and/or a nonfunctional mismatch repair pathway have been blamed for much of the observed resistance to TMZ [13], but it has also been suggested that the repair of O6-methylG by MGMT is not the sole mechanism responsible for resistance to alkylating agents [14–17].
DNA double-strand breaks (DSBs) occur in wild-type cells and in other cell culture systems in response to treatments with methylating agents [18,19]. Since DSBs are likely to be the final trigger leading to cell death, it would thus be expected that cells defective in DSB repair would be more sensitive to methylating agents. DSBs are repaired through the homologous recombination (HR) and non-homologous end joining (NHEJ) [20] pathways. In human cells, the proteins involved in HR include members of the MRN complex (meiotic recombination 11 (MRE11)/radiation sensitive mutant 50 (Rad50)/Nijmegen breakage syndrome 1 (NBS1)), Rad51, the Rad51 paralogs (Rad51B, Rad51C, Rad51D), X-ray repair cross-complementing group 2 (XRCC2) and group 3 (XRCC3), Rad54, and Rad54B [20]. The proteins involved in the NHEJ pathway include Ku70/80, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), Ligase IV (Lig4) and XRCC4 [20].
The work described here was designed to lead to a better understanding of details in the DSB repair pathways which contribute to TMZ sensitivity. The activity of components of HR repair (XRCC2 and Rad54) and NHEJ repair (Lig4) leading to TMZ-induced DNA damage were assessed using clonogenic survival assays. A panel of p53 tumor suppressor gene knockout mouse embryonic fibroblast cell lines (MEFs) was used which contained cells which were defective in specific components in the repair pathways (XRCC2, Rad54 and Lig4).
Next, to test whether the resulting observations were applicable to glioma cells, targeted repair pathways were down regulated using small interference RNAs (siRNA), and the sensitivity of human glioblastoma A172 cells to TMZ was measured. In order to determine if DSBs were formed in response to TMZ, the expression of γH2AX was monitored at different times following treatment with TMZ in cells deficient in specific repair pathway components, and in the corresponding parental cells. Hopefully, the DNA repair mechanism which was identified as contributing to TMZ resistance in this study will be able to provide tools which can be utilized to improve drug efficacy.
2. Materials and methods
2.1. Cell lines
The cell lines used in these studies were the MEF cell lines XRCC2−/− p53−/− (XRCC2−/−); XRCC2+/+p53−/− (XRCC2+/+); Rad54−/−Lig4+/+p53−/− (Rad54−/−); Rad54+/+Lig4−/− p53−/− (Lig4−/−); Rad54−/−Lig4−/− p53−/− (Rad54−/−Lig4−/−); and Rad54+/+Lig4+/+p53−/− (Rad54+/+Lig4+/+). Human glioblastoma A172 cells were purchased from the American Type Culture Collection of Cell Cultures (Manassas, VA). Cells were cultured in DMEM-10 (Dulbecco’s modified Eagle’s medium) containing 10% (v/v) fetal bovine serum, 20 mM 2-[4-(2-hydroxyethyl)-1-piperazinyl] ethanesulfonic acid, penicillin (50 units/ml), streptomycin (50 μg/ml), and kanamycin (50 μg/ml). Cells were cultured at 37°C in a conventional humidified CO2 incubator.
2.2. Drugs and drug treatments
TMZ (LKT Laboratories Inc. St. Paul, MN) was dissolved at a stock concentration of 100 mM in dimethylsulfoxide (DMSO). TMZ stock solutions were stored at −80°C until used. Cells were treated with medium containing TMZ at various concentrations for 3 h and then rinsed twice with PBS.
2.3. Colony forming assays
Cell survival was measured using a standard clonogenic survival assay. Three flasks were used for each point, and three independent experiments were repeated for each point. Colonies obtained after 5–10 days were fixed with methanol and stained with a 2% Giemsa solution. Microscopic colonies composed of more than approximately 50 cells were counted as having arisen from single surviving cells.
2.4. Flow cytometry
After treatment, cells were fixed with cold 70% methanol and kept at 4°C for up to 1 week before analysis. Cells were centrifuged and rinsed with Tris-PBS (TPBS). The cells were blocked with bovine serum for 15 min at room temperature and rinsed with TPBS. Cells were then incubated with anti-phospho-H2AX (Ser 139) monoclonal antibody (JBW301; Millipore, Billerica, MA) at a 300-fold dilution for 60 min at room temperature, rinsed with TPBS, incubated with an AlexaFluor 488-conjugated anti-mouse IgG second antibody (Invitrogen, Carlsbad, CA) at a 400-fold dilution for 60 min at room temperature, and then rinsed in TPBS. Before flow cytometric analysis, the samples were filtered through a 35 mm nylon mesh. Samples were analyzed using a flow cytometer (Becton Dickinson, San Jose, CA).
2.5. siRNA transfection
The siRNA sequences used for human Lig4 and its unspecific negative control were GCUAGAUGGUGAACGUAUG [21] and TATTCGCGCGTATAGCGGTTT [22], respectively. The siRNA duplexes were synthesized by Japan Bio Services Co., Ltd. (Saitama, Japan) and provided as a purified and annealed duplex. Transfections were carried out using Lipofectamine RNAiMAX in accordance with the manufacturer’s instructions (Invitrogen). Briefly, cells were seeded at 1–5×104 cells per 6 cm plate for 16–24 h without antibiotics. The siRNA was diluted in Opti-MEM I (Invitrogen) to produce a final siRNA concentration of 50 nM in a 1 ml final transfection volume. In a separate tube, 10 μl of Lipofectamine RNAiMAX was added to 490 μl of Opti-MEM I. The Lipofectamine RNAiMAX dilution was mixed with the diluted siRNA and incubated at room temperature for 15 min. The complex was then added drop-wise onto the cells. The cells were incubated for 48 h before further processing. These cells were then trypsinized for colony forming assays.
2.6. Statistical analysis
Data were compared statistically using the two-tailed Student’s t test.
3. Results
3.1. The role of repair genes in the presence of TMZ-induced DNA damage
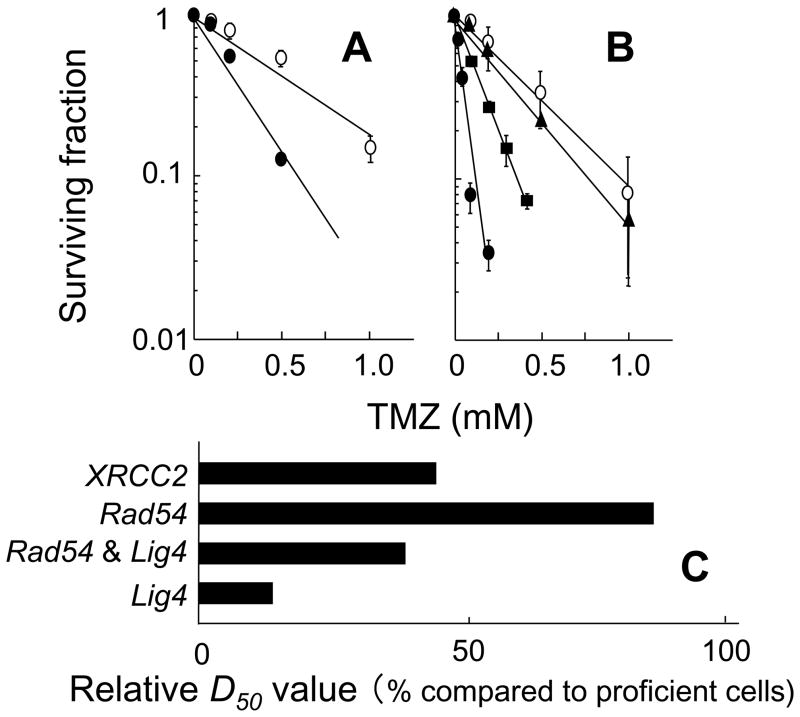
In this study, in order to understand the relative contributions of the HR and NHEJ repair pathways, cellular responses to TMZ were examined using clonogenic survival assays after a 3 h exposure to TMZ. In these studies, XRCC2 defective cells (Fig. 1A), Rad54 and/or Lig4 defective cells (Fig. 1B) were used. The sensitivity of each cell line was assessed from its D50 value, i.e. from the TMZ dose which reduced cell survival to 50% (Table 1). Each D50 value was calculated from the cell survival data shown in Figs. 1A and B. XRCC2−/− cells, Rad54−/−Lig4−/− cells and Lig4−/− cells were more sensitive to TMZ than the corresponding proficient cells (Figs. 1A and B). However, the sensitivity of Rad54−/− cells to TMZ was comparable to that of proficient cells (Fig. 1B). In addition, Rad54−/−Lig4−/− cells were less sensitive to TMZ than Lig4−/− cells (Fig. 1B).
Fig. 1.
Contributions of HR and NHEJ DSB repair to cellular survival following TMZ treatment. A,XRCC2+/+ cells (open circles) and XRCC2−/− cells (closed circles). B, Lig4+/+Rad54+/+ cells (open circles), Rad54−/− cells (closed triangles), Lig4−/−Rad54−/− cells (closed squares), Lig4−/− cells (closed circles). Each point represents the mean of at least three independent experiments; bars indicate the SD. C, relative D50 value (% compared to proficient cells).
Table 1.
The sensitivity of each cell line to TMZ was assessed by its D50 values: i.e. the dose that reduces cell survival to 50%.
| Genes | D50 (μM)* | |
|---|---|---|
| Proficient cells | Deficient cells | |
| XRCC2 | 406 | 178 |
| Rad54 | 284 | 234 |
| Rad54 & Lig4 | 284 | 107 |
| Lig4 | 284 | 38 |
Each D50 value was calculated from results of the cell survival data shown in Figs. 1A and B.
In order to accurately compare TMZ sensitivities in the repair defective cell lines, the relative D50 values were normalized using the D50 value of the corresponding proficient cell lines. The relative D50 values listed sequentially in the order in which they increase (reflecting decreasing sensitivities to TMZ) are: Lig4−/− cells < Rad54−/−Lig4−/− cells < XRCC2−/− cells < Rad54−/− cells (Fig. 1C). In summary, the relative D50 value of the Lig4 defective cells was the smallest after treatment with TMZ reflecting their high sensitivity to TMZ.
3.2. Lig4 activity in repair of DSBs induced by TMZ
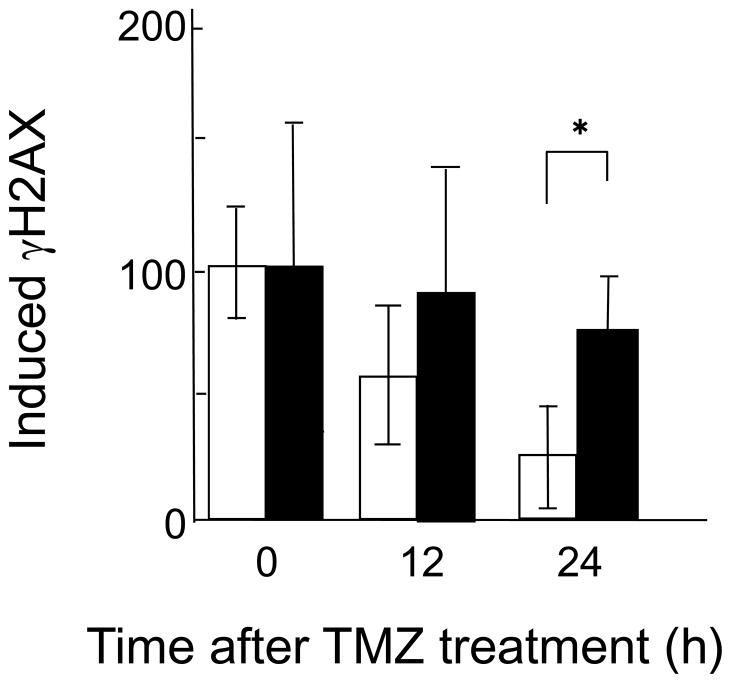
To determine whether DSBs are formed in response to TMZ, and how many DSBs are formed, the overall levels of phosphorylated H2AX (γH2AX) were measured with flow cytometry. Phosphorylated H2AX (γH2AX) is formed in response to the presence of DSBs [23–25] in Lig4−/− cells and in the corresponding proficient cells, and the levels of γH2AX are different at different times following treatment with TMZ. In proficient cells, γH2AX levels decreased to 25% of their initial levels at 24 h after TMZ treatment. However, in Lig4−/− cells, γH2AX levels were still present at up to 80% of their initial levels at 24 h after TMZ treatment. At 24 h after treatment with TMZ, there was a significant difference in γH2AX levels between these two cell lines (Fig. 2).
Fig. 2.
Phosphorylation of H2AX following treatment with medium containing 300 μM TMZ for 3 h in Lig4+/+ cells (open columns) or Lig4−/− cells (close columns) at the indicated time points. The relative inducible γH2AX levels at different time points were normalized against the γH2AX levels measured immediately after treatment. For each cell line, the value of the γH2AX levels measured immediately after treatment was set as 100. Columns show the means of at least three independent experiments; the bars indicate the SD. *, Difference is statistically significant (P < 0.05).
3.3. Effect of silencing Lig4 on cellular sensitivity to TMZ in A172 glioblastoma cells
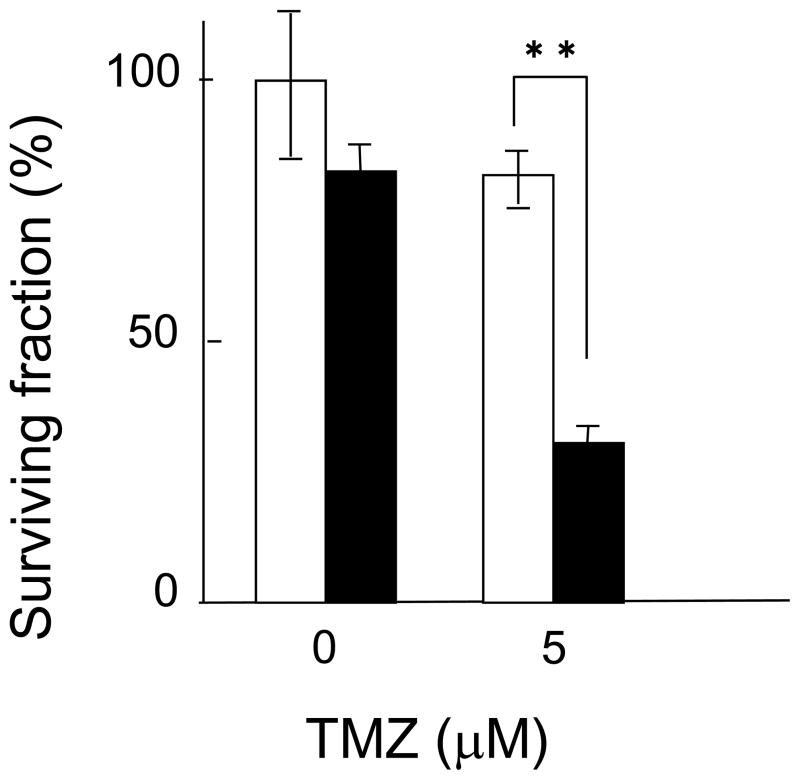
To test whether this result was pertinent to chemotherapy used against glioblastoma, Lig4 expression was silenced in A172 glioblastoma cells using siRNA, and clonogenic survival assays were then performed on the silenced cells. Lig4 silencing caused a 20% reduction in colony formation when compared to cells transfected with negative control siRNA. In addition, after TMZ treatment, Lig4 silencing caused a 62.5% reduction in colony formation when compared to cells transfected with negative control siRNA. In A172 glioblastoma cells Lig4 silencing increased cellular sensitivity to TMZ approximately three times (Fig. 3).
Fig. 3.
Effect of siRNA silencing of Lig4 in glioblastoma A172 cells. Closed columns, Lig4 siRNA; open columns, negative control RNA. Columns show the means of at least three independent experiments; the bars indicate the SD. **, Difference is statistically significant (P < 0.01).
4. Discussion
The data in this paper provide the first evidence that NHEJ and specifically, the NHEJ protein, Lig4, play a prominent role in the repair of TMZ-mediated DNA damage. In agreement with this, human glioblastoma cells harboring a mutated DNA-PKcs also showed hypersensitivity towards TMZ when compared to the corresponding wild-type cells. In this case, the relative D50 value was 19.5 as low as that observed in Lig4 defective cells (data not shown). Observations of the relative D50 values also support this idea. The parental cells of the Rad54−/− and Lig4−/− cells had the same genetic background, and the relative D50 values showed a significant difference: the relative D50 value for Rad54−/− was 85.7, and that for Lig4−/− was 13.5 (Fig. 1C). In addition, HR and NHEJ double knockout cells which were Rad54−/−Lig4−/− were less sensitive to TMZ than single NHEJ knockout Lig4−/− cells (Figs. 1B and C). These results clearly eliminate the importance of HR in the repair of TMZ-induced DNA damage. These results are in agreement with previous studies which have revealed a hypersensitivity in NHEJ mutants to other DSB-causing agents, such as ionizing radiation, and etoposide (VP-16) [26,27].
In contrast, Roos et al. [19] reported HR defective cells, but not NHEJ defective cells, are hypersensitive to TMZ. This discrepancy might be explained by the fact that different cell lines were used in their study. p53 knockout MEFs defective in HR and/or NHEJ were used here, while Roos et al [19] used mutant type (mt) Chinese hamster cell lines with defects in these functions. As mentioned before, the parental cell genetic background was the same for the HR (Rad54) and NHEJ (Lig4) proficient cells. In their study, the parental cells had different genetic backgrounds for the HR and NHEJ proficient cells. Thus, an accurate comparison between HR and NHEJ might be difficult. Furthermore, in their clonogenic survival assays, the maximum concentration of TMZ used was 20 μM for the NHEJ mt cell lines, while 100 μM was used for the HR mt cell lines. If higher concentrations of TMZ had also been tested in NHEJ mt cell lines, it is possible that they might have shown a higher TMZ sensitivity.
Although the HR and NHEJ repair pathways can both act to remove DSBs induced by TMZ, the number of DSBs present in a cell or the initial binding of repair factors to the break sites may affect which system is used. Several reports suggest that poly (ADP ribose) polymerase-1 (PARP-1) might have a role in regulating the balance between HR and NHEJ activity by decreasing the affinity of Ku for DSBs [28,29], and thus favoring access for HR factors. Other work suggests that DNA polymerase μ interacts with NHEJ components, and that this may favor this repair pathway [30].
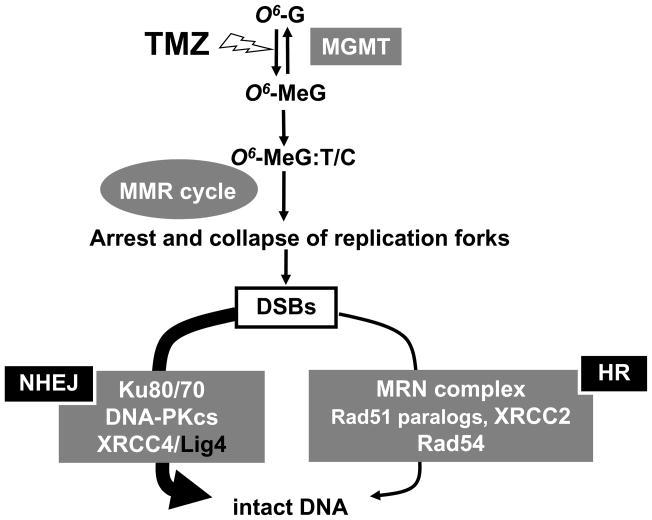
DNA repair pathways of TMZ-induced DNA damage are summarized in Fig. 4. TMZ–induced O6-methylG residues are repaired by MGMT [7]. Takagi et al. described the sensitivity of MGMT−/− cells to TMZ [31], and the relative D50 value of MGMT deficient cells was found to be 33.3 here (data not shown). In newly diagnosed glioblastomas, MGMT sometimes shows low levels of activity! partly! because p53 mutations suppress MGMT expression [32–34] or MGMT promoter hypermethylation results in gene silencing of MGMT [35]. Therefore, if MGMT is targeted with O6-benzylG, improvements in therapeutic efficacy are considered to be limited. !If O6-meG is not repaired, replication over unrepaired O6-meG:C will result in an O6-meG:T mismatch (or possibly an O6-meG:C ambiguous pair) (Fig. 4) [36]. In the next round of replication this would result in an A:T transition mutation, and again to an O6-meG:C pair, or in an O6-meG:T mismatch [36]. The O6-meG:T or C pair is recognized by the MutSα complex (hMSH2 and hMSH6) which initiates mismatch repair (MMR) and this can create a gapped duplex after incision of the newly replicated strand (Fig. 4) [36]. Since O6-meG remains in the template, this process may be repeated in a “futile repair loop” which eventually results in highly toxic DSBs that are intermediates in DSB repair pathways (Fig. 4) [36]. The XRCC2 protein plays a role in HR via its interaction with Rad51 [37]. Therefore, it may be argued that XRCC2 must be required for the Rad51 mediated strand invasion step to occur during HR. Without strand invasion of the homologous DNA, tolerance of the replication blocking lesion cannot occur. The Rad54 protein interacts with Rad51 directly during the HR process after the induction of DNA damage in mammalian cells [38]. In the NHEJ pathway, after DSB formation, the Ku70/80 heterodimer binds to the damaged DNA ends. This facilitates the recruitment of the DNA-PKcs to the DSB. The sequential binding of these proteins activates the phosphorylation function of the DNA-PKcs which then phosphorylates itself, the Ku heterodimer, and other proteins involved in cell cycle regulation [39]. It has been suggested that Ku70/80 might also function as an alignment factor which binds DSB ends, and can thus provide ready access for, and greatly stimulate the functioning of the Lig4-XRCC4 complex. This can increase the efficiency and accuracy of NHEJ [40]. The Lig4-XRCC4 complex then rejoins the juxtaposed DNA ends. In conclusion, NHEJ, and in particular Lig4, play an important role in the repair of TMZ-mediated DNA damage. The degree of HR and NHEJ contributions towards the repair of TMZ-induced DNA damage are indicated by the width of the arrows in Fig. 4, and it is suggested that Lig4 could provide a new molecular target for TMZ chemotherapy.
Fig. 4.
DNA repair pathways active in the repair of TMZ-induced DNA damage.
The data presented here suggests that Lig4 can generate cellular resistance to TMZ exposure by repairing lesions which trigger the activation of DNA damage response cascades. H2AX, a histone protein, is rapidly phosphorylated at Ser139 when DNA breaks are introduced in mammalian cell DNA in response to damage and replication fork collapse [23–25]. Many of the early components in the DNA damage response pathway co-localize with γH2AX at sites of DNA breaks [41–44]. Therefore, the detection and quantitation of γH2AX is a useful tool to monitor the induction of DNA damage response signaling pathways. DSBs induced by TMZ are repaired in 24 h in Lig4 proficient cells. However, in Lig4−/− cells DSBs remained because they were not repaired (Fig. 2). Considering that the relative D50 value in Lig4−/− cells was low (13.5) (Fig. 1C), this model appears to be reasonable.
In addition, it was found that the down regulation of Lig4 by siRNA increased the sensitivity of glioblastoma A172 cells to TMZ (Fig. 3). It was also confirmed that A172 cells exhibit very low levels of MGMT activity [45]. This suggests that Lig4 down regulation could potentially be a useful strategy for augmenting the therapeutic effects of TMZ in glioma treatments.
In TMZ chemotherapy, we focus on DSBs which might be induced by unrepaired O6-meG (Fig. 4), and it was demonstrated that the depression of DSB repair can enhance the sensitivity of glioma cells to TMZ (Figs. 2 and 3). In this study, it was found that Lig4 could provide a new molecular target for TMZ. In view of the work shown here, it is proposed that Lig4 contributes significantly towards the repair of TMZ-induced DSBs and that modulating Lig4 activity could enhance sensitivity to chemotherapeutic agents such as TMZ.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This work was also funded in part by a grant from the Central Research Institute of the Electric Power Industry in Japan, and by a grant for Exploratory Research for Space Utilization from the Japan Space Forum.
The authors thank Dr. G. Iliakis (University Duisburg-Essen Medical School, Essen, Germany) and Dr. Y. Takagi (Fukuoka Dental College, Fukuoka, Japan) for kindly providing cell lines used in this work.
References
- 1.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat Rev Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 2.Chaney SG, Sancar A. DNA repair: enzymatic mechanisms and relevance to drug response. J Natl Cancer Inst. 1996;88:1346–1360. doi: 10.1093/jnci/88.19.1346. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups, National Cancer Institute of Canada Clinical Trials Group, Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Yung WK, Albright RE, Olson J, Fredericks R, Fink K, Prados MD, Brada M, Spence A, Hohl RJ, Shapiro W, Glantz M, Greenberg H, Selker RG, Vick NA, Rampling R, Friedman H, Phillips P, Bruner J, Yue N, Osoba D, Zaknoen S, Levin VA. A phase II study of temozolomide vs. procarbazine in patients with glioblastoma multiforme at first relapse. Br J Cancer. 2000;83:588–593. doi: 10.1054/bjoc.2000.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yung WK, Prados MD, Yaya-Tur R, Rosenfeld SS, Brada M, Friedman HS, Albright R, Olson J, Chang SM, O’Neill AM, Friedman AH, Bruner J, Yue N, Dugan M, Zaknoen S, Levin VA. Multicenter phase II trial of temozolomide in patients with anaplastic astrocytoma or anaplastic oligoastrocytoma at first relapse. Temodal Brain Tumor Group. J Clin Oncol. 1999;17:2762–2771. doi: 10.1200/JCO.1999.17.9.2762. [DOI] [PubMed] [Google Scholar]
- 6.Tentori L, Graziani G. Pharmacological strategies to increase the antitumor activity of methylating agents. Curr Med Chem. 2002;9:1285–1301. doi: 10.2174/0929867023369916. [DOI] [PubMed] [Google Scholar]
- 7.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 8.Day RS, 3rd, Ziolkowski CH, Scudiero DA, Meyer SA, Lubiniecki AS, Girardi AJ, Galloway SM, Bynum GD. Defective repair of alkylated DNA by human tumour and SV40-transformed human cell strains. Nature. 1980;288:724–727. doi: 10.1038/288724a0. [DOI] [PubMed] [Google Scholar]
- 9.Yarosh DB, Foote RS, Mitra S, Day RS., 3rd Repair of O6-methylguanine in DNA by demethylation is lacking in Mer-human tumor cell strains. Carcinogenesis. 1983;4:199–205. doi: 10.1093/carcin/4.2.199. [DOI] [PubMed] [Google Scholar]
- 10.Friedman HS, Keir S, Pegg AE, Houghton PJ, Colvin OM, Moschel RC, Bigner DD, Dolan ME. O6-benzylguanine-mediated enhancement of chemotherapy. Mol Cancer Ther. 2002;1:943–948. [PubMed] [Google Scholar]
- 11.Barvaux VA, Ranson M, Brown R, McElhinney RS, McMurry TB, Margison GP. Dual repair modulation reverses Temozolomide resistance in vitro. Mol Cancer Ther. 2004;3:123–127. [PubMed] [Google Scholar]
- 12.Tserng KY, Ingalls ST, Boczko EM, Spiro TP, Li X, Majka S, Gerson SL, Willson JK, Hoppel CL. Pharmacokinetics of O6-benzylguanine (NSC637037) and its metabolite, 8-oxo-O6-benzylguanine. J Clin Pharmacol. 2003;43:881–893. doi: 10.1177/0091270003256060. [DOI] [PubMed] [Google Scholar]
- 13.Kokkinakis DM, Bocangel DB, Schold SC, Moschel RC, Pegg AE. Thresholds of O6-alkylguanine-DNA alkyltransferase which confer significant resistance of human glial tumor xenografts to treatment with 1,3-bis(2-chloroethyl)-1-nitrosourea or temozolomide. Clin Cancer Res. 2001;7:421–428. [PubMed] [Google Scholar]
- 14.Bobola MS, Tseng SH, Blank A, Berger MS, Silber JR. Role of O6- methylguanine-DNA methyltransferase in resistance of human brain tumor cell lines to the clinically relevant methylating agents temozolomide and streptozotocin. Clin Cancer Res. 1996;2:735–741. [PubMed] [Google Scholar]
- 15.Bobola MS, Blank A, Berger MS, Silber JR. Contribution of O6-methylguanine-DNA methyltransferase to monofunctional alkylating-agent resistance in human brain tumor-derived cell lines. Mol Carcinog. 1995;13:70–80. doi: 10.1002/mc.2940130203. [DOI] [PubMed] [Google Scholar]
- 16.Bobola MS, Berger MS, Silber JR. Contribution of O6-methylguanine-DNA methyltransferase to resistance to 1,3-(2-chloroethyl)-1-nitrosourea in human brain tumorderived cell lines. Mol Carcinog. 1995;13:81–88. doi: 10.1002/mc.2940130204. [DOI] [PubMed] [Google Scholar]
- 17.Bocangel DB, Finkelstein S, Schold SC, Bhakat KK, Mitra S, Kokkinakis DM. Multifaceted resistance of gliomas to temozolomide. Clin Cancer Res. 2002;8:2725–2734. [PubMed] [Google Scholar]
- 18.Ochs K, Kaina B. Apoptosis induced by DNA damage O6-methylguanine is Bcl-2 and caspase-9/3 regulated and Fas/caspase-8 independent. Cancer Res. 2000;60:5815–5824. [PubMed] [Google Scholar]
- 19.Roos WP, Nikolova T, Quiros S, Naumann SC, Kiedron O, Zdzienicka MZ, Kaina B. Brca2/Xrcc2 dependent HR, but not NHEJ, is required for protection against O6-methylguanine triggered apoptosis, DSBs and chromosomal aberrations by a process leading to SCEs. DNA Repair (Amst) 2009;8:72–86. doi: 10.1016/j.dnarep.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Windhofer F, Wu W, Iliakis G. Low levels of DNA ligases III and IV sufficient for effective NHEJ. J Cell Physiol. 2007;213:475–483. doi: 10.1002/jcp.21120. [DOI] [PubMed] [Google Scholar]
- 22.Ohnishi K, Nagata Y, Takahashi A, Taniguchi S, Ohnishi T. Effective enhancement of X-ray-induced apoptosis in human cancer cells with mutated p53 by siRNA targeting XIAP. Oncol Rep. 2008;20:57–61. [PubMed] [Google Scholar]
- 23.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi A, Ohnishi T. Does γH2AX foci formation depend on the presence of DNA double strand breaks? Cancer Lett. 2005;229:171–179. doi: 10.1016/j.canlet.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 25.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 26.Adachi N, Ishino T, Ishii Y, Takeda S, Koyama H. DNA ligase IV-deficient cells are more resistant to ionizing radiation in the absence of Ku70: Implications for DNA double-strand break repair. Proc Natl Acad Sci USA. 2001;98:12109–12113. doi: 10.1073/pnas.201271098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adachi N, Suzuki H, Iiizumi S, Koyama H. Hypersensitivity of nonhomologous DNA end-joining mutants to VP-16 and ICRF-193: implications for the repair of topoisomerase II-mediated DNA damage. J Biol Chem. 2003;278:35897–35902. doi: 10.1074/jbc.M306500200. [DOI] [PubMed] [Google Scholar]
- 28.Li B, Navarro S, Kasahara N, Comai L. Identification and biochemical characterization of a Werner’s syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J Biol Chem. 2004;279:13659–13667. doi: 10.1074/jbc.M311606200. [DOI] [PubMed] [Google Scholar]
- 29.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol Cell Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takagi Y, Hidaka M, Sanada M, Yoshida H, Sekiguchi M. Different initial steps of apoptosis induced by two types of antineoplastic drugs. Biochem Pharmacol. 2008;76:303–311. doi: 10.1016/j.bcp.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Blough MD, Zlatescu MC, Cairncross JG. O6-methylguanine-DNA methyltransferase regulation by p53 in astrocytic cells. Cancer Res. 2007;67:580–584. doi: 10.1158/0008-5472.CAN-06-2782. [DOI] [PubMed] [Google Scholar]
- 33.Russell SJ, Ye YW, Waber PG, Shuford M, Schold SC, Jr, Nisen PD. p53 mutations, O6-alkylguanine DNA alkyltransferase activity, and sensitivity to procarbazine in human brain tumors. Cancer. 1995;75:1339–1342. doi: 10.1002/1097-0142(19950315)75:6<1339::aid-cncr2820750616>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 34.Anker L, Ohgaki H, Ludeke BI, Herrmann HD, Kleihues P, Westphal M. p53 protein accumulation and gene mutations in human glioma cell lines. Int J Cancer. 1993;55:982–987. doi: 10.1002/ijc.2910550618. [DOI] [PubMed] [Google Scholar]
- 35.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 36.Margison GP, Santibañez-Koref MF. O6-Alkylguanine-DNA alkyltransferase: role in carcinogenesis and chemotherapy. Bioessays. 2002;24:255–266. doi: 10.1002/bies.10063. [DOI] [PubMed] [Google Scholar]
- 37.Shim KS, Schmutte C, Tombline G, Heinen CD, Fishel R. hXRCC2 enhances ADP/ATP processing and strand exchange by hRAD51. J Biol Chem. 2004;279:30385–30394. doi: 10.1074/jbc.M306066200. [DOI] [PubMed] [Google Scholar]
- 38.Tan TL, Essers J, Citterio E, Swagemakers SM, de Wit J, Benson FE, Hoeijmakers JH, Kanaar R. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- 39.Weterings E, van Gent DC. The mechanism of non-homologous end-joining: a synopsis of synapsis. DNA Repair (Amst) 2004;3:1425–1435. doi: 10.1016/j.dnarep.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Thode S, Schafer A, Pfeiffer P, Vielmetter W. A novel pathway of DNA end-to-end joining. Cell. 1990;60:921–928. doi: 10.1016/0092-8674(90)90340-k. [DOI] [PubMed] [Google Scholar]
- 41.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 42.Chen HT, Bhandoola A, Difilippantonio MJ, Zhu J, Brown MJ, Tai X, Rogakou EP, Brotz TM, Bonner WM, Ried T, Nussenzweig A. Response to RAG-mediated VDJ cleavage by NBS1 and γ-H2AX. Science. 2000;290:1962–1965. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rappold I, Iwabuchi K, Date T, Chen J. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J Cell Biol. 2001;153:613–620. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi A, Yamakawa N, Kirita T, Omori K, Ishioka N, Furusawa Y, Mori E, Ohnishi K, Ohnishi T. DNA damage recognition proteins localize along heavy ion induced tracks in the cell nucleus. J Radiat Res (Tokyo) 2008;49:645–652. doi: 10.1269/jrr.08007. [DOI] [PubMed] [Google Scholar]
- 45.Hermisson M, Klumpp A, Wick W, Wischhusen J, Nagel G, Roos W, Kaina B, Weller M. O6-methylguanine DNA methyltransferase and p53 status predict temozolomide sensitivity in human malignant glioma cells. J Neurochem. 2006;96:766–776. doi: 10.1111/j.1471-4159.2005.03583.x. [DOI] [PubMed] [Google Scholar]