Summary
Borrelia burgdorferi (Bb), the Lyme disease spirochete, encodes a potential ferric uptake regulator (Fur) homologue, BosR (BB0647). Thus far, a role for BosR in Bb metabolism, gene regulation, or pathogenesis has not been determined, largely due to the heretofore inability to inactivate bosR in low-passage, infectious Bb isolates. Herein, we report the generation of the first bosR-deficient mutant in a virulent strain of Bb. Whereas the bosR mutant persisted normally in ticks, the mutant was unable to infect mice, indicating that BosR is essential for Bb infection of a mammalian host. Moreover, transcriptional profiling of the bosR mutant showed that a number of genes were either positively or negatively influenced by BosR deficiency, suggesting that BosR may function both as a global repressor and activator in Bb. Strikingly, our study showed that BosR controls the expression of two major virulence-associated Bb lipoproteins, OspC and DbpA, likely via an influence on the alternative sigma factor, RpoS. This study thus not only has elucidated another key virulence gene of Bb, but also provides new insights into a previously unknown layer of gene regulation governing RpoS in Bb.
Introduction
Lyme disease is the most common arthropod-borne disease in the United States. Borrelia burgdorferi (Bb), the causative agent of Lyme disease, survives in nature through a complex life cycle including an arthropod vector (Ixodes tick) and a variety of mammalian hosts (Burgdorfer et al., 1982; Steere et al., 1983). In order to maintain its life cycle in nature, Bb must adapt to and transit between these two disparate environments by altering its gene expression profile in response to various environmental stimuli. Studies have shown that certain signals, including temperature, pH, cell density and other unknown host factors, modulate Bb gene expression (Akins et al., 1998; Anguita et al., 2003; Brooks et al., 2003; Burtnick et al., 2007; Caimano et al., 2005; Caimano et al., 2007; Hyde et al., 2007; Lybecker and Samuels, 2007; Ojaimi et al., 2003; Revel et al., 2002; Schwan, 2003; Schwan et al., 1995; Singh and Girschick, 2004; Stevenson et al., 1995; Tokarz et al., 2004; Yang et al., 2000). Moreover, Bb controls its major outer membrane lipoproteins, such as outer surface (lipo)protein C (OspC) and decorin-binding (lipo)protein A (DbpA), through a central regulatory pathway consisting of a putative response regulator Rrp2 and two alternative sigma factors RpoN and RpoS (Boardman et al., 2008; Caimano et al., 2004; Caimano et al., 2007; Hubner et al., 2001; Lybecker and Samuels, 2007; Ouyang et al., 2008; Smith et al., 2007; Yang et al., 2003a; Fisher et al., 2005).
In addition to the Rrp2-RpoN-RpoS pathway, the Bb genome also encodes a putative transcriptional regulator, BB0647 (Boylan et al., 2003; Fraser et al., 1997; Katona et al., 2004). This protein has been predicted to belong to the ferric uptake regulator (Fur) family. However, its role in Bb biology has remained obscure. In a wide variety of microorganisms, Fur functions principally as a global repressor to control gene expression in response to iron availability (Carpenter et al., 2009; Lee and Helmann, 2007). When intracellular iron supply is abundant, Fur forms a complex with its co-factor (Fe2+), and the complex binds to a Fur box (with a consensus sequence of GATAATGATAATCATTATC) (Escolar et al., 1999; Lee and Helmann, 2007) located in the promoter regions of Fur-regulated genes, thereby blocking gene transcription. In contrast, when the iron supply is limited, Fur dissociates from Fe2+ and the Fur box, leading to de-repression. Fur also can serve as an activator to positively regulate genes, probably via an indirect effect in which Fur represses RhyB, a small regulatory RNA that blocks gene expression by binding to and degrading target mRNAs (Lee and Helmann, 2007). In addition, Fur can regulate gene expression in its apo form without binding to co-factors. Not surprisingly, by sensing the intracellular iron levels, Fur modulates the expression of genes involved in iron acquisition. Fur also controls genes unrelated to iron transport. For example, in E. coli, the Fur-Fe2+ complex represses the expression of many ‘non-iron’ genes, such as cyoA, flbB, fumC, gpmA, metH, nohB, nrdH, purR and sodA, with functions in respiration, flagellar chemotaxis, the TCA cycle, glycolysis, methionine biosynthesis, phage-DNA packaging, DNA synthesis, purine metabolism, and redox stress resistance (Lee and Helmann, 2007). Moreover, some Fur homologues, such as the regulators that sense zinc (Zur), manganese (Mur), nickel (Nur) and peroxide stress (PerR), also regulate the expression of genes involved in the uptake of zinc, manganese, or nickel, as well as the peroxide stress response (Jacquamet et al., 2009; Lee and Helmann, 2007).
Given the notion that Bb may not accumulate or rely on iron (Posey and Gherardini, 2000), it seems unlikely that BB0647 regulates iron homeostasis in this pathogen. However, BB0647 may regulate Bb genes involved in non-iron functions, such as the acquisition of metal ions (e.g., zinc and manganese), or even oxidative stress responses. In a previous report, by employing a lacZ reporter vector (napAP/O-lacZ, generated by fusing the bb0690 [napA/dps] promoter to a promoterless lacZ gene), and an isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible BB0647 expression construct (pJAB3), Boylan et al. (Boylan et al., 2003) reported that BB0647 (expressed from pJAB3) activated transcription of napAP/O-lacZ in E. coli and proposed that BB0647 positively regulated napA/dps expression in Bb. napA/dps is a homologue of dps (DNA-binding protein from starved cells) implicated in being important for Bb to protect itself against oxidative stress (Boylan et al., 2003; Li et al., 2007). Thus, BB0647 was presumed to regulate genes involved in the oxidative stress response in Bb and, consequently, BB0647 was renamed as BosR (Borrelia oxidative stress regulator) (Boylan et al., 2003). However, this regulation effect has not yet been widely corroborated, mainly due to the inability to obtain a bosR-deficient mutant in low-passage (LP), infectious strains of Bb. To date, the only reported bosR mutant was created in the high-passage (HP), non-infectious Bb isolate CHP100 (Seshu et al., 2004). Although interesting data were acquired by using this latter mutant, the data obtained may be equivocal for several reasons. First, the HP, noninfectious Bb isolate CHP100 was deficient in a few plasmids (Seshu et al., 2004). It also contained unidentified mutations (Seshu et al., 2004). Third, the strain differs dramatically from LP, infectious Bb in resistance to oxidizing agents, in which CHP100 is nearly 105-fold more sensitive to t-butyl peroxide (Seshu et al., 2004). Fourth, the bosR allele (bosRR39K) in the HP strain possessed a critical point mutation relative to the wild-type (WT) bosR, leading to a substantial alteration in the biochemical activity of BosR (Seshu et al., 2004). All of these observations suggest that the functions of HP BosRR39K and WT BosR are distinctly different. As such, studies on BosRR39K might not be applicable to WT BosR. To more directly explore the role of BosR in Bb, we created a bosR-deficient mutant in virulent Bb strain B31MI. This allowed us to assess the role(s) of BosR in the tick/mammalian life cycle of Bb, including its effects on Bb's ability to colonize ticks, infect mice, and cause disease. The mutant also was strategic for revealing other Bb genes controlled by BosR, thereby establishing it as another global regulator of virulence expression in Bb.
Results
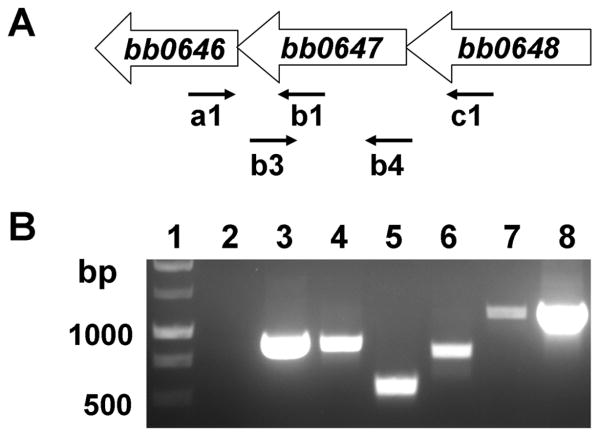
bosR is co-transcribed with bb0646 and bb0648
In the Bb B31 genome, bb0646, bosR, and bb0648 are proposed to be oriented in the same transcriptional direction (Fraser et al., 1997); bosR is separated from bb0648 and bb0646 by 100 bp and 3 bp, respectively (Fig. 1A). bosR encodes a Fur homologue, whereas bb0646 is predicted to encode a hypothetical protein belonging to the α/β hydrolase family (Fraser et al., 1997). In addition, bb0648 encodes a putative serine/threonine kinase (Fraser et al., 1997). To determine whether bb0646, bosR, and bb0648 constitute a single transcript, RNA was isolated from low-passage B31, reverse-transcribed to cDNA, and analyzed by PCR amplification using specific primers (Table S1). As shown in Fig. 1B, a fragment was amplified using the primer pair spanning the junction of either bb0646/bosR (lane 4) or bosR/bb0648 (lane 6). Further, an amplicon spanning bb0646, bosR, and bb0648 was amplified using primers complementary to bb0646 or bb0648 (lane 7). Positive controls were conducted by using genomic DNA (Fig. 1B, lanes 3 and 8) or cDNA (lane 5) as templates, and PCR amplification using RNA only as the template (lane 2) was conducted as a negative control. These data support that these three genes likely form an operon and are co-transcribed as a single transcriptional unit.
Fig. 1.
bb0646, bb0647 (bosR), and bb0648 are co-transcribed. (A) Schematic representation of the putative bb0646-bb0648 operon in Bb. (B) Results from RT-PCR. Lane 1, molecular weight markers; lane 2, primer pair a1 and b1 in control PCR using RNA as template (no reverse transcriptase [RT]); lane 3, primer pair a1 and b1 in ordinary PCR using genomic DNA as template; lane 4, primer pair a1 and b1; lane 5, primer pair b3 and b4; lane 6, primer pair b3 and c1; lane 7, primer pair a1 and c1; lane 8, primer pair a1 and c1 using genomic DNA as template. Arrows indicate the approximate positions of the oligonucleotide primers used for subsequent PCR analyses.
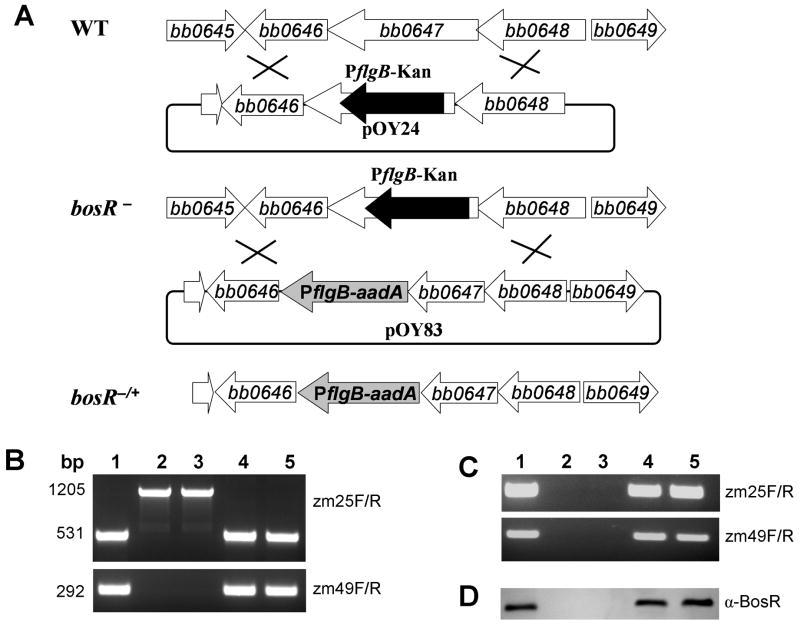
Inactivation and complementation of bosR in Bb
A bosR mutant was constructed by introducing the suicide plasmid pOY24 into strain B31. Through allelic exchange, a 469-bp internal fragment of bosR was replaced with the 1143-bp PflgB-kan cassette (Fig. 2A). The insertion and the orientation of the PflgB-kan cassette within the disrupted bosR were confirmed by sequence analysis. The PflgB-kan cassette was inserted in bosR in the same direction as bosR transcription, in order to allow bb0646 expression from PflgB (Fig. 2A). Following transformation, two kanamycin-resistant transformants, OY10/D12 and OY10/H3, were isolated. To complement the bosR mutant, the suicide vector pOY83 was created by linking the bb0649-bosR fragment to the PflgB-aadA cassette (Fig. 2A). In pOY83, the PflgB-aadA cassette was located between bosR and bb0646 in the same direction as bosR-bb0646 transcription, which also allows bb0646 expression from PflgB (and thus the same level of expression of bb0646 in the bosR mutants and the complemented strains). Upon electroporating pOY83 into OY10/D12 or OY10/H3, two corresponding complemented clones, OY34/E6 and OY34/C4, were created. The inactivation and complementation of bosR in these strains were confirmed using PCR amplification (Fig. 2B). Using primers ZM25F and ZM25R, a 531-bp fragment was amplified in both the WT B31 and the complemented strains, whereas a 1205-bp fragment was amplified in the bosR mutants. This is consistent with the replacement of a 469-bp internal fragment of bosR with the 1143-bp PflgB-kan cassette.
Fig. 2.
Construction of bb0647 (bosR) disruption mutants (bosR-) and complemented strains (bosR-/+). (A) Schematic representation of the bb0645-bb0649 genes in the Bb chromosome, insertion of the PflgB-kan gene cassette by homologous recombination, and the relevant complemented strain. (B) PCR analysis of WT B31, bosR mutants, and the complemented strains. The bosR-specific primer pairs used in PCR are indicated on the right. Lane 1, WT B31; lane 2, bosR- OY10/D12; lane 3, mutant OY10/H3; lane 4, bosR-/+ OY34/E6; lane 5, bosR-/+ OY34/C4. RT-PCR (C) and immunoblot analyses (D) were employed to determine the expression of BosR. α-BosR: rat polyclonal antibody against BosR. Lanes and primer pair designations are as in (B).
To detect the expression of BosR in WT B31, the bosR mutants and the complemented strains, RT-PCR employing primers specific for bosR was performed to detect bosR transcripts. As expected, bosR transcripts were detected in both B31 and the complemented strains, but not in the mutants (Fig. 2C). This observation was further confirmed by detecting BosR using immunoblot employing a specific polyclonal antibody against BosR, which demonstrates that the mutant does not express the BosR protein (Fig. 2D). In addition, we also examined the expression of bb0646 and bb0648 in the bosR mutants and complemented strains using RT-PCR. Results showed that transcripts of both genes were readily detected in all Bb isolates (Fig. S1). As intended from the cloning and recombination strategy employed, the expression of bb0646 in the bosR mutants was desired and promoted via the insertion of PflgB just upstream of bb0646 (Fig. 2A).
To confirm that all essential plasmids, such as lp25, lp28-1, lp54, lp36 and cp26 (Jewett et al., 2007; Labandeira-Rey and Skare, 2001; Purser and Norris, 2000; Stewart et al., 2005), were retained in the bosR mutant and complemented strains, PCR-based plasmid profiling (Purser and Norris, 2000) was performed. As shown in Fig. S2, the bosR mutant OY10/H3 lost only cp9 and cp32-9, but retained all essential plasmids required for Bb virulence. In addition, the bosR mutant clone OY10/D12 and the complemented strains (OY34/E6 and OY34/C4) also contained the same plasmid profile as that of OY10/H3 (data not shown). No obvious differences were observed among WT B31, the bosR mutants, and the complemented strains when spirochete morphology and motility were examined using dark-field microscopy. The effect of the inactivation of bosR on Bb in vitro growth was assessed by determining the growth curves of Bb strains grown in Barbour-Stoenner-Kelly (BSK)-II medium. As shown in Fig. S3, no discernable effect on growth was observed when bosR was inactivated. The bosR mutants displayed similar growth to WT B31 and the complemented strains.
bosR is required by Bb to infect mice via needle inoculation
To investigate the functional importance of bosR to Bb biology, C3H/HeN mice were challenged intradermally via needle inoculation with WT B31, bosR mutants, or complemented strains. After four weeks, mice were sacrificed and assessed for Bb infection by culturing mouse skin, heart and joint specimens in BSK-II medium. Cultures were monitored continually for 4 weeks for spirochete growth. Whereas motile spirochetes were recovered from all cultures from mice inoculated with 104 spirochetes per mouse of WT B31 or the complemented strains (OY34/E6 and OY34/C4), no bacterial growth was observed in cultures from mice infected with 107 spirochetes per mouse of the bosR mutants (OY10/D12 and OY10/H3) (Table 1). These data suggest that bosR is essential for Bb to establish infection in mice.
Table 1.
Infectivity of B. burgdorferi B31 clones in mice.a
| Strain, clone | Description | Dose | No. of cultures positive / total No. of specimens examined | No. of mice infected / total No. of mice | |||
|---|---|---|---|---|---|---|---|
| Heart | Joint | Skin | All sites | ||||
| B31 | wild type B. burgdorferi | 104 | 6/6 | 6/6 | 6/6 | 18/18 | 6/6 |
| OY10/D12 | B31, bosR−b | 107 | 0/4 | 0/4 | 0/4 | 0/12 | 0/4 |
| OY10/H3 | B31, bosR− | 107 | 0/7 | 0/7 | 0/7 | 0/21 | 0/7 |
| OY34/E6 | OY10/D12 with pOY83, bosR−/+b | 104 | 3/3 | 3/3 | 3/3 | 9/9 | 3/3 |
| OY34/C4 | OY10/H3 with pOY83, bosR−/+ | 104 | 3/3 | 3/3 | 3/3 | 9/9 | 3/3 |
Data were collected from two independent experiments.
bosR−: bosR mutant; bosR−/+, bosR complemented strain.
bosR is not essential for Bb persistence in ticks
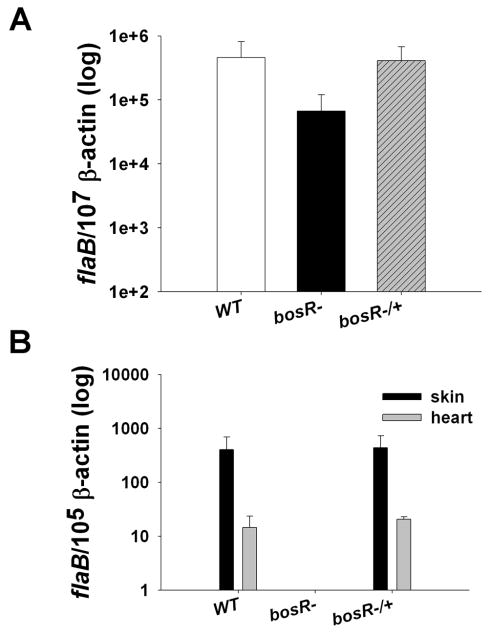
The inability to infect mice with a bosR mutant via needle inoculation precluded us from assessing whether the mutant could be naturally acquired by feeding ticks. Thus, as a surrogate system to examine the mutant's ability to colonize ticks, sustain itself in tick midguts, and be transmitted to naïve mice, Ixodes scapularis nymphs were first loaded with the bosR mutant via a widely used microinjection technique (Pal et al., 2004). We initially examined the survival of spirochetes in injected unfed ticks using an immunofluorescence assay (IFA), which revealed that spirochetes were readily detected in ticks 4 days after injection with all three isolates (WT B31, the bosR mutant OY10/H3, or the complemented strain OY34/C4) (Fig. S4). We then investigated the survival of Bb isolates during tick engorgement. For each isolate, 36 microinjected nymphs were allowed to feed on 3 naïve mice. Three ticks from each group were forcibly removed after 24h and 72h of feeding, whereas the remaining nymphs were allowed to feed to repletion (collected at 24h post-feeding). To determine the spirochete burden in ticks, nymphs were then analyzed by detecting Bb flaB transcripts using quantitative RT-PCR (qRT-PCR) and data were normalized using tick β-actin. As shown in Fig. 3A, regardless of the Bb isolates used, comparable levels of spirochetes were detected in ticks (collected after 72h of feeding) injected with WT B31, the bosR mutant OY10/H3, or the complemented strain OY34/C4. Similar results were obtained for ticks collected at 24h feeding or 24h post-feeding, showing that the spirochete burden in ticks injected with the bosR mutant was not significantly below the spirochete burden in ticks injected with either WT B31 or the complemented strain (data not shown). These data demonstrated that the bosR mutant was able to colonize and survive in ticks and that bosR likely is dispensable for Bb to infect and survive in ticks.
Fig. 3.
Survival of the bosR mutant in ticks. I. scapularis nymphs microinjected with various Bb strains were allowed to feed on naive mice. Ticks were collected at various times; infected mice were sacrificed at 7d post-feeding and multiple mouse tissues also were collected. Spirochete burdens in ticks after 72h of feeding (A) or mouse samples collected at 7d post-feeding (B) were examined using qRT-PCR. The experiments were replicated twice, and bars represent the mean measurements ± SEM from six representative qPCR measurements.
The bosR mutant is defective in establishing tick-borne murine infection
We then assessed whether the bosR mutant was able to be transmitted from ticks to naïve murine hosts. To achieve this, nymphs were microinjected with either WT B31, the bosR mutant OY10/H3, or the complemented strain OY34/C4 and the nymphs were allowed to feed on naïve mice. One week after tick feeding, mice were sacrificed and infection was determined by culture and qRT-PCR. As shown in Table 2, no spirochetes were cultivated from multiple samples of mice infested with nymphs harboring the bosR mutant. In contrast, spirochetes were recovered from tissues from mice infested with ticks containing WT B31 or the complemented strain. This was further confirmed by qRT-PCR analyses, which demonstrated that bosR mutants remained virtually undetectable in mice (Fig. 3B). The combined data indicate that a bosR mutant is unable to establish infection in mice when transmitted by ticks.
Table 2.
Recovery of B. burgdorferi from mice infected by microinjected ticksa
| Strain, clone | No. of cultures positive / total No. of specimens examined | No. of mice infected / total No. of mice | |||
|---|---|---|---|---|---|
| Heart | spleen | Skin | All sites | ||
| wild type B. burgdorferi B31 | 3/5 | 3/6 | 4/4 | 10/15 | 5/6b |
| OY10/D12, bosR−c | 0/6 | 0/6 | 0/6 | 0/18 | 0/6 |
| OY34/E6, bosR−/+c | 5/5 | 4/6 | 5/6 | 15/17 | 5/6b |
Data were acquired from two independent experiments. For each strain, totally 6 mice were infected and 18 samples were collected
Culture data from several samples were not determined due to contamination
bosR−: bosR mutant; bosR−/+, bosR complemented strain
Identification of BosR-regulated genes in Bb using microarray analysis
Although the above data that bosR plays an important role in Bb virulence were compelling, the precise function of this protein remains unknown. Inasmuch as it was plausible that bosR encodes some type of regulatory protein, we sought to assess the potential effect of bosR deficiency on the spirochete transcriptome. A 70-mer B31-based oligonucleotide array (Ouyang et al., 2008; Terekhova et al., 2006) was employed to compare the gene expression profiles in the bosR mutant relative to WT B31. Our data showed that 199 genes were differentially regulated from 2.09- to 116.7 fold by BosR (relative to the mutant deficient in bosR, 137 and 62 genes were up- or down-regulated, respectively, in WT B31) (Table S2 and Table S3). Of these genes, 67 (34%), 23 (12%) and 19 (10%) genes were located on the chromosome, lp54, or lp28-2, respectively (Fig. 4); 119 (60%) genes had unknown function, categorized as either hypothetical (77 genes) or hypothetically conserved (42 genes), and 80 (40%) genes had some predicted functions.
Fig. 4.
Genomic distribution of genes (see Table S2 and S3) differentially regulated by BosR. Black bars indicate the numbers of genes that are activated by BosR, whereas grey bars indicate the number of genes down-regulated.
To validate the results of the microarray analysis, qRT-PCR was utilized to examine expression of 19 genes differentially regulated by BosR. As shown in Fig. S5, a strong and significant linear correlation (r=0.94) was found when the log-transformed ratios determined by qRT-PCR and microarray were compared. When examining the absolute levels of gene expression for each given gene, similar trends were observed between these experimental approaches (Table 3). These data indicate that the differences observed in mRNA expression levels obtained by qRT-PCR correlated well with those obtained from microarray-based transcriptome analysis.
Table 3.
Validation of microarray results using quantitative RT-PCR
| Gene | Function | WT/bosR mutant | |
|---|---|---|---|
| microarray | qRT-PCR | ||
| bbb19 | outer surface protein C (ospC) | 87.76 | 38.96 |
| bbg27 | conserved hypothetical protein | 35.84 | 42.86 |
| bba24 | decorin binding protein A (dbpA) | 14.54 | 42.86 |
| bbo39 | ErpL protein (erpL) | 7.04 | 9.38 |
| bba48 | hypothetical protein | 5.14 | 1.25 |
| bbe31 | antigen, P35, putative | 4.37 | 4.05 |
| bbj26 | ABC transporter, ATP-binding protein | 4.32 | 4.11 |
| bb0565 | purine-binding chemotaxis protein (cheW-2) | 3.46 | 4.29 |
| bbf01 | ErpD protein, putative | 3.02 | 8.82 |
| bbr41 | conserved hypothetical protein | 2.87 | 2.24 |
| bb0071 | RNA polymerase sigma factor (rpoS) | 2.81 | 3.66 |
| bb0690 | neutrophil activating protein (napA) | a-2.11 | -1.57 |
| bb0763 | response regulatory protein (rrp-2) | -2.16 | -1.32 |
| bb0104 | periplasmic serine protease DO (htrA) | -2.18 | -1.49 |
| bb0153 | superoxide dismutase (sodA) | -2.25 | -1.37 |
| bba62 | lipoprotein | -2.33 | -2.07 |
| bb0032 | hypothetical protein | -2.53 | -2.04 |
| bb0401 | glutamate transporter, putative | -2.72 | -1.15 |
| bb0646 | hydrolase, alpha/beta fold family | -3.03 | -2.29 |
indicates the fold change of genes down-regulated in WT B31 relative to the bosR mutant.
Notably, our transcriptome comparison revealed that genes activated by the alternative sigma factor RpoS in Bb, such as ospC, dbpB, dbpA and bb0844, and even rpoS itself, were up-regulated in WT B31 relative to the bosR mutant (Table S2). When comparing the bosR microarray data with our previous transcriptional profiling data in one rpoS mutant (Ouyang et al., 2008), we found that, of those 137 genes up-regulated in WT B31, 87 genes including ospC, dbpB, dbpA and etc, were also affected by RpoS, whereas 50 genes were only regulated by BosR, but not by RpoS. Moreover, 9 Erp outer surface lipoproteins encoding genes (Fraser et al., 1997), including bbf01, bbl39 (erpA), bbo39 (erpL), bbo40 (erpM), bbq47 (erpX), bbr40 (erpH), bbr41 (ospE), bbr42 (erpY or ospF), and bbs41 (erpG or ospG), were found to be induced by BosR. These Erp proteins have been proposed to bind to the complement inhibitory factor H proteins in a variety of mammalian hosts and are important for Bb to establish infection (Alitalo et al., 2001; Brissette et al., 2008; Brissette et al., 2009; Bykowski et al., 2007; Coleman et al., 2008; Hartmann et al., 2006; Hefty et al., 2001; McDowell et al., 2003). Additionally, of the 62 genes down-regulated in WT B31 relative to the bosR mutant, only 5 genes, including bb0076 (ftsY), bb0646, bba62, bba74 (oms28) and bbu05, were affected by both BosR and RpoS, whereas a majority (57/62) of genes were affected only by BosR, not by RpoS. Of these 57 genes repressed only by BosR, superoxide dismutase A (bb0153, sodA) and bb0690 (napA/dps) are important for Bb to protect itself against oxidative stress (Esteve-Gassent et al., 2009; Li et al., 2007). Additional genes of interest in this category include: (i) bb0763, which encodes the response regulator Rrp2; (ii) bb0184 encoding a putative carbon storage regulator; (iii) many genes involved in physiological responses: bb0240 (glpF), bb0401 (encodes a putative glutamate transporter), bb0812 (dfp, encodes a pantothenate metabolism flavoprotein), bb0730 (pgi, encodes a glucose-6-phosphate isomerase), bb0152 (nagB, encodes a glucosamine-6-phosphate isomerase), and oligopeptide ABC transporter encoding genes bb0333 (oppC-1) and bb0335 (oppF).
BosR controls the expression of OspC and DbpA in Bb
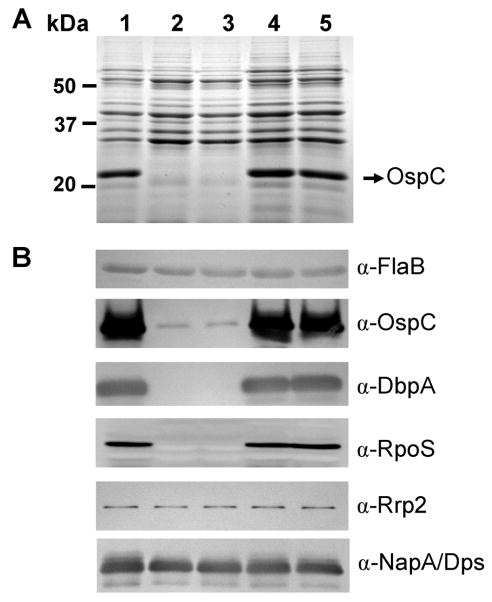
The above microarray analysis suggested that ospC, dbpA and rpoS were regulated by BosR. To further substantiate the role of BosR in the regulation of Bb genes, WT B31, the bosR mutants and the complemented strain were cultured at 37°C in BSK-H media at pH6.8 or pH7.6. pH6.8 was chosen for one of the Bb culture conditions because the RpoS regulon is highly expressed under this condition (Ouyang et al., 2008; Yang et al., 2000). Cells were harvested at late-log phase and whole-cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. As shown in Fig. 5A, when Bb was grown at pH6.8, the expression of ospC essentially was abolished in the bosR mutants. This result was further confirmed using immunoblot analysis, showing that the expression of ospC was dramatically diminished in bosR mutants (Fig. 5B). Moreover, immunoblot data also revealed that the expression of another major outer membrane lipoprotein in Bb, DbpA, was abolished when bosR was inactivated (Fig. 5B). To verify the reduction of OspC and DbpA expression in the bosR mutant was due solely to the inactivation of bosR, we investigated the expression of both OspC and DbpA in the complemented strains (OY34/E6 and OY34/C4). As shown in Fig. 5A and 5B, the expression of both OspC and DbpA were fully restored when the bosR mutation was complemented. Similar data were also obtained when Bb was grown in BSK-H at pH7.6 (data not shown). These data indicate that BosR plays a prominent role in controlling the expression of major outer membrane lipoproteins, such as OspC and DbpA.
Fig. 5.
BosR controls the expression of OspC and DbpA in Bb. Bb was inoculated into BSK-H medium (pH6.8) at 1000 spirochetes/ml and grown at 37°C. Spirochetes were collected at late-log phase. A volume of whole-cell lysates equivalent to 4 × 107 cells was loaded per gel lane and Bb gene expression was assessed by SDS-PAGE (A) and immunoblot (B) analyses. Approximate molecular masses are indicated at the left in kDa. The arrow in (A) indicates the position of OspC in SDS-PAGE. Specific antibodies, indicated as α-, used in the immunoblot (B) are indicated on the right. Lane 1, WT B31; lane 2, bosR- OY10/D12; lane 3, mutant OY10/H3; lane 4, bosR-/+ OY34/E6; lane 5, bosR-/+ OY34/C4.
Previous studies suggested that both OspC and DbpA were controlled by the central Rrp2-RpoN-RpoS pathway (Boardman et al., 2008; Caimano et al., 2004; Caimano et al., 2007; Fisher et al., 2005; Hubner et al., 2001; Lybecker and Samuels, 2007; Ouyang et al., 2008; Smith et al., 2007; Yang et al., 2003a). More specifically, RpoS directly controls OspC through an RpoS-dependent promoter (Eggers et al., 2004; Yang et al., 2005), although it is still unclear how RpoS controls DbpA. In this study, we examined the expression of RpoS in the bosR mutants and the complemented strains. Immunobloting revealed that the expression of RpoS was abolished in the bosR mutants but restored in the complemented strains (Fig. 5B). These data were further verified by RT-PCR analysis (Fig. S6). In addition, we also examined the expression of Rrp2 and RpoN in Bb variants. Consistent with the notions that both Rrp2 and RpoN are constitutively expressed in Bb, expression of Rrp2 (Fig. 5B, Fig. S6) and RpoN (Fig. S6) were not significantly altered when bosR was inactivated.
Discussion
Although BosR has been the focus of a number of previous studies (Boylan et al., 2003; Katona et al., 2004; Seshu et al., 2004), prior attempts to disrupt bosR within a low-passage, infectious strain of Bb were unsuccessful (Seshu et al., 2004). In this study, we successfully created a Bb mutant lacking BosR in low-passage, virulent Bb strain B31. Inasmuch as bosR constitutes an operon with bb0648 and bb0646, three critical strategies were employed to ensure that the phenotypes of the bosR mutant observed in this study were due solely to the inactivation of bosR (and not indirect effects). First, in the bosR mutants, the PflgB-Kan cassette was inserted into bosR in the direction of bosR-bb0646 transcription, thereby promoting the transcription of bb0646 from PflgB. Second, in the complemented strains, the PflgB-aadA cassette was inserted between bosR and bb0646 and oriented in the same direction as bb0646 transcription, which allowed bb0646 expression (from PflgB, and at the same level in both the bosR mutant and the complemented strain). Finally, the bosR mutation was complemented in cis using a construct harboring the native promoter of bosR, thereby facilitating a similar WT-level of expression of BosR in the complemented strain.
Bb growth in vitro was not affected by bosR inactivation. Similarly, bosR was dispensable for Bb persistence in ticks. However, our data revealed that inactivation of bosR rendered Bb completely noninfectious for mice, irrespective of needle inoculation or tick challenge, whereas genetic complementation fully restored the WT phenotype. This demonstrated that BosR is essential for Bb infectivity of a mammalian host. In this regard, an association between fur mutation and decreased bacterial virulence has been observed for many other pathogens (Carpenter et al., 2009), such as Campylobacter jejuni, Listeria monocytogenes, Helicobacter pylori, Staphylococcus aureus, Actinobacillus pleuropneumoniae, Bacillus cereus, and Vibrio cholerae, although the precise underlying mechanism(s) remain unclear. Given that iron is essential to the physiology and pathogenesis of these bacteria, aberrations in iron homeostasis are strongly implicated. Alternatively, a decrease in the virulence of fur mutants might also result from the variation in expression of specific virulence determinants. Because Bb does not appear to accumulate iron when cultivated in vitro, and no iron uptake systems have been predicted or identified in Bb (Posey and Gherardini, 2000), it seems less likely that the loss of infectivity for mice by the bosR mutant is due to a generalized effect in Bb on iron homeostasis. It is thus more plausible that other unknown virulence factors controlled by BosR contribute to Bb's mammalian infectivity phenotype.
Microarray analysis was employed to identify Bb genes potentially regulated, either directly or indirectly, by BosR. Our data revealed that BosR influences the expression of two essential Bb outer membrane lipoproteins, OspC and DbpA, which was substantiated by SDS-PAGE and immunoblotting. It is well established that OspC and DbpA are important for Bb virulence (Blevins et al., 2008; Grimm et al., 2004; Pal et al., 2004; Shi et al., 2008; Stewart et al., 2006; Tilly et al., 2006; Weening et al., 2008), and they both are governed by the central Rrp2-RpoN-RpoS pathway (Boardman et al., 2008; Burtnick et al., 2007; Caimano et al., 2004; Caimano et al., 2007; Fisher et al., 2005; Hubner et al., 2001; Lybecker and Samuels, 2007; Ouyang et al., 2008; Smith et al., 2007; Yang et al., 2003a). In this pathway, under stimulation by various environmental factors including pH, temperature, cell density, and other unknown mammalian factors, the putative response regulator Rrp2 is likely activated via phosphorylation (via an unidentified histidine kinase). Activated Rrp2 then activates RpoN which, in turn, promotes the expression of the alternative sigma factor RpoS. RpoS then allows the expression of a number of virulence-associated proteins, including OspC and DbpA. Additionally, in response to changes in temperature, a small RNA molecule, DsrABb, also activates RpoS (Lybecker and Samuels, 2007). In our current study, the expression of OspC and DbpA was markedly decreased or abolished in the bosR mutant, suggesting that expression of both OspC and DbpA are activated by BosR. Thus far, it remains unknown how BosR controls the expression of OspC and DbpA. Although it remains possible that BosR controls ospC and dbpA directly, BosR more likely is somehow involved in regulating the expression of RpoS which, in turn, influences ospC and dbpA, inasmuch as the expression of RpoS was also abolished in the bosR mutant. BosR may influence rpoS directly by binding to a sequence proximal to the rpoS minimal promoter (Burtnick et al., 2007; Smith et al., 2007), or alternatively influence the phosphorylation of Rrp2 by modulating the unknown cognate histidine kinase required for Rrp2 activation. In addition, BosR may also be involved in the pathway(s) of environmental signals that induce the Rrp2-RpoN-RpoS pathway. All of these possibilities warrant further investigation. Regardless, this is the first study to reveal another important layer of gene regulation, which somehow involves BosR, exerted on the central Rrp2-RpoN-RpoS pathway. Interference with the putative regulatory activity of BosR may lead to new strategies to interrupt the spirochete's life cycle.
Previous studies have indicated that the RpoS regulon is induced at higher temperature (35°C or 37°C), but is repressed at lower temperature (23°C) (Burtnick et al., 2007; Caimano et al., 2004; Caimano et al., 2007; Yang et al., 2000). Nonetheless, BosR is expressed efficiently at both 23°C or 35°C (Katona et al., 2004), suggesting that BosR may also participate in gene regulation by means other than inducing the RpoS regulon. In agreement with this possibility, transcriptome profiling of the bosR mutant revealed that, in addition to the genes regulated by RpoS, other genes were found to be induced only by BosR. Moreover, an additional group of 62 genes was found to be down-regulated in WT B31 (relative to the bosR mutant), suggesting that these genes are repressed by BosR. Among these genes, a majority were repressed only by BosR, but not by RpoS. However, considering that the fold-changes in the expression of these genes were modest (<4-fold), it remains possible that BosR may not regulate these 62 genes; the modest changes in the expression levels of these genes could be due to subtle, but generalized, metabolic effects. On the other hand, the in vitro culture conditions for Bb used in this study may not accurately mimic the in vivo conditions that Bb encounters in its life cycle. Under the culture conditions employed in this study (37°C), or in mammalian hosts, BosR may function in Bb principally as an activator to induce the RpoS regulon. However, under other conditions, such as when Bb is cultivated at 23°C or when Bb encounters a metal deficiency or oxidative stress, BosR may adopt another molecular configuration or cooperative structure that exerts different regulatory effects on those 62 genes. Future studies on the structural aspects of BosR may be strategic for addressing these possibilities.
The 62 genes repressed by BosR included sodA and napA/dps. In Bb, sodA ostensibly encodes a superoxide dismutase A; napA/dps has been annotated as a neutrophil activating protein A (Fraser et al., 1997) that has homology to the Dps proteins. In many bacteria, Dps proteins protect DNA during starvation and oxidative stress. Studies have showed that NapA/Dps is important for spirochete persistence in ticks during the inter-molt period (Li et al., 2007), and SodA is required by Bb to contend against oxidative stress (Esteve-Gassent et al., 2009). Moreover, others have reported that purified recombinant BosR binds to the promoters of sodA and napA/dps (Boylan et al., 2003; Katona et al., 2004; Seshu et al., 2004). These independent observations thus provide credence for the validity of our microarray data. Boylan et al. (Boylan et al., 2003) reported that LacZ expression from the napA/dps promoter-lacZ transcriptional fusion vector was activated by hyper-expression of BosR in E. coli, suggesting that BosR activates napA/dps. However, our data showed that the expression of napA/dps was up-regulated in the bosR mutant (Table 3, Table S3, and Fig. 5B), suggesting that napA/dps is actually repressed, rather than induced, by BosR, under the experimental conditions examined. The explanation for this apparent discrepancy remains unclear. One possibility is that BosR may adopt different configurations in E. coli vs. Bb. In other Fur homologues, the proteins typically contain two metal binding sites in the C-terminal domain, consisting of a Zn2+ structural site and a regulatory site usually occupied by Fe2+ (Lee and Helmann, 2007). In E. coli, BosR might prefer iron as a co-factor and regulate genes directly or indirectly. In Bb, BosR may utilize other metal ions, such as zinc or manganese, as cofactors. Of note, Bb appears not to accumulate iron (Boylan et al., 2003; Posey and Gherardini, 2000), but does accumulate manganese and zinc, probably through the manganese transporter BmtA (Ouyang et al., 2009) and one or more unidentified zinc transporters, respectively. Or, BosR may influence Bb gene expression in its apo form. Given these possibilities, E. coli may not be an ideal heterologous system for studying Bb gene expression and regulation, especially for those genes affected by BosR.
Our study clearly demonstrated that BosR influences Bb virulence. In addition to the key lipoproteins OspC and DbpA, a large number of genes with unknown functions and genes involved in a variety of putative functions were also positively or negatively regulated by BosR. It is plausible that some of these BosR-regulated genes also are important for Bb virulence. Thus, BosR likely plays multifaceted roles in Bb pathogenesis by coordinating the synthesis of Bb virulence factors with the appropriate in vivo signals, and in Bb global gene regulation.
Our study prompts a number of new and important questions. First, given the fact that bb0646, bosR, and bb0648 form an operon, what roles do the other two members of the bosR operon play in Bb pathogenesis and gene regulation? Inactivation of each gene of the operon may shed additional light on this issue. Second, it is unclear how BosR exerts its regulatory effect in Bb. Does it employ manganese or zinc as a co-factor, as previously assumed (Boylan et al., 2003; Posey and Gherardini, 2000), or does it regulate gene expression in its apo form? Finally, it remains unknown how bosR is expressed in Bb. bosR was reported to be expressed in Bb grown at either 23°C or 35°C, suggesting that expression of BosR is not influenced by temperature (Katona et al., 2004). However, the expression of BosR was induced when Bb was grown under anaerobic conditions, inferring that the dissolved CO2 level in the medium influences bosR expression (Hyde et al., 2007). How may dissolved CO2 influence BosR expression? Does Bb express bosR in response to other environmental stimuli, such as intracellular metal levels, pH, or cell density? Continued efforts are warranted to address these salient questions.
Experimental procedures
Strains and culture conditions
Infectious Bb strain B31MI (Fraser et al., 1997) was used as the WT strain (referred as B31) throughout this study. Isogenic Bb strains, including bosR mutants and bosR-complemented strains, were generated as described below. Bb was routinely cultured at 37°C and 5% CO2 in either BSK-II medium or BSK-H medium (Sigma) (Pollack et al., 1993) supplemented with 6% rabbit serum (Pel-Freeze). When appropriate, supplements were added to media at following concentrations: kanamycin, 160 μg/ml; streptomycin, 150 μg/ml. E. coli was grown at 37°C in Luria-Bertani (LB) broth or on LB agar plates.
Construction of bosR mutants and complemented strains
bosR was inactivated through homologous recombination using a suicide vector (pOY24). All constructs were confirmed using PCR amplification, restriction digestion, and sequence analysis. The 1261-bp 5′ arm for creating pOY24 was PCR-amplified using primers ZM22.2F and ZM22R, whereas the 1579-bp 3′ arm was amplified using ZM23F and ZM23R (Table S1). The 5′ DNA fragment was cloned into pGEM-Teasy vector (Promega), yielding pOY18. After digestion with BssHII, the 3′ arm was ligated into pOY18 that was digested with AscI, creating pOY22. The PflgB-Kan cassette, excised from pJD55 (Frank et al., 2003; Stewart et al., 2001; Blevins et al., 2008) using AscI, was then ligated into pOY22 at the AscI site. In the resulting construct, pOY24, the PflgB-Kan cassette was inserted into bosR in the same direction as transcription of bb0648-0646 (Fraser et al., 1997), which allows the expression of bb0646 from the PflgB. Bb transformation was performed as previously described (Yang et al., 2005). Transformants were selected using kanamycin and confirmed by PCR amplification.
To complement the bosR mutation, a suicide plasmid (pOY83) was constructed. Briefly, the 3772-bp DNA (bb0649-bosR) containing bosR and 3241-bp upstream of bosR, was PCR-amplified using primers ZM82 and ZM83, digested with XmaI, and ligated into pJD54 (Frank et al., 2003; Stewart et al., 2001), which yielded pOY82. Next, the bb0649-bosR-PflgB-aadA cassette was excised from pOY82 using AscI, and ligated into pOY22, creating pOY83. The resulting construct, pOY83, was electroporated into Bb OY10, which created OY34. Transformants were selected using 150 μg/ml of streptomycin, and all transformants and constructs were confirmed by PCR. Plasmid profiling for all Bb strains were performed as previously described (Purser and Norris, 2000).
Recombinant BosR expression and generation of rat polyclonal anti-BosR antibody
Recombinant BosR was produced in E. coli using the bacterial expression vector pPROEX-HTB (Invitrogen). Briefly, bosR was amplified using primers ZM26F and ZM26R. The resultant PCR fragment contained a BamHI site at the 5′-end and an EcoRI site at the 3′-end. After digestion with BamHI and EcoRI, the PCR product was ligated into the corresponding polylinker sites of the vector pPROEX-HTB, creating pOY21. The resulting construct, pOY21, was then transformed into E. coli strain BL21-DE3. After induction with 1mM IPTG (Sigma), recombinant BosR was purified using a Ni-NTA spin column according to the manufacturer's instruction (Qiagen). Rat polyclonal antibody against the purified BosR, Ab-BosR, was generated as previously described (Yang et al., 2003b).
SDS-PAGE and immunoblot analysis
SDS-PAGE and immunoblot analysis were carried out as previously described (Yang et al., 2003b). Briefly, a volume of whole cell lysate equivalent to 4 × 107 bacteria was loaded per lane on a 12.5% acrylamide gel. Resolved proteins were either stained with Coomassie brilliant blue or transferred to nitrocellulose membrane for immunoblot analysis. BosR was detected using the anti-BosR rat polyclonal antibody, Ab-BosR. Rrp2, RpoS, OspC, DbpA and NapA/Dps were detected using anti-Rrp2 monoclonal antibody 5B8-100-A1, anti-RpoS monoclonal antibody 6A7-101, anti-OspC monoclonal antibody 1B2-105A, anti-DbpA monoclonal antibody 6B3, or a polyclonal antibody against NapA/Dps, respectively. To confirm equal loading of bacteria in each lane, immunoblotting for the flagellar core protein (FlaB) was performed using a chicken IgY anti-FlaB antibody. Immunoblots were developed colorimetrically using 4-chloro-1-napthol as the substrate or by chemiluminescence using ECL Plus Western Blotting Detection system (Amersham Biosciences).
Microarray and qRT-PCR analyses
To compare gene expression profiles between Bb WT strain B31 and its isogenic bosR mutant OY10/H3, microarray analysis was performed essentially as described (Ouyang et al., 2008). Briefly, B31 and OY10/H3 were grown in triplicate in BSK-H medium at 37°C and 5% CO2. Spirochetes were harvested when the bacterial growth reached a density of 5×107 cells per ml. Total RNA was isolated using Trizol (Invitrogen) according to the instructions. After genomic DNA was digested using RNase-free DNase I (GenHunter Technology), RNA was further purified using RNeasy Mini Kit (Qiagen). cDNA was synthesized from 10 μg of total RNA using Amino Allyl cDNA Labeling Kit (Ambion) and then applied to the pre-hybridized 70-mer Bb array slides. Hybridization and slide scanning on an Axon 4000B microarray scanner (using GenePix Pro 6.1; Molecular Devices) were performed as previously described (Ouyang et al., 2008). The data were analyzed using the professional microarray data analysis program Acuity 4.0 according to the manufacturer's instructions (Molecular Devices). Briefly, raw data were first normalized using a ratio-based normalization method to equalize the means and medians of the features to 1. Additionally, the features that were designated by the software as “bad”, “absent”, or “not found” were also excluded from further analysis. Statistical analyses were performed using a one-sample t-test in the Acuity program. Differentially expressed genes were identified by both fold change (≥ 2-fold) and statistical significance (P<0.05). Fully processed and the raw microarray data from the microarray experiments were deposited in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number of GSE17422. qRT-PCR was employed to validate selected data from microarray experiments, using the relative quantification method (ΔΔCT) as described (Ouyang et al., 2008).
Bb infection of mice via needle inoculation
The infectivity of the bosR mutants and complemented strains was assessed using the murine needle-challenge model of Lyme borreliosis (Akins et al., 1998; Barthold et al., 1993). All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at UT Southwestern Medical Center. Prior to infection, bacteria were enumerated by dark-field microscopy. C3H/HeN mice (Charles River Laboratories) were infected via intradermal injection with various concentrations of Bb. At 4 weeks post inoculation, mice were sacrificed and skin, heart, and joint tissues were collected and cultured in BSK-II medium supplemented with 1× Borrelia antibiotic mixture (BAM, Sigma). The outgrowth of spirochetes in these cultures was assessed using dark-field microscopy.
Microinjection of Bb into nymphal ticks and transmission studies
I. scapularis nymphs were reared and maintained at the University of Maryland, College Park. A microinjection procedure was used to introduce spirochetes into the gut of I. scapularis nymphs as previously described (Pal et al., 2004). Briefly, B31, OY10/H3 and OY34/C4 grown in BSK-II medium containing appropriate antibiotics were harvested by centrifugation and injected into nymphs (∼4.2 × 104 spirochetes/tick). For each Bb strain, approximately 50 ticks were injected. After injection, 36 ticks were placed onto naïve C3H/HeN mice (3 animals/group, 12 ticks/mouse). After 24h or 72 h of feeding, 3 ticks were forcibly collected at each time-point, and at 24h post-feeding, 6 additional ticks were collected. The spirochete burden in ticks was analyzed by qRT-PCR as described previously (Coleman et al., 2008), and statistical analysis was performed by the Student's t-test. At day 4 after injection, the remaining unfed ticks were dissected and spirochetes were detected in the gut using IFA confocal microscopy. To examine whether injected ticks were able to transmit Bb, tick-infested mice were sacrificed at 7d post-feeding and murine tissue samples including skin, heart, and spleen were collected. Transmission of Bb from microinjected ticks to mice were determined by recovering spirochetes from murine tissues cultured in BSK-II medium and by assessing Bb flaB transcripts in infected murine skin and heart samples using qRT-PCR (Coleman et al., 2008). Two independent experiments were performed.
Immunofluorescence assays
Spirochetes were detected in ticks using confocal IFAs as detailed (Pal et al., 2001). Briefly, nymphal guts were isolated from ticks, air-dried, and fixed in acetone for 10 min. Slides were blocked at room temperature for 30min with blocking solution (PBS/0.05% Tween 20 with 5% goat serum) in a humidified chamber, and then incubated for 1 h with BacTrace flourescein isothiocyanate (FITC)-conjugated goat anti-Bb antibody (KPL Inc.). After two washes in PBS/0.05% Tween 20, slides were counterstained with 20 μg/ml of propidium iodide in PBS for 5 min, washed twice with PBS/0.05% Tween 20, and then treated with the SlowFade Antifade Kit (Invitrogen). Samples were imaged using a Zeiss LSM 510 scanning laser confocal microscope equipped with an argon/krypton laser.
Supplementary Material
Acknowledgments
We thank Erol Fikrig and Sukanya Narasimhan for providing antibody against NapA/Dps. This work was supported by Public Health Service Grant AI-059062 from the NIAID (NIH).
Abbreviations
- Bb
Borrelia burgdorferi
- BSK medium
Barbour-Stoenner-Kelly medium
- Dbp
decorin-binding protein
- HP
High-passage
- IFA
immunofluorescence assay
- LP
low-passage
- Osp
outer surface protein
- qRT-PCR
quantitative RT-PCR
- RT
reverse transcriptase
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- WT
wild-type
References
- Akins DR, Bourell KW, Caimano MJ, Norgard MV, Radolf JD. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J Clin Invest. 1998;101:2240–2250. doi: 10.1172/JCI2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alitalo A, Meri T, Ramo L, Jokiranta TS, Heikkila T, Seppala IJ, et al. Complement evasion by Borrelia burgdorferi: serum-resistant strains promote C3b inactivation. Infect Immun. 2001;69:3685–3691. doi: 10.1128/IAI.69.6.3685-3691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguita J, Hedrick MN, Fikrig E. Adaptation of Borrelia burgdorferi in the tick and the mammalian host. FEMS Microbiol Rev. 2003;27:493–504. doi: 10.1016/S0168-6445(03)00036-6. [DOI] [PubMed] [Google Scholar]
- Barthold SW, de Souza MS, Janotka JL, Smith AL, Persing DH. Chronic Lyme borreliosis in the laboratory mouse. Am J Pathol. 1993;143:959–971. [PMC free article] [PubMed] [Google Scholar]
- Blevins JS, Hagman KE, Norgard MV. Assessment of decorin-binding protein A to the infectivity of Borrelia burgdorferi in the murine models of needle and tick infection. BMC Microbiol. 2008;8:82. doi: 10.1186/1471-2180-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman BK, He M, Ouyang Z, Xu H, Pang X, Yang XF. Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect Immun. 2008;76:3844–3853. doi: 10.1128/IAI.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan JA, Posey JE, Gherardini FC. Borrelia oxidative stress response regulator, BosR: a distinctive Zn-dependent transcriptional activator. Proc Natl Acad Sci U S A. 2003;100:11684–11689. doi: 10.1073/pnas.2032956100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissette CA, Cooley AE, Burns LH, Riley SP, Verma A, Woodman ME, et al. Lyme borreliosis spirochete Erp proteins, their known host ligands, and potential roles in mammalian infection. Int J Med Microbiol. 2008;298 1:257–267. doi: 10.1016/j.ijmm.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissette CA, Haupt K, Barthel D, Cooley AE, Bowman A, Skerka C, et al. Borrelia burgdorferi infection-associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infect Immun. 2009;77:300–306. doi: 10.1128/IAI.01133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CS, Hefty PS, Jolliff SE, Akins DR. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect Immun. 2003;71:3371–3383. doi: 10.1128/IAI.71.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. Lyme disease-a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- Burtnick MN, Downey JS, Brett PJ, Boylan JA, Frye JG, Hoover TR, et al. Insights into the complex regulation of rpoS in Borrelia burgdorferi. Mol Microbiol. 2007;65:277–293. doi: 10.1111/j.1365-2958.2007.05813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykowski T, Woodman ME, Cooley AE, Brissette CA, Brade V, Wallich R, et al. Coordinated expression of Borrelia burgdorferi complement regulator-acquiring surface proteins during the Lyme disease spirochete's mammal-tick infection cycle. Infect Immun. 2007;75:4227–4236. doi: 10.1128/IAI.00604-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Eggers CH, Gonzalez CA, Radolf JD. Alternate sigma factor RpoS is required for the in vivo-specific repression of Borrelia burgdorferi plasmid lp54-borne ospA and lp6.6 genes. J Bacteriol. 2005;187:7845–7852. doi: 10.1128/JB.187.22.7845-7852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Eggers CH, Hazlett KR, Radolf JD. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect Immun. 2004;72:6433–6445. doi: 10.1128/IAI.72.11.6433-6445.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, et al. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol. 2007;65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter BM, Whitmire JM, Merrell DS. This is not your mother's repressor: the complex role of fur in pathogenesis. Infect Immun. 2009;77:2590–2601. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman AS, Yang X, Kumar M, Zhang X, Promnares K, Shroder D, et al. Borrelia burgdorferi complement regulator-acquiring surface protein 2 does not contribute to complement resistance or host infectivity. PLoS One. 2008;3:3010e. doi: 10.1371/journal.pone.0003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Radolf JD. Analysis of promoter elements involved in the transcriptional initiation of RpoS-dependent Borrelia burgdorferi genes. J Bacteriol. 2004;186:7390–7402. doi: 10.1128/JB.186.21.7390-7402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Gassent MD, Elliott NL, Seshu J. sodA is essential for virulence of Borrelia burgdorferi in the murine model of Lyme disease. Mol Microbiol. 2009;71:594–612. doi: 10.1111/j.1365-2958.2008.06549.x. [DOI] [PubMed] [Google Scholar]
- Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, Rosa PA, et al. Borrelia burgdorferi sigma54 is required for mammalian infection and vector transmission but not for tick colonization. Proc Natl Acad Sci U S A. 2005;102:5162–5167. doi: 10.1073/pnas.0408536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank KL, Bundle SF, Kresge ME, Eggers CH, Samuels DS. aadA confers streptomycin resistance in Borrelia burgdorferi. J Bacteriol. 2003;185:6723–6727. doi: 10.1128/JB.185.22.6723-6727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Grimm D, Tilly K, Byram R, Stewart PE, Krum JG, Bueschel DM, et al. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc Natl Acad Sci U S A. 2004;101:3142–3147. doi: 10.1073/pnas.0306845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann K, Corvey C, Skerka C, Kirschfink M, Karas M, Brade V, et al. Functional characterization of BbCRASP-2, a distinct outer membrane protein of Borrelia burgdorferi that binds host complement regulators factor H and FHL-1. Mol Microbiol. 2006;61:1220–1236. doi: 10.1111/j.1365-2958.2006.05318.x. [DOI] [PubMed] [Google Scholar]
- Hefty PS, Jolliff SE, Caimano MJ, Wikel SK, Radolf JD, Akins DR. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect Immun. 2001;69:3618–3627. doi: 10.1128/IAI.69.6.3618-3627.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A. 2001;98:12724–12729. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JA, Trzeciakowski JP, Skare JT. Borrelia burgdorferi alters its gene expression and antigenic profile in response to CO2 levels. J Bacteriol. 2007;189:437–445. doi: 10.1128/JB.01109-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquamet L, Traore DA, Ferrer JL, Proux O, Testemale D, Hazemann JL, et al. Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol Microbiol. 2009;73:20–31. doi: 10.1111/j.1365-2958.2009.06753.x. [DOI] [PubMed] [Google Scholar]
- Jewett MW, Lawrence K, Bestor AC, Tilly K, Grimm D, Shaw P, et al. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol Microbiol. 2007;64:1358–1374. doi: 10.1111/j.1365-2958.2007.05746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona LI, Tokarz R, Kuhlow CJ, Benach J, Benach JL. The fur homologue in Borrelia burgdorferi. J Bacteriol. 2004;186:6443–6456. doi: 10.1128/JB.186.19.6443-6456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labandeira-Rey M, Skare JT. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect Immun. 2001;69:446–455. doi: 10.1128/IAI.69.1.446-455.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals. 2007;20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- Li X, Pal U, Ramamoorthi N, Liu X, Desrosiers DC, Eggers CH, et al. The Lyme disease agent Borrelia burgdorferi requires BB0690, a Dps homologue, to persist within ticks. Mol Microbiol. 2007;63:694–710. doi: 10.1111/j.1365-2958.2006.05550.x. [DOI] [PubMed] [Google Scholar]
- Lybecker MC, Samuels DS. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol Microbiol. 2007;64:1075–1089. doi: 10.1111/j.1365-2958.2007.05716.x. [DOI] [PubMed] [Google Scholar]
- McDowell JV, Wolfgang J, Tran E, Metts MS, Hamilton D, Marconi RT. Comprehensive analysis of the factor h binding capabilities of borrelia species associated with lyme disease: delineation of two distinct classes of factor h binding proteins. Infect Immun. 2003;71:3597–3602. doi: 10.1128/IAI.71.6.3597-3602.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour A, et al. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect Immun. 2003;71:1689–1705. doi: 10.1128/IAI.71.4.1689-1705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Blevins JS, Norgard MV. Transcriptional interplay among the regulators Rrp2, RpoN and RpoS in Borrelia burgdorferi. Microbiology. 2008;154:2641–2658. doi: 10.1099/mic.0.2008/019992-0. [DOI] [PubMed] [Google Scholar]
- Ouyang Z, He M, Oman T, Yang XF, Norgard MV. A manganese transporter, BB0219 (BmtA), is required for virulence by the Lyme disease spirochete, Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2009;106:3449–3454. doi: 10.1073/pnas.0812999106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal U, Montgomery RR, Lusitani D, Voet P, Weynants V, Malawista SE, et al. Inhibition of Borrelia burgdorferi-tick interactions in vivo by outer surface protein A antibody. J Immunol. 2001;166:7398–7403. doi: 10.4049/jimmunol.166.12.7398. [DOI] [PubMed] [Google Scholar]
- Pal U, Yang X, Chen M, Bockenstedt LK, Anderson JF, Flavell RA, et al. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J Clin Invest. 2004;113:220–230. doi: 10.1172/JCI19894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack RJ, Telford SR, 3rd, Spielman A. Standardization of medium for culturing Lyme disease spirochetes. J Clin Microbiol. 1993;31:1251–1255. doi: 10.1128/jcm.31.5.1251-1255.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey JE, Gherardini FC. Lack of a role for iron in the Lyme disease pathogen. Science. 2000;288:1651–1653. doi: 10.1126/science.288.5471.1651. [DOI] [PubMed] [Google Scholar]
- Purser JE, Norris SJ. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2000;97:13865–13870. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revel AT, Talaat AM, Norgard MV. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc Natl Acad Sci U S A. 2002;99:1562–1567. doi: 10.1073/pnas.032667699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan TG. Temporal regulation of outer surface proteins of the Lyme-disease spirochaete Borrelia burgdorferi. Biochem Soc Trans. 2003;31:108–112. doi: 10.1042/bst0310108. [DOI] [PubMed] [Google Scholar]
- Schwan TG, Piesman J, Golde WT, Dolan MC, Rosa PA. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci U S A. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshu J, Boylan JA, Hyde JA, Swingle KL, Gherardini FC, Skare JT. A conservative amino acid change alters the function of BosR, the redox regulator of Borrelia burgdorferi. Mol Microbiol. 2004;54:1352–1363. doi: 10.1111/j.1365-2958.2004.04352.x. [DOI] [PubMed] [Google Scholar]
- Shi Y, Xu Q, McShan K, Liang FT. Both decorin-binding proteins A and B are critical for overall virulence of Borrelia burgdorferi. Infect Immun. 2008;76:1239–1246. doi: 10.1128/IAI.00897-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Girschick HJ. Molecular survival strategies of the Lyme disease spirochete Borrelia burgdorferi. Lancet Infect Dis. 2004;4:575–583. doi: 10.1016/S1473-3099(04)01132-6. [DOI] [PubMed] [Google Scholar]
- Smith AH, Blevins JS, Bachlani GN, Yang XF, Norgard MV. Evidence that RpoS (sigmaS) in Borrelia burgdorferi is controlled directly by RpoN (sigma54/sigmaN) J Bacteriol. 2007;189:2139–2144. doi: 10.1128/JB.01653-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere AC, Grodzicki RL, Kornblatt AN, Craft JE, Barbour AG, Burgdorfer W, et al. The spirochetal etiology of Lyme disease. N Engl J Med. 1983;308:733–740. doi: 10.1056/NEJM198303313081301. [DOI] [PubMed] [Google Scholar]
- Stevenson B, Schwan TG, Rosa PA. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PE, Byram R, Grimm D, Tilly K, Rosa PA. The plasmids of Borrelia burgdorferi: essential genetic elements of a pathogen. Plasmid. 2005;53:1–13. doi: 10.1016/j.plasmid.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Stewart PE, Thalken R, Bono JL, Rosa P. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol Microbiol. 2001;39:714–721. doi: 10.1046/j.1365-2958.2001.02256.x. [DOI] [PubMed] [Google Scholar]
- Stewart PE, Wang X, Bueschel DM, Clifton DR, Grimm D, Tilly K, et al. Delineating the requirement for the Borrelia burgdorferi virulence factor OspC in the mammalian host. Infect Immun. 2006;74:3547–3553. doi: 10.1128/IAI.00158-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terekhova D, Iyer R, Wormser GP, Schwartz I. Comparative genome hybridization reveals substantial variation among clinical isolates of Borrelia burgdorferi sensu stricto with different pathogenic properties. J Bacteriol. 2006;188:6124–6134. doi: 10.1128/JB.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, Bueschel D, et al. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect Immun. 2006;74:3554–3564. doi: 10.1128/IAI.01950-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R, Anderton JM, Katona LI, Benach JL. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect Immun. 2004;72:5419–5432. doi: 10.1128/IAI.72.9.5419-5432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weening EH, Parveen N, Trzeciakowski JP, Leong JM, Hook M, Skare JT. Borrelia burgdorferi lacking DbpBA exhibits an early survival defect during experimental infection. Infect Immun. 2008;76:5694–5705. doi: 10.1128/IAI.00690-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Goldberg MS, Popova TG, Schoeler GB, Wikel SK, Hagman KE, et al. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol Microbiol. 2000;37:1470–1479. doi: 10.1046/j.1365-2958.2000.02104.x. [DOI] [PubMed] [Google Scholar]
- Yang XF, Alani SM, Norgard MV. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2003a;100:11001–11006. doi: 10.1073/pnas.1834315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Hubner A, Popova TG, Hagman KE, Norgard MV. Regulation of expression of the paralogous Mlp family in Borrelia burgdorferi. Infect Immun. 2003b;71:5012–5020. doi: 10.1128/IAI.71.9.5012-5020.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Lybecker MC, Pal U, Alani SM, Blevins J, Revel AT, et al. Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. J Bacteriol. 2005;187:4822–4829. doi: 10.1128/JB.187.14.4822-4829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.