Abstract
RAD51, a key protein in the homologous recombinational DNA repair (HRR) pathway, is the major strand-transferase required for mitotic recombination. An important early step in HRR is the formation of single-stranded DNA (ss-DNA) coated by RPA (a ss-DNA-binding protein). Displacement of RPA by RAD51 is highly regulated and facilitated by a number of different proteins known as the ‘recombination mediators’. To assist these recombination mediators, a second group of proteins also is required and we are defining these proteins here as ‘recombination co-mediators’. Defects in either recombination mediators or co-mediators, including BRCA1 and BRCA2, lead to impaired HRR that can genetically be complemented for (i.e. suppressed) by overexpression of RAD51. Defects in HRR have long been known to contribute to genomic instability leading to tumor development. Since genomic instability also slows cell growth, precancerous cells presumably require genomic re-stabilization to gain a growth advantage. RAD51 is overexpressed in many tumors, and therefore, we hypothesize that the complementing ability of elevated levels of RAD51 in tumors with initial HRR defects limits genomic instability during carcinogenic progression. Of particular interest, this model may also help explain the high frequency of TP53 mutations in human cancers, since wild-type p53 represses RAD51 expression.
INTRODUCTION
Homologous recombinational DNA repair plays an important role in cancer
The central protein involved in the homologous recombinational DNA repair (HRR) pathway is RAD51, a strand transfer protein. Although no mutations in the RAD51 open reading frame have been found in cancers, defects in other HRR genes have been shown to play an important role in carcinogenesis, and particularly in breast cancer. Inherited heteroallelic mutations in BRCA1 and BRCA2 each result in a breast cancer predisposition syndrome, and both of these genes are involved in HRR (1). In addition, eight more breast cancer predisposition syndromes have been identified, and all of these also involve defects in HRR-related genes (2). There have been several excellent and comprehensive reviews in recent years that describe the connections between HRR and cancer (particularly breast cancer) (1,3), the role of TP53 mutations in recombination (4,5) and the observation that RAD51 is overexpressed in many tumors (6). What seems to be missing from these and other reviews is an overall hypothesis explaining the relationship between these three different components of carcinogenesis. Here, we review what we feel is the missing link that helps tie these components together. This missing link is the observation that overexpression of RAD51 partially complements (i.e. suppresses) defects in a number of different HRR genes that encode recombination mediator and co-mediator proteins, including BRCA1 and BRCA2. Recombination mediators and co-mediators are proteins that assist RAD51 in displacing the single-strand DNA (ss-DNA)-binding protein RPA from ss-DNA, thus initiating the formation of the RAD51 filament. Complementation by RAD51 overexpression, which has not been reviewed before, is the central focus of this article.
RAD51 expression is up-regulated in many cancers
For many years it has been observed that the levels of the RAD51 protein are greatly elevated (∼2–7-fold) in many cancer cell lines and in primary tumors [reviewed by (6)]. The wild-type p53 protein plays an important role both in suppressing the transcriptional expression of RAD51 and in down-regulating RAD51 protein activity (4,5,7,8). The advantage for cancer cells of overexpressing RAD51 has yet to be adequately explained, but it has been suggested that the high levels of RAD51 are involved in tumor progression by destabilizing the genome (9). Another possible explanation is that elevated RAD51 confers a DNA replication advantage during the more rapid cell divisions that follow the activation of oncogenes and inactivation of tumor suppressors. Each of these explanations, at least in their basic form, suffers from the fact that, in general, overexpression of RAD51 is deleterious and slows cell growth in vitro, and therefore would also be likely to hinder the growth of cancer cells in vivo. Based on published reports about the ability of overexpressed RAD51 to suppress defects in HRR genes, two new alternative hypotheses are presented at the end of this article that may explain why RAD51 is up-regulated in many human cancers.
RECOMBINATION MEDIATORS AND CO-MEDIATORS
The HRR pathway is composed of a highly orchestrated network of proteins, many of which presumably still remain to be discovered. HRR frequently is initiated by a double-strand break, either due to endogenous lesions at the replication fork, or induced by different DNA damaging agents, including cisplatin, mitomycin C (MMC), camptothecin (CPT) and ionizing radiation. First, endonucleolytic activity produces a long 3′ ss-DNA overhang that is rapidly coated with RPA, the heterotrimeric ss-DNA-binding protein. Displacement of RPA by RAD51 is critical and highly regulated, and initiates the strand transfer process (Figure 1). The proteins directly involved in the displacement of RPA are named ‘recombination mediators’ (Table 1) (10,11) (here sometimes referred to as just ‘mediators’). Recombination mediator activity is ideally assayed by biochemical methods (10,12–15). In place of biochemical evidence, reductions in RAD51 foci formation are frequently utilized as an indirect indicator of a mediator defect (16–18), if the defective/missing protein is not required for the formation of RPA-coated ss-DNA. It is significant that mutations in most of these mediators in mammalian cells result in high levels of genomic instability, as has been demonstrated for cells with deletions of RAD51D in both Chinese hamster ovary cells (CHO) and murine embryonic fibroblast cells (MEFs) (19,20). Very high levels of mutagenesis were observed in Rad51d deletion CHO cells (19), and increased mutagenesis seems likely in other mediator defective cell lines as well.
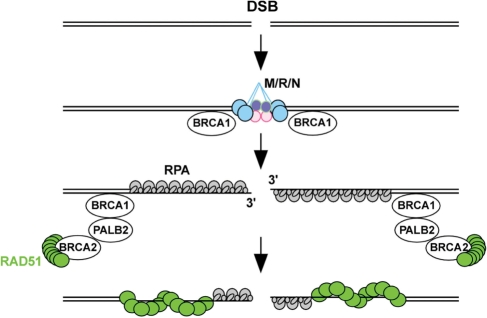
Figure 1.
Simplified schematic to depict the mediator step of recombination: displacement of RPA by RAD51. A critical step in homologous re-combinational repair is the displacement of RPA, the trimeric ssDNA-binding protein, by the RAD51 strand transfer protein. This step is highly regulated by cells to ensure that potentially deleterious events are avoided. Many different proteins are involved in assisting RAD51 to displace RPA at this stage, and the proteins that are directly involved are known as the ‘recombination mediators’ (Table 1). There are also additional proteins that function to assist the mediators or assist in their localization to DNA damage, and in this review, the proteins assisting the mediators are defined as ‘recombination co-mediators’. Human BRCA2 is a mediator that interacts directly with approximately eight RAD51 molecules and transports them to the site of ss-DNA bound by RPA, while PALB2 and BRCA1 are co-mediators that directly and indirectly, respectively, interact with BRCA2 and assist in localizing it, with RAD51 bound, to the sites of DNA damage (for references, see Table 1). DSB, double-strand DNA break; M/R/N, MRE11-RAD50-NBS1 complex.
Table 1.
Human recombination mediator and co-mediator proteins
| Involved in cancera | Suppressed by RAD51 overexpression in DT40 or human cells | Referencesb | |||
|---|---|---|---|---|---|
| Recombination mediators | |||||
| BRCA2/FANCD1 | Yes | Yes | (13,15) | ||
| RAD51 paralogs | |||||
| RAD51B/RAD51L1 | No evidence yet | Yes | (14,18,56,57) | ||
| RAD51C | No evidence yet | Yes | (14,18,56,57) | ||
| RAD51D/RAD51L3 | No evidence yet | Yes | (18,56,57) | ||
| XRCC2 | No evidence yet | Yes | (18,56,57) | ||
| XRCC3 | No evidence yet | Yes | (18,56–58) | ||
| RAD52 | No evidence yet | ntc | (12,59) | ||
| Recombination co-mediators | |||||
| BRCA1 | Yes | Yes | (24,60) | ||
| CHK2 | Yes | nt | (61) | ||
| PALB2/FANCN | Yes | nt | (60,62,63) | ||
| SWS1 | No evidence yet | nt | (64) | ||
| Potential mediators/co-mediators | Main interacting protein partner | References |
|---|---|---|
| BARD1 | BRCA1 | (65) |
| BCCIPα&β | BRCA2 | (66) |
| BRIP/FANCJ/BACH1 | BRCA1 | (67) |
| DSS1/SHFM1 | BRCA2 | (68) |
aDirect involvement in cancer, since inherited mutations result in a pre-disposition towards increased cancer, particularly breast and ovarian (1–3), and some mutant forms are also found in sporadic cancers (3).
bTop part: recent references to recombination mediator and co-mediator functions; for additional information/references see review articles (1,2,11,63,69,70); bottom part: references to protein interactions with BRCA1 or BRCA2.
cnt, not tested yet.
There are also a number of proteins that function to assist these mediators or localize them to the site of the DNA break. These proteins have not previously been given a general name, but we propose to call them ‘recombination co-mediators’. Many of the mediators and co-mediators are known tumor suppressor proteins, and mutant versions frequently can be suppressed by overexpression of RAD51, as discussed below. This makes considerable biological sense, since these proteins function to assist RAD51, but in their absence, RAD51 overexpression can in some cases partially compensate. It is still unknown whether defects in other HRR proteins (i.e. non-mediators such as MRE11 and RAD54) can also be suppressed by RAD51 overexpression. Also, only some of the mediators/co-mediators have been tested for suppression by RAD51 overexpression, and there are additional proteins that may also be mediators/co-mediators, since they interact with BRCA1 or BRCA2 (i.e. BARD1, BCCIPα and β, BRIP1/FANCJ/BACH1 and SHFM1/DSS1). With the use of the RNAi technique, it should be relatively easy to test these other potential human mediators/co-mediators for complementation by RAD51 overexpression. Ideally these experiments should be done in human cell lines that initially express low levels of endogenous RAD51.
FUNCTIONAL SUPPRESSION BY RAD51 OVEREXPRESSION
Functional suppression in model organisms
Overexpression of RAD51 has been shown to partially suppress defects in recombination mediators and co-mediators in yeast, avian, rodent and human cells, although suppression of the HRR defect by exogenous RAD51 is generally not as complete as the complementation by a wild-type copy of the HRR gene that is mutated. In addition, in these experiments many different end-points were assessed. For each end-point, the relative degree of complementation by exogenous RAD51 differs, for unknown reasons.
Yeast. In Saccharomyces cerevisiae, two different groups observed that cells with deletion mutations in RAD55 and RAD57, the two yeast RAD51 paralogs, are partially suppressed by overexpression of RAD51 (21,22). In addition, overexpression of RAD52 resulted in partial suppression of both X-ray sensitivity and gene conversion defects, and simultaneous overexpression of both RAD51 and RAD52 gave near complete suppression for both phenotypes (21). Furthermore, although most mutations in RAD52 are not suppressed by RAD51 overexpression, there is one mutation in RAD52 that is suppressed (23). Since S. cerevisiae lacks both BRCA1 and BRCA2 genes, it is not possible to test defects in these genes for complementation by RAD51 overexpression.
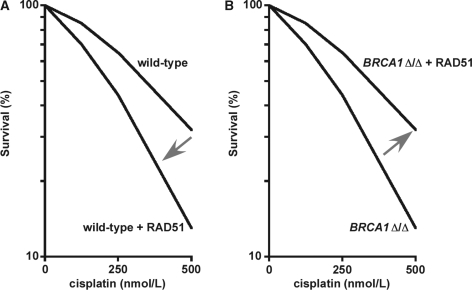
Chicken DT40 cells. The chicken DT40 cell line (p53 deficient) has been extensively used to characterize DNA repair defects. Each of the five RAD51 paralog genes has been separately knocked out in DT40 cells, and each knockout is partially complemented by overexpression of human RAD51 (16,17). The complementation of RAD51B−/− by hRAD51 overexpression was the most extensively studied, and very interestingly, the authors observed significant complementation for MMC, cisplatin and X-ray sensitivity and for genomic instability, but not for homologous integration events (16). This result argues that in DT40 cells and under conditions where a RAD51 paralog is lost, overexpression of human RAD51 largely, but not fully, ameliorates the HRR-deficiency. Interestingly, this complementation occurred in RAD51B−/− cells in which the level of the hRAD51 was approximately equal to the level of the endogenous avian RAD51, suggesting that simply doubling the level of RAD51 was sufficient for such suppression in these cells. In addition, RAD51 overexpression partially complemented a BRCA1 deletion, since overexpression of human RAD51 (∼15-fold increase relative to the endogenous protein) in BRCA1Δ/Δ DT40 cells almost completely rescued defects in cell proliferation (i.e. slow growth) and in DNA damage survival (Figure 2), and also partially rescued the defect in gene targeting frequency (24). These investigators also showed that human tumors with BRCA1 mutations frequently exhibit elevated RAD51 transcripts, as well as elevated transcripts of RAD51AP1 and RAD54, encoding two relatively late acting HRR proteins (24). This is indirect evidence that these BRCA1-deficient tumors had undergone selection for overexpression of RAD51, RAD51AP1 and RAD54, and the authors suggested a somewhat similar hypothesis to the one presented here: ‘The growth and viability defects of BRCA1-deficient cells are difficult to reconcile with the deregulated growth of BRCA1-deficient tumors. The findings we present here suggest that one mechanism by which this apparent paradox is resolved is via genetic suppression of BRCA1 phenotypes by up-regulation of HR. The fact that the majority of BRCA1-deficient tumors examined exhibit significantly up-regulated expression of HR genes suggests either that there is a selection for such up-regulation during tumor development or that a high level of expression is a precondition for tumorigenesis of BRCA1-deficient cells.’ (24)
Figure 2.
Effects of RAD51 overexpression in isogenic wild-type and BRCA1-deleted DT40 cells. (A) In cells with wild-type HRR, overexpression of RAD51 results in a reduced ability to repair cisplatin-induced DNA damage. Such increased sensitivity to a DNA cross-linking agent frequently coincides with genomic instability in untreated HRR-defective cells, presumably due to unrepaired spontaneous damage (16,17,44). (B) Alternatively, in cells with a BRCA1 homozygous deletion, the HRR defect is complemented by RAD51 overexpression. The complemented cells also presumably have increased genomic stability, a concept incorporated into Model I. Importantly, comparing just the cells overexpressing RAD51 (in both panels), the BRCA1 deletions actually result in increased resistance to cisplatin, and presumably also increased genomic stability, a concept incorporated into Model II. This figure is adapted directly from the data in Figure 2D of (24), with permission from Douglas K. Bishop and AACR.
Functional suppression in mammalian cells
In the first direct studies in mammalian cells, two recent independent publications report that BRCA2 defects are suppressed by RAD51 overexpression. In one study, the BRCA2 mutant Capan-1 human pancreatic carcinoma cell line (with mutant p53) was shown to be partially complemented by overexpression of RAD51, assaying for cell survival after exposure to X-rays or cisplatin (25). In addition, complementation was better, and nearly complete, when a mutant form of RAD51 that is resistant to caspase-3 cleavage was expressed. In the same study, similar results on complementation by overexpression of wild-type RAD51 were also observed for Brca2 mutant MEFs. In the second study, mouse hybridoma cell lines depleted for Brca2, using stable expression of siRNA, were studied for the effect of overexpression of the mouse Rad51, Rad52 and Rad54 proteins (26). In mouse hybridoma cells with low levels of Brca2, the reduced frequency of targeted integration and DNA damage-induced Rad51 foci was partially complemented by overexpression of Rad51, but not by overexpression of either Rad52 or Rad54.
We have found only one relevant report that demonstrated a lack of complementation by human RAD51, and these experiments were carried out in Xrcc3-deficient CHO cells (27). In this study, complementation by RAD51 was assessed using an integrated recombinational reporter plasmid. It is possible that, if these Xrcc3-deficient CHO cells had been tested for the cellular sensitivity to MMC or cisplatin, a different result with respect to RAD51 complementation would have been observed, as discussed earlier for the DT40 RAD51B−/− cell line (16). It is worth noting that RAD51 is one of the most highly conserved proteins known. Consequently, the fact that the heterologous human RAD51 protein was expressed in Xrcc3-deficient CHO cells is unlikely to have biased the results reported by Pierce and collaborators (27). There is a Brca2 mutant hamster V79 cell line (with mutant Trp53) (28), which would provide a unique tool to study whether or not wild-type human RAD51 can complement Brca2-deficiency in a hamster background.
EFFECTS OF ECTOPIC OVEREXPRESSION OF RAD51
Since the effects of RAD51 overexpression in many different cellular systems have recently been extensively reviewed (6), the results of only some studies will be discussed here. In chicken DT40 cells with wild-type HRR, overexpression of human RAD51 resulted in increased cellular sensitivity to cisplatin, and this enhanced sensitivity of wild-type DT40 cells was equal to the cellular sensitivity of BRCA1Δ/Δ DT40 cells that did not express ectopic RAD51 (24). On the other hand, when RAD51 was overexpressed in the BRCA1Δ/Δ DT40 cells, near complete complementation was observed for cell survival after both cisplatin and X-rays. These experiments suggest that overexpression of RAD51 in cells with normal HRR can be deleterious, while overexpression in cells with an HRR defect can be beneficial (Figure 2). (Note that the authors did not see X-ray sensitization of wild-type DT40 cells following RAD51 overexpression, but this test is not very sensitive, since non-homologous end-joining is the major repair pathway for X-ray damage.)
In wild-type (i.e. HRR-proficient) mammalian cells, the effects of ectopic RAD51 overexpression are somewhat contradictory, but this may relate to the end-points assessed, the levels of RAD51 overexpression achieved, the fraction of RAD51 that is cytoplasmic versus nuclear and the p53 status of the cell lines utilized. In CHO cells, overexpression of hamster RAD51 resulted in increased HRR, while a second study reported that overexpression of human RAD51 in CHO cells reduced homologous recombination at a double-strand break (29,30). In mouse ES cells, RAD51 overexpression led to aneuploidy and chromosomal rearrangements (9). In the human fibrosarcoma HT1080 cell line, ectopic overexpression of human RAD51 was very deleterious to cells, resulting in a slow growth phenotype and increased levels of apoptosis (31).
UP-REGULATION OF RAD51 IN TUMORS AND CANCER-DERIVED CELL LINES
Many different cancer-derived cell lines, as well as many tumors have been shown to have higher than normal levels of RAD51, as recently reviewed (6). The exact cause of this overexpression is not known, but there are important clues. For example, wild-type p53 has been reported to suppress the transcriptional regulation of RAD51, and TP53 deletions and some TP53 point mutations have been shown to up-regulate the expression of RAD51 (7). In addition to its role in transcriptional repression of RAD51, the wild-type p53 protein directly interacts with the RAD51 protein, inhibiting its activity (8).
There are also a number of additional factors that appear to play a role in RAD51 regulation. There is evidence that transcriptional activator protein 2 (AP2), in combination with p53, functions to down-regulate RAD51 transcription (32). In addition, there is evidence that the oncogenic fusion tyrosine kinase BCR/Abl, the result of translocations, increases RAD51 expression (33) and indirect evidence that c-Abl is also involved in up-regulating RAD51 transcription, since imatinib (Gleevec), a relatively specific inhibitor of c-Abl, reduced the elevated levels of RAD51 in some cancer cell lines (34,35). Recently, it has also been reported that unlike p53, wild-type p63 and p73 act as inducers of both basal and DNA-damage-induced levels of RAD51 in MEFs (36). Although, many cancer cell lines and primary tumors overexpress RAD51 (6), chronic hypoxia has been shown to decrease the level of RAD51, leading to reduced levels of HRR in hypoxic tumors that also show increased sensitivity to DNA cross-linking agents (37,38). In general, a more complete understanding of how RAD51 is regulated may lead to better treatments for cancer patients.
TP53 MUTATIONS ARE UNDER REPRESENTED IN MISMATCH REPAIR-INITIATED CANCERS
Approximately 50% of all cancers have mutations in TP53. In particular, cancers initiated by general genomic instability frequently have mutations in TP53. Alternatively, cancers initiated by mutations in genes of the mismatch repair (MMR) pathway, and the subsequent microsatellite instability (MSI), are less frequently associated with mutations in TP53 (39,40). This disparate occurrence of TP53 mutations has never been satisfactorily explained. The hypothesis presented here, that one of the main functions of mutant p53 is to up-regulate RAD51, takes this disparity into account and provides an explanation for it, as discussed below. It is important to note that TP53 mutations are very heterogeneous (4,41), and that only a few mutant versions, other than the TP53 homozygous deletion, have been tested for their effects on up-regulating RAD51 expression.
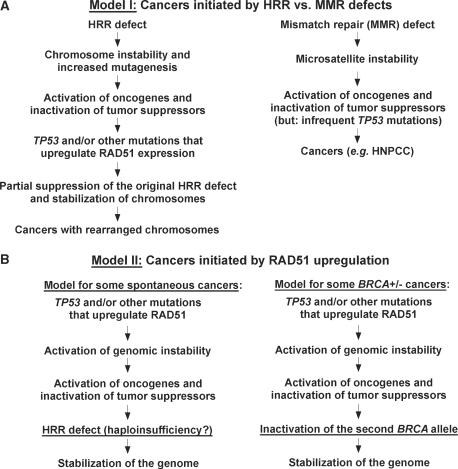
Model I: UP-REGULATION OF RAD51 SUPPRESSES HRR DEFECTS TO RESTABILIZE THE GENOME
We proposed that carcinogenesis is frequently initiated by defects in HRR genes (particularly in genes encoding recombination mediators or co-mediators) that result in genomic instability (Figures 3A and 4A). There are likely to be many ways for a normal cell to develop an HRR defect, since HRR is a complex DNA repair pathway with many known genes involved and presumably some genes yet to be discovered. In addition, there is evidence for haploinsufficiency for some HRR defects, and epigenetic silencing events also may account for compromised HRR ability. Interestingly, there is evidence that point mutations in known HRR genes are not common in either breast or colorectal cancers: in a study of these two cancers, most of the transcripts from 11 cancers of each type were sequenced and only a small number of mutations in a few HRR-related genes were identified in cancers of the breast (i.e. BCCIP, BRCA1, BRCA2, MRE11, FANCA and FANCM) and of the colon (FANCG) (42). Of these, only mutations in BRCA1 and BRCA2 are known to be suppressible by RAD51 overexpression.
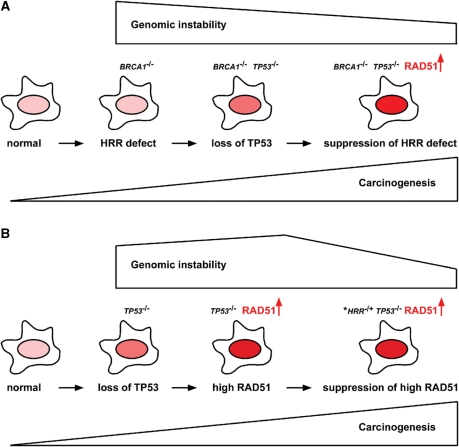
Figure 3.
Schemes to explain the order of events in the two proposed models. (A) Model I: normal cells that develop an HRR defect (e.g. BRCA1−/−) select for loss of TP53, which then acts to up-regulate RAD51 expression (darker nuclei represent higher levels of RAD51). Overexpression of RAD51 can in some cases lead to partial suppression of the original HRR defect, helping to stabilize the genome. (B) Model II: in some cancers, TP53 mutations are early events and these mutations may up-regulate RAD51 expression, resulting in increased genomic instability. Subsequent mutations/silencing in an HRR function partially suppresses the phenotype of RAD51 overexpression, reducing genomic instability. The asterisk indicates that for sporadic cancers many of these HRR mutations are likely to be in genes that exhibit haploinsufficiency, or the mutations themselves may be dominant-negative alleles.
Figure 4.
(A) Summary of Model I explaining cancer development initiated by HRR defects. The model suggests that many cancers with genomic instability are initiated by HRR defects, just as cancers with MSI are initiated by MMR defects. In cancers with HRR defects, selection for up-regulation of RAD51 acts to suppress the original HRR defect, partially restoring genomic stability and enhancing cell proliferation. TP53 mutations frequently result in the up-regulation of RAD51, and are selected for in cancers with a pre-existing HRR defect. HNPCC, hereditary non-polyposis colorectal cancer. (B) Summary of Model II explaining cancer development initiated by RAD51 up-regulation. In this model, inactivation of TP53 and up-regulation of RAD51 occur early in sporadic oncogenesis (i.e. cancers initiated by spontaneous non-inherited mutations) (left side). In some BRCA heterozygous cells (BRCA1+/− or BRCA2+/−), these events occur prior to functional loss of HRR resulting from a mutation or silencing of the second BRCA allele (right side). These are identical models, with the exception of the underlined phrases.
Genomic instability presumably facilitates both the activation of oncogenes and the inactivation of tumor suppressor genes (with the exception of TP53, which in the current model occurs as a later event). However, genomic instability initiated by defects in HRR is also deleterious due to the inhibitory effects on cell growth. Therefore, there is pressure to suppress genomic instability initiated by HRR deficiencies, and TP53 mutations and/or mutations in other genes are selected for partially because they lead to the up-regulation of RAD51, which then at least partially suppresses the initial HRR defect. It also seems likely that due to synergistic effects, some cancers may select for the overexpression of both RAD51 and HRR proteins that function later than the recombination mediators/co-mediators. [Note that there is evidence that RAD51AP1, which is not a RAD51 paralog, functions later in HRR than the recombination mediators (43,44), and RAD51AP1 has been shown to be up-regulated in a number of cancers (24,45,46)]. With regard to genomic rearrangements, there is evidence from cell lines derived from breast cancers, that genomic stability appears to be partially restored during tumor development (47), consistent with the proposed model. The concept that genomic instability might be restored by RAD51 overexpression during tumor progression, as previously suggested by the Bishop laboratory (24,48), has been expanded here.
We also propose that cancers initiated by MMR defects do not select for TP53 mutations. Such mutations might up-regulate RAD51, which would be deleterious in HRR-proficient cells. Besides, in such cancers there is no need to up-regulate RAD51 for suppression purposes, and the precancerous cells achieve the additional benefits of TP53 inactivation, such as down-regulation of both apoptosis and checkpoint functions, by selecting for mutations in other genes that do not affect RAD51 expression.
The model presented here, if correct, has profound implications for how cancers might be treated more effectively in the future. Our proposed hypothesis argues that many more cancers may arise from HRR defects than has previously been suggested. If so, restoration of RAD51 to normal levels (i.e. no longer suppressing the original HRR defect) may sensitize these tumor cells to genotoxic agents that kill HRR defective cells. Imatinib has been reported to suppress the overexpression of RAD51, while not strongly affecting the levels of RAD51 in non-cancerous cells (35). Imatinib sensitizes tumors to radiation and to chemotherapy (34,35), and the model presented here provides a new perspective on such cancer treatment, that is based on the pro-oncogenic activity of RAD51 overexpression. Recently, PARP inhibitors have been used to target tumors with HRR defects such as BRCA1 and BRCA2 (49). Since PARP inhibitors preferentially target HRR-defective tumor cells, we propose that a combination therapy using both imatinib-type compounds and PARP inhibitors may be highly potent in the treatment of RAD51 overexpressing cancers.
MODEL II: RAD51 UP-REGULATION LEADS TO GENOMIC INSTABILITY FIRST, FOLLOWED BY SELECTION FOR INACTIVATION OF AN HRR GENE TO RESTABILIZE THE GENOME
Overexpression of RAD51, like mutations in HRR, can destabilize the genome (9), as previously discussed. An alternative to Model I is that in some cancers the up-regulation of RAD51 is an early event (Figures 3B and 4B). Subsequently, as carcinogenesis proceeds, there is selection for an HRR defect to specifically counteract the effects of RAD51 overexpression. The consequence of each of these two proposed cancer developmental processes (i.e. Models I and II) is the same (i.e. cells with both an HRR defect and higher than normal levels of RAD51 protein), but the order of events is reversed. The alternative Model II helps to explain why in some BRCA+/− carriers, inactivation of the second BRCA allele is a late event (50). This model suggests that in cancers arising by this order of events, the BRCA heterozygosity has little to do with the early events of carcinogenesis, but is required to rescue highly unstable cancer cells from the RAD51 up-regulation. It is important to note that, although both models are mutually exclusive to explain the origin of a single tumor, each hypothesis may correctly explain the origin of some cancers. For both sporadic and inherited predisposition-related cancers, some may have evolved with an HRR defect first, while others may have evolved first with a TP53 mutation and/or other mutation that up-regulates RAD51.
Although there appears to be no cancer-related mutations in the RAD51 coding region, there is a single-nucleotide polymorphism (SNP) in the 5′-untranslated region of RAD51 that increases the likelihood of breast cancer in BRCA2+/− carriers (51). This SNP has been shown to significantly increase the relative levels of the SNP-containing RAD51 transcripts (∼2–3-fold) for a reporter construct expressed in the U2-OS osteosarcoma cell line (52). The increased cancer predisposition of BRCA2 carriers with this RAD51 SNP can be explained by either of the two different models presented. For each model, the up-regulation of RAD51 is a necessary event for carcinogenesis, and the reported RAD51 SNP may help to facilitate this up-regulation.
FUTURE RESEARCH FOR EXAMINING RAD51 OVEREXPRESSION AND SUPPRESSION
There are a number of questions that should be experimentally addressed to further examine RAD51 overexpression and complementation. (i) Which HRR genes/defects are suppressed by RAD51 overexpression, are all of these suppressible HRR defects in either recombination mediator or co-mediator genes, and is there suppression of mediators that also function at an additional stage of HRR? (ii) Are elevated levels of RAD51AP1 and/or RAD54 synergistic to RAD51 overexpression in this suppression? (iii) In addition to TP53, TP63, TP73, c-Abl and AP2, what other genes are involved in regulating the expression of RAD51? (iv) Is RAD51 expression and/or activity regulated by translational and/or post-translational mechanisms (presumably fruitful areas for further study)? (v) Do most tumors that overexpress RAD51 have suppressed HRR defects and also TP53 mutations? (vi) If epigenetic silencing is involved in inactivating HRR functions, can these silencing events be reversed by drugs, and if so, is this reversal lethal or semi-lethal to cancer cells with high levels of RAD51 protein, even in the absence of genotoxic agents?
There is also a great need for additional mechanisms to assay for the levels of RAD51 transcript, protein and foci in primary tumors. A recent study successfully analyzed RAD51 foci, as well as BRCA1 and FANCD2 foci, in sporadic breast cancer biopsies (treated with X-rays ex vivo), and the absence of such foci was closely correlated with likely defects in the BRCA1 pathway (53). Improved mechanisms for analyzing RAD51 levels and re-localization/DNA-damage foci should be useful in the analyses of primary tumors and help to determine potential treatment modalities.
CONCLUDING REMARKS
In summary, overexpression of RAD51 in a number of different organisms has been shown to partially suppress defects in recombination mediator and co-mediator functions. In addition, RAD51 is frequently up-regulated in cancer cell lines and in primary tumors, although RAD51 overexpression in the absence of any underlying HRR defect is frequently detrimental to growth of cells, at least in tissue culture.
Largely based on the published findings reviewed here, two closely related models are proposed that may help to explain the role of RAD51 up-regulation in cancers. It is worth remembering that cancer is an extremely complex set of diseases, and that cancer cells develop many different mechanisms to achieve the same phenotype of independent and uncontrolled growth (54,55). The models presented here strive to explain certain aspects of oncogenic progression, and if these models prove correct, they will point the way to how some cancers arise and to novel roles for HRR functions during oncogenesis.
FUNDING
National Cancer Institute/National Institutes of Health (grant R01-CA120315 to D.S.; P01-CA092584); National Aeronautics and Space Administration (grant NNJ05HI361 to C.W.); Low Dose Radiation Research Program of the Department of Energy. Funding for open access charge: National Institutes of Health (grant CA120315).
Conflict of interest statement. None declared.
ACKNOWLEDGMENTS
We would like to thank the researchers whose excellent work allowed us to come to the conclusions presented here. We would also like to thank Amy Kronenberg and Larry H. Thompson for their useful comments on this manuscript, Douglas K. Bishop for permission to include a figure based on data published by his laboratory and Patrick Sung for suggesting the term ‘recombination co-mediators’.
REFERENCES
- 1.Venkitaraman AR. Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Ann. Rev. Pathol. 2009;4:461–487. doi: 10.1146/annurev.pathol.3.121806.151422. [DOI] [PubMed] [Google Scholar]
- 2.Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11:103–105. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Ralhan R, Kaur J, Kreienberg R, Wiesmuller L. Links between DNA double strand break repair and breast cancer: accumulating evidence from both familial and nonfamilial cases. Cancer Lett. 2007;248:1–17. doi: 10.1016/j.canlet.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Gatz SA, Wiesmuller L. p53 in recombination and repair. Cell Death Differ. 2006;13:1003–1016. doi: 10.1038/sj.cdd.4401903. [DOI] [PubMed] [Google Scholar]
- 5.Lazaro-Trueba I, Arias C, Silva A. Double bolt regulation of Rad51 by p53: a role for transcriptional repression. Cell Cycle. 2006;5:1062–1065. doi: 10.4161/cc.5.10.2764. [DOI] [PubMed] [Google Scholar]
- 6.Klein HL. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair (Amst) 2008;7:686–693. doi: 10.1016/j.dnarep.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arias-Lopez C, Lazaro-Trueba I, Kerr P, Lord CJ, Dexter T, Iravani M, Ashworth A, Silva A. p53 modulates homologous recombination by transcriptional regulation of the RAD51 gene. EMBO Rep. 2006;7:219–224. doi: 10.1038/sj.embor.7400587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchhop S, Gibson MK, Wang XW, Wagner P, Sturzbecher HW, Harris CC. Interaction of p53 with the human Rad51 protein. Nucleic Acids Res. 1997;25:3868–3874. doi: 10.1093/nar/25.19.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardson C, Stark JM, Ommundsen M, Jasin M. Rad51 overexpression promotes alternative double-strand break repair pathways and genome instability. Oncogene. 2004;23:546–553. doi: 10.1038/sj.onc.1207098. [DOI] [PubMed] [Google Scholar]
- 10.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 11.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 12.Shi I, Hallwyl SC, Seong C, Mortensen UH, Rothstein R, Sung P. Role of the Rad52 amino-terminal DNA binding activity in DNA strand capture in homologous recombination. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.057752. [6 October 2009, Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shivji MK, Mukund SR, Rajendra E, Chen S, Short JM, Savill J, Klenerman D, Venkitaraman AR. The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange. Proc. Natl Acad. Sci. USA. 2009;106:13254–13259. doi: 10.1073/pnas.0906208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, Sung P. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 2001;15:3308–3318. doi: 10.1101/gad.935501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Q, Kojic M, Holloman WK. DNA-binding domain within the Brh2 N terminus is the primary interaction site for association with DNA. J. Biol. Chem. 2009;284:8265–8273. doi: 10.1074/jbc.M809226200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, Albala JS, Swagemakers SM, Kanaar R, Thompson LH, Takeda S. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell Biol. 2000;20:6476–6482. doi: 10.1128/mcb.20.17.6476-6482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarsounas M, Davies AA, West SC. RAD51 localization and activation following DNA damage. Philos. Trans. R Soc. Lond. B Biol. Sci. 2004;359:87–93. doi: 10.1098/rstb.2003.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinz JM, Tebbs RS, Wilson PF, Nham PB, Salazar EP, Nagasawa H, Urbin SS, Bedford JS, Thompson LH. Repression of mutagenesis by Rad51D-mediated homologous recombination. Nucleic Acids Res. 2006;34:1358–1368. doi: 10.1093/nar/gkl020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smiraldo PG, Gruver AM, Osborn JC, Pittman DL. Extensive chromosomal instability in Rad51d-deficient mouse cells. Cancer Res. 2005;65:2089–2096. doi: 10.1158/0008-5472.CAN-04-2079. [DOI] [PubMed] [Google Scholar]
- 21.Hays SL, Firmenich AA, Berg P. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl Acad. Sci. USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson RD, Symington LS. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schild D. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics. 1995;140:115–127. doi: 10.1093/genetics/140.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin RW, Orelli BJ, Yamazoe M, Minn AJ, Takeda S, Bishop DK. RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res. 2007;67:9658–9665. doi: 10.1158/0008-5472.CAN-07-0290. [DOI] [PubMed] [Google Scholar]
- 25.Brown ET, Holt JT. Rad51 overexpression rescues radiation resistance in BRCA2-defective cancer cells. Mol. Carcinog. 2009;48:105–109. doi: 10.1002/mc.20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SA, Roques C, Magwood AC, Masson JY, Baker MD. Recovery of deficient homologous recombination in Brca2-depleted mouse cells by wild-type Rad51 expression. DNA Repair (Amst.) 2009;8:170–181. doi: 10.1016/j.dnarep.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiegant WW, Overmeer RM, Godthelp BC, van Buul PP, Zdzienicka MZ. Chinese hamster cell mutant, V-C8, a model for analysis of Brca2 function. Mutat. Res. 2006;600:79–88. doi: 10.1016/j.mrfmmm.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Vispe S, Cazaux C, Lesca C, Defais M. Overexpression of Rad51 protein stimulates homologous recombination and increases resistance of mammalian cells to ionizing radiation. Nucleic Acids Res. 1998;26:2859–2864. doi: 10.1093/nar/26.12.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim PM, Allen C, Wagener BM, Shen Z, Nickoloff JA. Overexpression of human RAD51 and RAD52 reduces double-strand break-induced homologous recombination in mammalian cells. Nucleic Acids Res. 2001;29:4352–4360. doi: 10.1093/nar/29.21.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flygare J, Falt S, Ottervald J, Castro J, Dackland AL, Hellgren D, Wennborg A. Effects of HsRad51 overexpression on cell proliferation, cell cycle progression, and apoptosis. Exp. Cell Res. 2001;268:61–69. doi: 10.1006/excr.2001.5265. [DOI] [PubMed] [Google Scholar]
- 32.Hannay JA, Liu J, Zhu QS, Bolshakov SV, Li L, Pisters PW, Lazar AJ, Yu D, Pollock RE, Lev D. Rad51 overexpression contributes to chemoresistance in human soft tissue sarcoma cells: a role for p53/activator protein 2 transcriptional regulation. Mol. Cancer Ther. 2007;6:1650–1660. doi: 10.1158/1535-7163.MCT-06-0636. [DOI] [PubMed] [Google Scholar]
- 33.Slupianek A, Schmutte C, Tombline G, Nieborowska-Skorska M, Hoser G, Nowicki MO, Pierce AJ, Fishel R, Skorski T. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol. Cell. 2001;8:795–806. doi: 10.1016/s1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- 34.Choudhury A, Zhao H, Jalali F, Al Rashid S, Ran J, Supiot S, Kiltie AE, Bristow RG. Targeting homologous recombination using imatinib results in enhanced tumor cell chemosensitivity and radiosensitivity. Mol. Cancer Ther. 2009;8:203–213. doi: 10.1158/1535-7163.MCT-08-0959. [DOI] [PubMed] [Google Scholar]
- 35.Russell JS, Brady K, Burgan WE, Cerra MA, Oswald KA, Camphausen K, Tofilon PJ. Gleevec-mediated inhibition of Rad51 expression and enhancement of tumor cell radiosensitivity. Cancer Res. 2003;63:7377–7383. [PubMed] [Google Scholar]
- 36.Lin YL, Sengupta S, Gurdziel K, Bell GW, Jacks T, Flores ER. p63 and p73 Transcriptionally Regulate Genes Involved in DNA Repair. PLoS Genet. 2009;5:e1000680. doi: 10.1371/journal.pgen.1000680. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat. Rev. Cancer. 2008;8:180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 38.Chan N, Koritzinsky M, Zhao H, Bindra R, Glazer PM, Powell S, Belmaaza A, Wouters B, Bristow RG. Chronic hypoxia decreases synthesis of homologous recombination proteins to offset chemoresistance and radioresistance. Cancer Res. 2008;68:605–614. doi: 10.1158/0008-5472.CAN-07-5472. [DOI] [PubMed] [Google Scholar]
- 39.Samowitz WS, Holden JA, Curtin K, Edwards SL, Walker AR, Lin HA, Robertson MA, Nichols MF, Gruenthal KM, Lynch BJ, et al. Inverse relationship between microsatellite instability and K-ras and p53 gene alterations in colon cancer. Am. J. Pathol. 2001;158:1517–1524. doi: 10.1016/S0002-9440(10)64102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soreide K, Janssen EA, Soiland H, Korner H, Baak JP. Microsatellite instability in colorectal cancer. Br. J. Surg. 2006;93:395–406. doi: 10.1002/bjs.5328. [DOI] [PubMed] [Google Scholar]
- 41.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 42.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 43.Modesti M, Budzowska M, Baldeyron C, Demmers JA, Ghirlando R, Kanaar R. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol. Cell. 2007;28:468–481. doi: 10.1016/j.molcel.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 44.Wiese C, Dray E, Groesser T, San Filippo J, Shi I, Collins DW, Tsai MS, Williams GJ, Rydberg B, Sung P, et al. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol. Cell. 2007;28:482–490. doi: 10.1016/j.molcel.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henson SE, Tsai SC, Malone CS, Soghomonian SV, Ouyang Y, Wall R, Marahrens Y, Teitell MA. Pir51, a Rad51-interacting protein with high expression in aggressive lymphoma, controls mitomycin C sensitivity and prevents chromosomal breaks. Mutat. Res. 2006;601:113–124. doi: 10.1016/j.mrfmmm.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 46.Song H, Xia SL, Liao C, Li YL, Wang YF, Li TP, Zhao MJ. Genes encoding Pir51, Beclin 1, RbAp48 and aldolase b are up or down-regulated in human primary hepatocellular carcinoma. World J. Gastroenterol. 2004;10:509–513. doi: 10.3748/wjg.v10.i4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin RW, Connell PP, Bishop DK. The Yin and Yang of treating BRCA-deficient tumors. Cell. 2008;132:919–920. doi: 10.1016/j.cell.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 49.Lord CJ, Ashworth A. Targeted therapy for cancer using PARP inhibitors. Curr. Opin. Pharmacol. 2008;8:363–369. doi: 10.1016/j.coph.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 50.Goggins M, Hruban RH, Kern SE. BRCA2 is inactivated late in the development of pancreatic intraepithelial neoplasia: evidence and implications. Am. J. Pathol. 2000;156:1767–1771. doi: 10.1016/S0002-9440(10)65047-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antoniou AC, Sinilnikova OM, Simard J, Leone M, Dumont M, Neuhausen SL, Struewing JP, Stoppa-Lyonnet D, Barjhoux L, Hughes DJ, et al. RAD51 135G–>C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am. J. Hum. Genet. 2007;81:1186–1200. doi: 10.1086/522611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hasselbach L, Haase S, Fischer D, Kolberg HC, Sturzbecher HW. Characterisation of the promoter region of the human DNA-repair gene Rad51. Eur. J. Gynaecol. Oncol. 2005;26:589–598. [PubMed] [Google Scholar]
- 53.Willers H, Taghian AG, Luo CM, Treszezamsky A, Sgroi DC, Powell SN. Utility of DNA repair protein foci for the detection of putative BRCA1 pathway defects in breast cancer biopsies. Mol. Cancer Res. 2009;7:1304–1309. doi: 10.1158/1541-7786.MCR-09-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 55.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodrigue A, Lafrance M, Gauthier MC, McDonald D, Hendzel M, West SC, Jasin M, Masson JY. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J. 2006;25:222–231. doi: 10.1038/sj.emboj.7600914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yonetani Y, Hochegger H, Sonoda E, Shinya S, Yoshikawa H, Takeda S, Yamazoe M. Differential and collaborative actions of Rad51 paralog proteins in cellular response to DNA damage. Nucleic Acids Res. 2005;33:4544–4552. doi: 10.1093/nar/gki766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forget AL, Bennett BT, Knight KL. Xrcc3 is recruited to DNA double strand breaks early and independent of Rad51. J. Cell Biochem. 2004;93:429–436. doi: 10.1002/jcb.20232. [DOI] [PubMed] [Google Scholar]
- 59.Deng X, Prakash A, Dhar K, Baia GS, Kolar C, Oakley GG, Borgstahl GE. Human replication protein A-Rad52-single-stranded DNA complex: stoichiometry and evidence for strand transfer regulation by phosphorylation. Biochemistry. 2009;48:6633–6643. doi: 10.1021/bi900564k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol. Cancer Res. 2009;7:1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, Willers H, Feng Z, Ghosh JC, Kim S, Weaver DT, Chung JH, Powell SN, Xia F. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol. Cell Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl Acad. Sci. USA. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 64.Martin V, Chahwan C, Gao H, Blais V, Wohlschlegel J, Yates JR, 3rd, McGowan CH, Russell P. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 2006;25:2564–2574. doi: 10.1038/sj.emboj.7601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simons AM, Horwitz AA, Starita LM, Griffin K, Williams RS, Glover JN, Parvin JD. BRCA1 DNA-binding activity is stimulated by BARD1. Cancer Res. 2006;66:2012–2018. doi: 10.1158/0008-5472.CAN-05-3296. [DOI] [PubMed] [Google Scholar]
- 66.Wray J, Liu J, Nickoloff JA, Shen Z. Distinct RAD51 associations with RAD52 and BCCIP in response to DNA damage and replication stress. Cancer Res. 2008;68:2699–2707. doi: 10.1158/0008-5472.CAN-07-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumaraswamy E, Shiekhattar R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol. Cell Biol. 2007;27:6733–6741. doi: 10.1128/MCB.00961-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J, Zou C, Bai Y, Wazer DE, Band V, Gao Q. DSS1 is required for the stability of BRCA2. Oncogene. 2006;25:1186–1194. doi: 10.1038/sj.onc.1209153. [DOI] [PubMed] [Google Scholar]
- 69.Livingston DM. Cancer. Complicated supercomplexes. Science. 2009;324:602–603. doi: 10.1126/science.1174839. [DOI] [PubMed] [Google Scholar]
- 70.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Ann. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]