Abstract
The bacterial activator protein NorR binds to enhancer-like elements, upstream of the promoter site, and activates σ54-dependent transcription of genes that encode nitric oxide detoxifying enzymes (NorVW), in response to NO stress. Unique to the norVW promoter in Escherichia coli is the presence of three enhancer sites associated with a binding site for σ54-RNA polymerase. Here we show that all three sites are required for NorR-dependent catalysis of open complex formation by σ54-RNAP holoenzyme (Eσ54). We demonstrate that this is essentially due to the need for all three enhancers for maximal ATPase activity of NorR, energy from which is used to remodel the closed Eσ54 complex and allow melting of the promoter DNA. We also find that site-specific DNA binding per se promotes oligomerisation but the DNA flanking the three sites is needed to further stabilise the functional higher order oligomer of NorR at the enhancers.
INTRODUCTION
Nitric oxide (NO) is a highly reactive radical species that is toxic to micro-organisms. It can be encountered exogenously, as a consequence of pathogen invasion, or endogenously through the process of respiratory denitrification (1,2). Escherichia coli is known to possess at least three enzymes capable of directly detoxifying NO, by utilising either NO reductase or NO dioxygenase activities (3–5). One of these systems comprises the enzyme flavorubredoxin (encoded by norV) and its associated NADH oxidoreductase (encoded by norW), which reduces the NO radical to nitrous oxide under anaerobic conditions (4,6).
Immediately upstream of the norVW genes is the divergently transcribed norR gene, which encodes an NO sensing σ54-dependent transcriptional regulator (7,8). This protein is essential for activating transcription of the norVW operon and shares ∼40% sequence homology with Ralstonia eutropha NorR, which is responsible for the transcriptional regulation of heme b3-iron NO reductase in response to NO (9). NorR has a modular domain architecture typical of bacterial σ54-dependent enhancer binding proteins (bEBPs) (10) and consists of three key domains: an N-terminal regulatory GAF (for cGMP-specific and cGMP-regulated cyclic nucleotide phosphodiesterase, Anabaena Adenylyl cyclase and E. coli transcription factor FhlA) domain containing a mononuclear ferrous iron centre that detects NO (11,12), a central AAA+ (for ATPase associated with various cellular activities) domain that interacts with σ54 and couples ATP hydrolysis to promoter DNA melting by RNA polymerase, and a C-terminal helix-turn-helix DNA-binding domain.
Previously we reported that E. coli NorR binds to three sites upstream of the norV promoter that contain inverted repeats with core consensus GT-(N7)-AC (13). These NorR-binding sites are conserved amongst the proteobacteria and are found upstream of genes encoding NorV (e.g. Salmonella typhimurium), Hmp (e.g. Pseudomonas aeruginosa) and NorA (Ralstonia eutropha). In E. coli, integration host factor (IHF) binds to the region upstream of norVW between the norV transcription start site and the NorR enhancer sites (13). In common with other σ54-dependent systems, the bending of DNA induced by IHF at this location may encourage interactions between NorR and σ54-RNA polymerase by DNA looping (14,15).
bEBPs function by coupling the energy yielded from ATP hydrolysis to the isomerisation of σ54-RNA polymerase from the closed promoter complex to the open promoter complex that is competent for transcription initiation (16–18). Oligomerisation of the AAA+ domain of bEBPs to form hexameric rings is required for the formation of a functional ATPase (19–21) but not for ATP binding (18,22). In some cases the ATPase activity of bEBPs is not only regulated in response to environmental signals but is also responsive to interaction with specific enhancers (23–25).
We previously demonstrated that the ATPase activity of NorR is enhancer DNA-dependent in vitro (11). Here we report that each of the three NorR enhancer sites upstream of the norV promoter is essential for transcriptional activation. We demonstrate that this is a consequence of the stringent requirement for the three enhancers for maximal ATPase activity of NorR through promoting and stabilising a functional higher order oligomer of the activator. We use a GAF domain deleted form of NorR (NorRΔGAF), which can activate transcription in the absence of NO, to correlate ATP hydrolysis with higher order oligomer formation. We also suggest that the NorR oligomer formed at the three enhancers is stabilised by the wrapping of flanking DNA around the higher order oligomer identified in negative-stain electron microscopy images.
MATERIALS AND METHODS
Protein purification
E. coli NorRΔGAF was over-expressed and purified as described previously (11). NorR178–452 AAA+ domain was generated from plasmid pNorR (13) that expresses NorR residues from 178 to 452 with a N-terminal 6-histidine tag in pET28b. The protein was purified by nickel affinity chromatography and gel filtration. The His-tag was removed by thrombin cleavage for 3 h at 23°C (38). Purified proteins were stored in buffer containing 100 mM Tris–HCl pH 8, 150 mM NaCl, 4 mM DTT and 5% glycerol at −80°C. Protein concentrations were determined by the Folin-Lowry method (26).
Construction of plasmids and site-directed mutagenesis
The pNPTfus series plasmids designed for use with lacZ fusion experiments contain either a wild type or a mutagenised norR/norV intergenic region cloned into the SmaI site of pUC19 (Table 1). Mutations were generated using a PCR based mutagenesis method (27). Wild type constructs were PCR amplified using the external primers only:
norRfusion (external) 5′-GGCGCTGAAAACGATCCTGG-3′,
norVfusion (external) 5′-TCACGCACTTCCCAGTCACG-3′.
Table 1.
Strains and plasmids
| Description | Reference/source | |
|---|---|---|
| Plasmid | ||
| pRS551 | Vector for construction of lacZ promoter fusions. | (28) |
| pNorR | Derivative of pET21a expressing norR | (13) |
| pNPTfusV | pUC19 carrying the norRV intergenic region for norV promoter fusions | This work |
| pNPTfus1V | As pNPTfusV but NorR site 1 mutated to GG(N7)CC | This work |
| pNPTfus2V | As pNPTfusV but NorR site 2 mutated to GG(N7)CC | This work |
| pNPTfus3V | As pNPTfusV but NorR site 3 mutated to GG(N7)CC | This work |
| pRS551-wtV | Fusion vector constructed from pNPTfusV | This work |
| pRS551-1V | Fusion vector constructed from pNPTfus1V | This work |
| pRS551-2V | Fusion vector constructed from pNPTfus2V | This work |
| pRS551-3V | Fusion vector constructed from pNPTfus3V | This work |
| Strain | ||
| DH10B | mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ- rpsL nupG | Invitrogen |
| MC1000 | araD139 Δ (araABC-leu)7679 galU galK Δ(lac)X74 rpsL | (44) |
| MH1003 | DH10B norR::cat λnorV-lacZ | (7) |
| MC100V | MC1000 λnorV-lacZ | This work |
| MC101V | MC1000 λnorV-lacZ NorR site 1 mutant | This work |
| MC102V | MC1000 λnorV-lacZ NorR site 2 mutant | This work |
| MC103V | MC1000 λnorV-lacZ NorR site 3 mutant | This work |
Mutant constructs were amplified using combinations of the external primers and the following mutagenic internal primers:
Site1− 5′-TAATGAGTAGGCAAAATGCCTATCAATC-3′,
Site1+5′-GATTGATAGGCATTTTGCCTACTCATTA-3′,
Site2− 5′-ATCAAATGGGCGATATGCCAATATCT-3′,
Site2+5′-AGATATTGGCATATCGCCCATTTGAT-3′,
Site3−5′-ATCTATAGGCAAATTGCCAGTGAGGCAAAG-3′,
Site3+5′-CTTTGCCTCACTGGCAATTTGCCTATAGAT-3′.
To construct lacZ fusions in the E. coli chromosome, wild type and mutant norR-norV intergenic region constructs were cloned from pNPTfus series plasmids into pRS551 (28) using EcoRI and BamHI to generate lacZ fusions. Derivatives of pRS551 were transformed into E. coli strain MC1000. The lacZ fusion constructs were then crossed into phage λRS45 by homologous recombination and transferred into the MC1000 chromosome at the phage λ attachment site as described previously (28).
β-galactosidase assays
Derivatives of E. coli strain MC1000 containing either the wild type promoter or mutant promoters upstream of the norV-lacZ reporter (Table 1) were grown either in LB medium aerobically or in LB medium supplemented with 1% glucose when grown anaerobically. The cultures were grown to an OD600 of ∼0.6 nm before being induced with potassium nitrite to a final concentration of 4 mM. Cultures were then grown for a further 2 h to allow for expression of the norV-lacZ promoter fusion construct.
Methylation protection footprinting
DNA fragments for footprinting reactions were prepared as described previously (13) but using plasmids pNPTfusV, pNPTfus1V, pNPTfus2V or pNPTfus3V. Binding reactions were carried out in DMS buffer (50 mM sodium cacodylate, 1 mM EDTA) with 0.5 μg of 5′ end-labelled EcoRI–BamHI restriction fragments and the indicated concentration of NorRΔGAF in a final volume of 200 μl. Salmon sperm DNA 2 μg was also present in reaction. Reactions were incubated for 10 min at room temperature, then 5 μl of 10% dimethyl sulphate (Sigma, in ethanol) was added to each binding reaction and incubation continued for a further 5 min. Reactions were stopped with 50 μl of ice cold DMS stop buffer (1 M β-mercaptoethanol, 1.5 M sodium acetate, 1 mg/ml glycogen) and the DNA was then precipitated by the addition of 750 μl of ice cold ethanol, followed by centrifugation at 13 000 rpm in bench-top centrifuge. After a wash with 0.3 M sodium acetate, the DNA was subjected to a second ethanol precipitation and then treated with 1 M piperidine (Sigma) for 30 min at 100°C. The samples were then lyophilised and subjected to two cycles of resuspension in 20 μl of sterile water followed by further lyophilisation. After the final lyophilisation, 10 μl of formamide loading dye was added to each sample followed by resolution on a 6% polyacrylamide sequencing gel.
Coupled ATPase activity assay
For experiments with low protein concentrations, ATPase activity was measured using an assay in which production of ADP is coupled to the oxidation of NADH by lactate dehydrogenase and pyruvate kinase (29). The oxidation of NADH was monitored at 340 nm at 37°C. All reaction mixtures contained ATP (30 mM), phosphoenolpyruvate (1 mM), NADH (0.3 mM), pyruvate kinase (7 U, Roche), lactate dehydrogenase (23 U, Roche) in 50 mM Tris–HCl (pH 8.0), 100 mM KCl, 2 mM MgCl2 and 300 nM of NorRΔGAF. Either wild type pNPTfusV or a plasmid carrying a mutation in one of the NorR-binding sites were added to the reaction mixtures, since NorRΔGAF ATPase activity is enhancer-DNA dependent (30). ATPase activity was measured by observing the change in absorbance at 340 nm.
Open promoter complex and band-shift assays
Template DNA for open complex assays was obtained by digesting the plasmid pNPTfusV, pNPTfus1V, pNPTfus2V or pNPTfus3V with EcoRI and BamHI to yield a DNA fragment including the norVW promoter and upstream activator sequences. The DNA fragments were 5′ end-labelled with 32P as described above for use in gel retardation and DNA footprinting assays. Open complex formation was assayed in TAP buffer (50 mM Tris–acetate, 100 mM potassium acetate, 8 mM magnesium acetate, 3.5% polyethylene glycol 8000, 1 mM DTT, pH 7.9) and contained 1 nM template DNA, 200 nM core RNA polymerase (Epicentre Biotechnologies), 200 nM σ54, 130 nM IHF, 5 mM ATP and 0.5 mM CTP. The reaction components were pre-incubated for 10 min at 30°C, and reactions were initiated by adding NorRΔGAF to a final concentration of between 115 and 460 nM. After a further 20 min incubation at 30°C, samples were mixed with 3 µl of dye mixture containing 50% glycerol, 0.05% bromophenol blue, 0.1% xylene cyanol and 2 µg of heparin and immediately loaded onto a 4% (wt/vol) polyacrylamide gel (acrylamide/bisacrylamide ratio, 80 : 1) in 25 mM Tris–400 mM glycine, pH 8.6, which had been pre-run at 180 V at room temperature down to a constant power of 2 W. Gels were run at 150 V and were dried and exposed to autoradiograph film or a phosphorimager screen. NorRΔGAF-DNA band-shift experiments were carried out using either a 32P labelled 266 bp PCR product (primer norvfus 5′-GGCGCTGAAAACGATCCTGG-3′ and primer norRpromR 5′-GGTTGACCAACCCAATGAATG-3′) or a 66 bp fragment generated by hybridisation of oligonucleotide primers spanning the NorR-binding region (5′-TCACTGTCAATTTGACTATAGATATTGTCATATCGACCATTTGATTGATAGTCATTTTGACTACTC-3′ and its reverse complemented partner). Reactions were prepared in the same way as for the open complex assay but with NorRΔGAF and DNA only in TAP buffer. The same dilution series of a fresh preparation of NorRΔGAF was used for both experiments, which were run at the same time. Gels were quantified with a Fujix BAS1000 phosphorimager and the data was plotted using Graphpad Prism as described previously (13).
Radioactive ATPase activity assay
Reactions were performed in a 10 µl final volume in ATPase buffer (50 mM Tris–HCl pH 8.0, 50 mM NaCl, 15 mM MgCl2, 0.01 mM DTT) and different concentrations of NorRΔGAF in complex with DNA, where indicated. The mix was preincubated at 23°C for 10 min and the reaction was started by adding 3 µl of an ATP mixture [1 mM ATP and 0.6 µCi/µl of [α-32P]ATP (3000 Ci/mmol)] and incubated for different times at 37°C. We examined the enhancer DNA-dependency of NorRΔGAF ATPase using double-stranded (ds) DNA of three different lengths (21, 66 and 266 bp) containing different regions of the norR-norV intergenic region. The 21 bp fragment was generated by hybridisation of oligonucleotide primers spanning the NorR-binding site 1 (5′-GATAGTCATTTTGACTACTCA-3′ and its reverse complemented partner). Complexes tested were composed of different protein to DNA molar ratios (as indicated in figure legends). Reactions were stopped by adding five volumes of 2 M formic acid. [α-32P]ADP was separated from [α-32P]ATP using thin-layer chromatography (Polygram Cel 300 PEI). Radioactivity was detected by PhosphorImager (Fuji Bas-5000) and quantified using the AIDA image analyzer software, version 3.52 (Raytest, Straubenhardt, Germany). Reactions were stopped when ∼20% of total ATP was hydrolysed to ensure similar proportion of ADP in all reactions. The ATPase activity is expressed in turnover per minute for all experiments. The curves were fitted using the Origin 7.0 software (OriginLab Corp.). All experiments were performed at least in triplicate. Moreover, we established (data not shown) that the rate of ATP hydrolysis was linear under assay conditions.
Analytical gel filtration
NorRΔGAF (at different concentrations) was incubated with different DNA fragments where specified (at concentrations specific to the desired molar stoichiometry of the complex) for 10 min at 23°C in buffer containing 20 mM Tris–HCl pH 8.0, 100 mM NaCl, 15 mM MgCl2 and 1 mM ATP where indicated. In the presence of ATP, samples were incubated at 4°C. Samples (100 µl) were then injected onto a Superose 6 column (10 × 300 mm, 24 ml) (GE Healthcare) installed on an AKTA system (GE Healthcare), which was pre-equilibrated with the sample buffer. Chromatography was performed at 4°C at a flow rate of 0.5 ml min−1, and the column was calibrated with globular proteins: apoferritin (443 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa) and carbonic anhydrase (29 kDa). NorR178–452 (100 µM) was filtered through a Superdex 200 column (10 × 300 mm, 24 ml; GE Healthcare) in the presence and absence of 1 mM ATP at 4°C. Chromatography conditions were similar to that of NorRΔGAF.
Negative-stain electron microscopy and image processing
Two microlitre of fractions containing the NorRΔGAF-266 bp DNA complex eluted from the gel filtration column (elution peak at 9.3 ml) was adsorbed onto glow-discharged continuous carbon grids (TAAB) and stained with 2% uranyl acetate. Data were collected at 50 000× magnification using a Phillips CM200 FEG electron microscope operating at 200 kV. Micrographs were recorded directly on a 4k × 4k CCD camera (F415 from Tietz Video and Imaging Processing GmbH), giving a pixel size of 1.76 Å. Digitised images were then coarsened by a factor of two giving a pixel size of 3.52 Å per pixel. Ten-thousand particles were picked automatically using the IMAGIC-5 software (31). Particles were windowed into 128 by 128 pixel boxes, extracted and band-pass filtered between 170 and 20 Å. Poor-quality particles were removed before reference free alignment to a total sum of the dataset was carried out. Initial class averages were generated by classification based on multi-variate statistical analysis (MSA). Strong class-averages were then used as references for multi-reference alignment (MRA) (32,33) using selected class averages as new references. The quality of the alignment was assessed by the class averages produced and the individual aligned images in each class. Multiple iterations of MRA, MSA and classification were performed with the selected new class-averages used as references for subsequent rounds of MRA. Poor-quality particles were removed throughout the alignment procedure based on alignment shifts and visual inspection of the particles within each class. The final class averages were generated from 5000 particles which were classified into 500 classes.
RESULTS
All three NorR enhancer sites are required for activation of norV expression in vivo
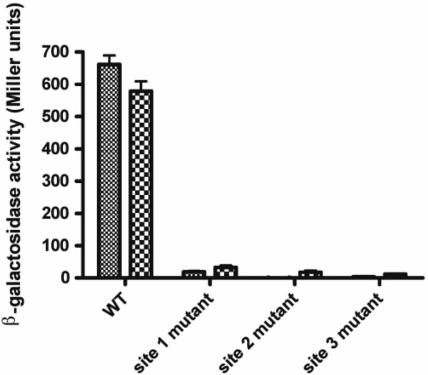
In order to assess the importance of each NorR-binding site, the enhancers were individually altered from the consensus sequence GT-(N7)-AC to GG-(N7)-CC and then introduced as norV-lacZ promoter fusions into the E. coli chromosome at the phage lambda insertion site. To activate NorR, cultures grown either under aerobic or anaerobic conditions were treated with potassium nitrite for 2 h to induce endogenous NO production. β-Galactosidase assays were then performed to determine the level of norV-lacZ expression. In agreement with previous microarray data (34) and the observation that NorR is competent to activate transcription in the presence of oxygen, we observed almost identical levels of norV expression in cultures grown either in aerobic or anaerobic conditions (Figure 1). Disruption of any one of the three NorR-binding sites was sufficient to completely prevent norVW expression. Thus, all three enhancer sites must be intact to facilitate NorR-dependent activation of norVW transcription in vivo.
Figure 1.
Effect of NorR-binding sites on activation of a norV-lacZ reporter. Strains MC100V, MC101V, MC102V and MC103V (Table 1) were grown under either aerobic or anaerobic conditions and induced with potassium nitrite (4 mM). norV-lacZ expression was then determined by measuring β-galactosidase activity. β-galactosidase activity was minimal in MC1000 lacking a promoter fusion construct.
NorR binding is disrupted at mutant, but not wild type enhancer sites in vitro
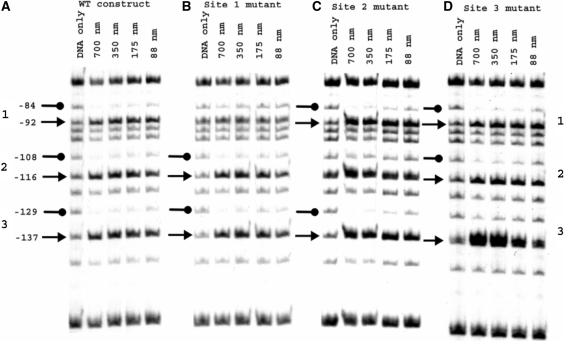
To determine the influence of the enhancer mutations on the interaction of NorR with the promoter, methylation protection footprinting was carried out with a 266 bp DNA fragment that contains all three NorR-binding sites. For these experiments and all subsequent biochemical assays described here, we used a truncated derivative of NorR (NorRΔGAF) that retains the AAA+ and the DNA-binding domains, but lacks the NO responsive GAF domain. We have demonstrated previously that NorRΔGAF activates norVW transcription in the absence of NO both in vitro and in vivo (11). The DNA-binding characteristics of NorRΔGAF are similar to that of wild-type NorR (data not shown). As observed previously, the interaction of NorR with the three enhancer sites in the wild-type promoter was manifested by protection and enhancement of G residues between position −137 and −84 relative to the norV transcript initiation site (13). In the case of the site 1 mutant, protection was observed at sites 2 and 3 but was absent at site 1 as anticipated (Figure 2B). Similarly, with either the site 2 or 3 mutant, protection by NorRΔGAF was lost at the mutant site but was maintained at the two remaining wild type enhancer sites (Figure 2C and D). However, some methylation enhancement was detectable in most cases at the mutant sites. If NorR binding to the three sites is strongly co-operative it would be expected that disruption of a single site would influence binding to the wild-type sites. As this was not evident from the methylation protection experiments, the mutations apparently prevent binding of NorR to each of the mutant sites, without discernable loss of binding to the remaining wild-type sites.
Figure 2.
Methylation protection footprinting of wild type and mutant norR-norV intergenic region constructs with NorRΔGAF. Footprinting reactions contained 32P-labelled 362-bp DNA fragments from pNPTfusV series plasmids (Table 1) spanning the norR-norV intergenic region 5′ end-labelled at the EcoRI end. Fragments encoded either the wild type promoter (A) or promoter constructs with NorR-binding site mutations at site 1 (B), site 2 (C) or site 3 (D). NorRΔGAF-DNA-binding reactions were treated with dimethyl sulphate. In each case, reactions carried out in the absence of protein are labelled ‘DNA only’. Other reactions contained between 88 and 700 nM NorRΔGAF as indicated. Residues are numbered relative to the norV transcript start site. Protected G residues are marked with lollipops on the left-hand side of each series. Arrows denote enhanced methylation at G residues.
All three enhancer sites are required for stimulating the ATPase activity of NorRΔGAF
Our previous investigations have demonstrated that activation of the ATPase activity of NorR not only requires the binding of NO to the Fe(II) centre in the GAF domain but also requires specific DNA containing the three enhancer sites in the norR-norV intergenic region [(11) and data unpublished]. This suggests that in the absence of NO, the GAF domain represses the ATPase activity of the AAA+ domain and that binding to specific DNA targets is also required for ATPase activation. In the case of the truncated NorRΔGAF protein, the requirement for NO-dependent signal activation is relieved, but the presence of DNA containing the norR-norV intergenic region is still required for significant ATPase activity (11). This further suggests that binding to the enhancer sites is necessary to activate the ATPase activity of NorR.
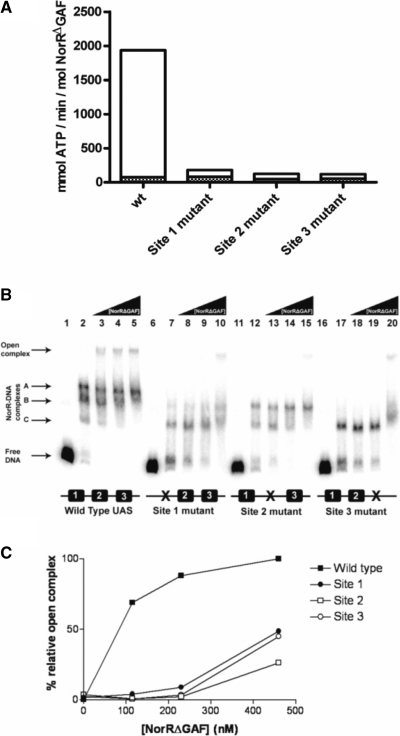
In order to investigate the role of individual enhancer sites in activation of the ATPase activity, we assayed ADP release in the presence and absence of DNA fragments carrying mutations in each of the NorR-binding sites. In the absence of promoter DNA, NorRΔGAF exhibited a low level of ATP hydrolysis (<50 mM ATP min−1 mol −1 NorR) (Figure 3A, solid bars). This activity was stimulated ∼40-fold in the presence of a DNA fragment containing all three enhancer sites as observed previously (11). However, the stimulation of the ATPase activity of NorRΔGAF was significantly reduced in the presence of DNA fragments carrying mutations in each of the NorR-binding sites (Figure 3A, open bars), and in each case the DNA stimulation was <2-fold. These results imply that under these conditions, at relatively low protein concentrations (300 nM), NorRΔGAF must be bound to all three enhancer binding sites to catalyse ATP hydrolysis.
Figure 3.
ATPase and open complex stimulating activities of NorRΔGAF associated with wild type and mutant promoter DNA constructs. (A) Rates of ATP hydrolysis by NorRΔGAF were monitored at 340 nm for 20 min at 37°C (filled bars) after which 5 nM of the wild type norR-norVW fragment was added, and the rates were monitored for a further 20 min at 37°C (empty bars). ATPase activities of NorRΔGAF in the presence of DNA constructs carrying a mutant NorR-binding site were compared with the wild type construct. ATPase activities are expressed as specific activity relative to protein concentration in mmol ATP min−1 mol NorR−1. The bar representation is a sum of the values obtained without and with DNA. (B) Each open complex assay reaction contained 1 nM of a 32P-labelled DNA fragment encoding either the wild type or mutant norR-norVW intergenic region. All lanes contained the components required for open complex formation except lanes 1, 6, 11 and 16, which contained DNA only and lanes 2, 7, 12 and 17, which contained 115 nM NorRΔGAF but lacked σ54. NorRΔGAF concentrations were 115 nM (lanes 3, 8 13, 18), 230 nM (lanes 4, 9, 14, 19) and 460 nM (lanes 5, 10, 15, 20). Free DNA, NorRΔGAF/DNA and open complexes are indicated by the arrows to the left of the figure. (C) Heparin resistant open complex species were quantified using a Fujix BAS 1000 phosphorimager. Bands were quantified by their intensity relative to the open complex band formed with the wild type DNA construct at 460 nM NorRΔGAF, which was assumed to be 100%.
Influence of NorR-binding sites on the formation of open promoter complexes by σ54-RNA polymerase
Conformational changes in bEBPs derived from ATP hydrolysis are coupled to the restructuring of the σ54-RNA polymerase–promoter complex to drive the transition from closed to open complexes. In order to correlate the effects of enhancer-binding on ATP hydrolysis with the ability of NorRΔGAF to remodel the σ54-RNA polymerase, we performed open complex assays with wild-type and mutant norV promoter DNA templates. We assayed open promoter complex formation by NorRΔGAF on wild type and mutant promoter templates in the presence of σ54-RNA polymerase, IHF, CTP and ATP (11) and quantified the heparin resistant species resolved on non-denaturing polyacrylamide gels (35). Although the binding of NorR to enhancer binding sites is itself heparin resistant, the mobility of nucleoprotein complexes formed upon open complex formation is considerably slower and such complexes can be detected as a super-shifted species (11). At low concentrations of NorRΔGAF (115 and 230 nM), open complex formation was only observed on the DNA fragment with the three wild type NorR enhancer sites (Figure 3B, lanes 3 and 4). In contrast, reactions containing DNA fragments with mutant NorR sites only exhibited open complex formation at concentrations of 460 nM NorRΔGAF or above (Figure 3B, lanes 10, 15 and 20). In this case, activation may occur in trans since open complex formation is possible in the absence of IHF at relatively high NorRΔGAF concentrations (data not shown). In control reactions carried out with 460 nM NorRΔGAF, but lacking the σ54 subunit, no open complex formation was observed, as expected (Figure 3B, lanes 2, 7, 12 and 17).
Super-shifted species were quantified as a percentage of the open complexes formed at the wild type norV promoter at 460 nM NorRΔGAF (Figure 3C). This analysis confirmed that the level of open complexes formed at the mutant promoter constructs was minimal compared to the wild type at relatively low protein concentrations (<200 nM) but increased to 50% or less of the wild type at 460 nM NorRΔGAF. At this relatively high concentration it is possible that activation of promoter bound σ54-RNAP by NorRΔGAF occurs from solution as observed with other σ54-dependent activators (17,36).
Three shifted bands (other than the super-shifted species) were visible in reactions containing the wild type DNA fragment (Figure 3B, lanes 2–5). These bands were observed in the absence of IHF and σ54-RNA polymerase (data not shown) and are presumably heparin resistant NorR–DNA complexes. The presence of more than one band is indicative of partial occupancy of the enhancer sites at relatively low protein concentrations under these buffer conditions. We assume that the three bands result from the binding of NorR to one, two or all three sites (labelled as NorR–DNA complexes A, B or C in Figure 3B). It is notable that bands B and C were evident in reactions carried out with the site 2 mutant DNA fragment whereas band C was the main species visible with fragments containing the site 1 or site 3 mutations (Figure 3B, lanes 7–10, 12–15 and 17–20, respectively). Therefore the latter mutations apparently influence the binding of NorR to adjacent wild-type enhancer sites in these heparin challenge experiments, thus providing some evidence for cooperative binding.
NorR functions as an oligomer in vitro
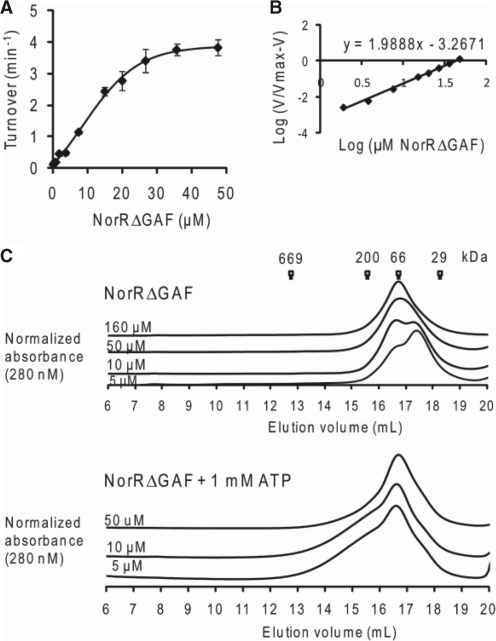
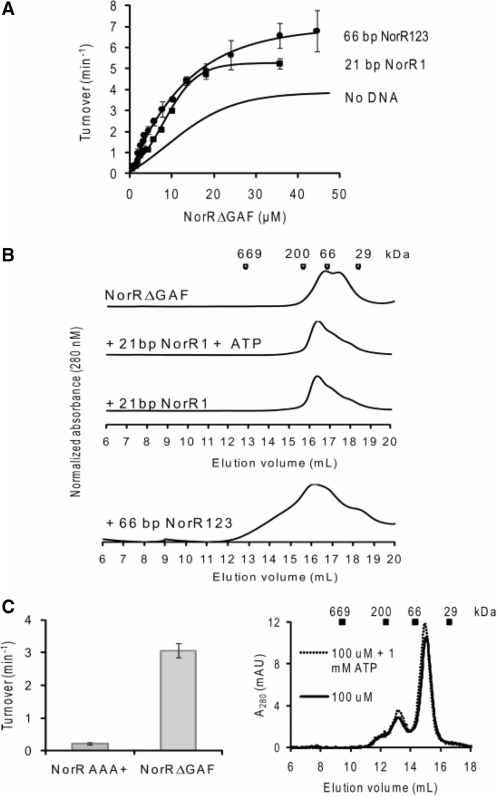
The prerequisite of all three enhancer sites for maximal ATP hydrolysis by NorR can be interpreted as the requirement of three functional dimers to form an active hexamer, given the dyad axes of symmetry of the individual sites. A number of well characterised bEBPs form hexameric ring assemblies in their active state, and this oligomerisation itself can depend upon nucleotide (ATP or ADP) binding or self-association at high protein concentrations (37). To determine if higher order oligomer formation is necessary for NorR ATPase activity, we assayed radioactively labelled ADP release from [α-32P]ATP at different concentrations of NorRΔGAF. The relationship between ATP turnover and the NorRΔGAF concentration is shown in Figure 4A. We obtained a concentration dependent sigmoidal activity curve (Figure 4A), with a Hill coefficient of 2 (Figure 4B), implying that cooperativity between monomers is required for maximum ATPase activity of NorRΔGAF. The maximum turnover (kcat), expressed in terms of NorRΔGAF monomer, was 3.8 min−1, and the amount of NorRΔGAF required to achieve half-maximal ATPase activity, Keff, was 12.5 μM (Figure 4A).
Figure 4.
ATPase activity and gel filtration profile of NorRΔGAF. (A) Plot of ATP turnover versus protein concentration, measured at a fixed concentration of ATP as substrate (1 mM). (B) Log of relative ATP hydrolysis rate V (compared to maximum rate Vmax at 47 μM) was plotted against the log of NorRΔGAF concentration. The slope of the linear regression slope was used to determine the Hill coefficient (1.98). (C) Different concentrations of NorRΔGAF were chromatographed at 4°C (upper) or preincubated with 1 mM ATP and chromatographed in the presence of 1 mM ATP at 4°C (lower). Corresponding molecular weight of standard globular proteins were indicated at their elution volume.
To examine the effect of physiological concentrations of ATP on oligomer formation, we performed analytical gel filtration experiments with various concentrations of NorRΔGAF in the presence and absence of 1 mM ATP at 4°C. Based on reference elution volumes obtained with different protein standards NorRΔGAF exhibited a concentration-dependent elution profile, with the dimer form predominating at high protein concentrations (Figure 4C, upper panel). However, in the presence of ATP, the dimer peak broadened towards higher oligomeric forms, independent of the protein concentration (Figure 4C, lower panel). These observations suggest that under these conditions, ATP binding promotes self association of NorRΔGAF as inferred from gel filtration profile and concentration-dependent ATPase activity (Figure 4).
Binding to a single enhancer site can stimulate NorRΔGAF ATPase activity in vitro
Our data so far demonstrate that the ATPase activity of NorRΔGAF is strongly stimulated by binding to DNA containing all three enhancer sites. This influence of DNA on ATP hydrolysis can be either due to DNA binding per se and/or due to increased protein concentration and propensity for stable oligomer formation at the enhancer sites.
To determine the effects of DNA binding per se on the state of association and ATPase activity of NorRΔGAF, we used a 21 bp oligonucleotide containing site 1 (NorR1), which apparently has a stronger binding affinity for NorR than either site 2 or site 3 (13). To determine the optimal molar ratio between NorRΔGAF and DNA, ATP turnover was measured at different protein:DNA molar ratios while maintaining NorRΔGAF at 35.6 μM and ATP at 1 mM (Supplementary Figure S1A, left panel). A 1:1 molar ratio gave maximal activity and was chosen for subsequent experiments in which we measured ATPase activity at various NorRΔGAF concentrations. In the presence of the NorR1 oligonucleotide, the ATPase activity displayed the same sigmoidal kinetics with respect to NorRΔGAF concentration (Figure 5A) while the Keff reduced from 12.5 µM with NorRΔGAF alone to 8.4 μM in the presence of the 21 bp DNA fragment (Figure 5A), suggesting that DNA binding increases the tendency of NorRΔGAF to self-associate. Interestingly, binding of NorRΔGAF to the 21 bp DNA fragment also increased the kcat from 3.8 to 5.2 min−1, implying allosteric stimulation of the intrinsic ATPase activity.
Figure 5.
Effect of individual enhancer sites on ATPase activity and oligomerisation states of NorRΔGAF. (A) Comparison of the ATPase activity of NorRΔGAF in the absence (cf. Fig 4A) and presence of NorR1 or NorR123 dsDNA. A molar ratio of 1: 1 and 3: 1 activator monomer: DNA was maintained for complexes formed with NorR1 and NorR123 fragments, respectively. (B) Gel filtration studies of NorRΔGAF (20 μM) in the absence or presence of dsDNA as in A. Complexes were also chromatographed in the presence of 1 mM ATP, with no significant effect observed on the elution profile. (C) left: ATPase activities of 26.5 μM NorRΔGAF and NorR178–452 AAA+ domain compared. The assays were performed at 37°C, and turnover in min−1 was calculated when [α-32P]ADP formed was ∼20 % of total radiolabeled nucleotide. Right: the elution profile of NorR AAA+ domain (100 µM) gel filtered through a Superdex 200 column in the presence and absence of 1 mM ATP at 4°C. Standard globular proteins were used for calibration: thyroglobulin (669 kDa), β-amylase (200 kDa), bovine serum albumin (66 kDa) and carbonic anhydrase (29 kDa).
To confirm whether binding to the 21 bp oligonucleotide indeed promotes self-association, we analysed the gel filtration profiles of NorRΔGAF:NorR1 complexes in the presence and absence of nucleotide. The presence of the 21 bp oligonucleotide shifted the NorRΔGAF associated peak towards higher molecular mass species, independent of the presence of ATP (Figure 5B), suggesting that binding to the single enhancer site promotes oligomerisation, in agreement with the increased ATPase activity observed in Figure 5A.
We next investigated if the DNA-binding domain of NorR controls the ATPase activity of the AAA+ domain in a manner analogous to the intramolecular repression exerted by the GAF domain. In this case the stimulatory effect on the ATPase upon DNA binding might simply be due to the removal of this inhibition, similar to the activation of the GAF domain by NO binding. We compared the ATPase activity of AAA+ domain alone with that of NorRΔGAF under the same protein concentrations. Interestingly, the isolated AAA+ domain of NorR had negligible ATPase activity compared to that of NorRΔGAF (Figure 5C, left panel). This result may suggest that DNA binding actively promotes ATPase activity (probably through promoting oligomerisation of AAA+ domain) rather than simply relieving an inhibition imposed by the DNA-binding domain. Indeed the AAA+ domain fails to form higher order oligomers at high concentration and in the presence of ATP (Figure 5C, right panel).
Binding to a 66 bp fragment containing three enhancer sites (NorR123) marginally increases the ATPase activity relative to binding to a single NorR1 site
Our data show that DNA binding per se promotes self-association and ATPase activity. We next tested whether availability of the three consecutive sites further enhances the ATPase activity of NorRΔGAF. We used a 66 bp DNA fragment (NorR123), which is the minimum DNA length containing the three NorR-binding sites based on previous DNA footprinting results. ATPase activity assays were performed with increasing NorRΔGAF concentrations while maintaining a 3 : 1 activator monomer: DNA molar stoichiometry in the complex. This optimal DNA to protein molar stoichiometry was determined as described above by fixing protein and ATP concentrations and varying DNA:protein ratios for optimal ATP turnover (Supplementary Figure S1A, right panel). In the presence of this 66 bp DNA fragment, ATPase activity increased marginally to a kcat of 6.8 min−1 when compared to that with the 21 bp NorR1 oligonucleotide (Figure 5A), although the Keff remained unchanged. Size exclusion chromatography indicated that the complexes elute as multiple peaks (Figure 5B), implying a highly heterogeneous population of oligomers. Our results so far indicate that the increased ATPase activity of NorRΔGAF upon binding to enhancer DNA is largely due to DNA binding per se rather than increased local protein concentration at the three enhancer sites.
Binding to a 266 bp DNA fragment, containing the three enhancer sites, strongly stimulates the ATPase activity of NorR
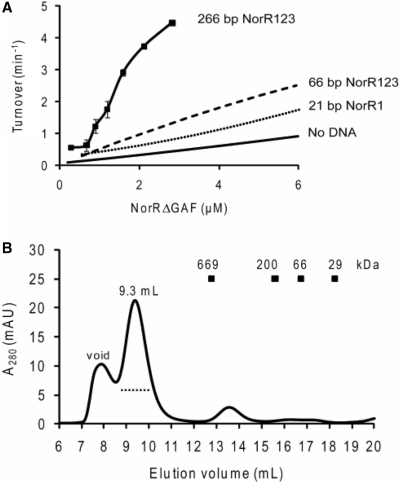
As stimulation of ATP hydrolysis by the 66 bp oligonucleotide was less than anticipated for a DNA fragment containing all three NorR enhancer sites (Figure 3A), we measured the ATPase activity of NorRΔGAF in complex with a longer (266 bp) DNA fragment. We performed ATPase assays with this fragment using a protein:DNA stoichoiometry of 6 : 1, which was again chosen based on the optimal ATPase activity at fixed protein and ATP concentrations (Supplementary Figure S1B). ATP turnover increased significantly at low protein concentrations and the Keff fell significantly to 1.4 μM when compared to that of protein alone or in complex with the 21 and 66 bp dsDNA (Figure 6A). Furthermore, the sigmoidal nature of the curve indicates clear evidence for cooperativity. Size exclusion chromatography of NorRΔGAF in complex with the 266 bp DNA fragment revealed a significant shift towards a higher molecular mass species (elution peak at 9.3 ml; Figure 6B), suggesting that this DNA fragment stabilises a higher order oligomeric form of NorRΔGAF (possibly a hexamer), in contrast to the complexes observed on the 21 and 66 bp oligonucleotides (compare Figures 5B with 6B). The presence of DNA in the peak fraction was confirmed by measuring the absorption at 280 and 260 nm wavelengths. We infer from these observations that stabilisation of a higher order oligomer on the 266 bp fragment may be responsible for the increased stimulation of the ATPase activity of NorRΔGAF.
Figure 6.
The ATPase activity and oligomerisation state of NorRΔGAF in the presence of the 266 bp dsDNA that contains all three enhancer sites. (A) Plot of ATP turnover versus NorRΔGAF concentration in the presence of 266 bp dsDNA, containing all three enhancer sites. Activity curves are also included for comparison of ATPase turnover at low concentrations in the presence and absence of shorter DNA fragments. (B) Gel filtration chromatography of 9 µM NorRΔGAF in complex with 0.75 µM 266 bp dsDNA (a molar ratio of 12 : 1 monomer: DNA) performed at 4°C using a Superose 6 column. The dotted line below the 9.3 ml elution peak represents the fractions analysed by negative-stain electron microscopy.
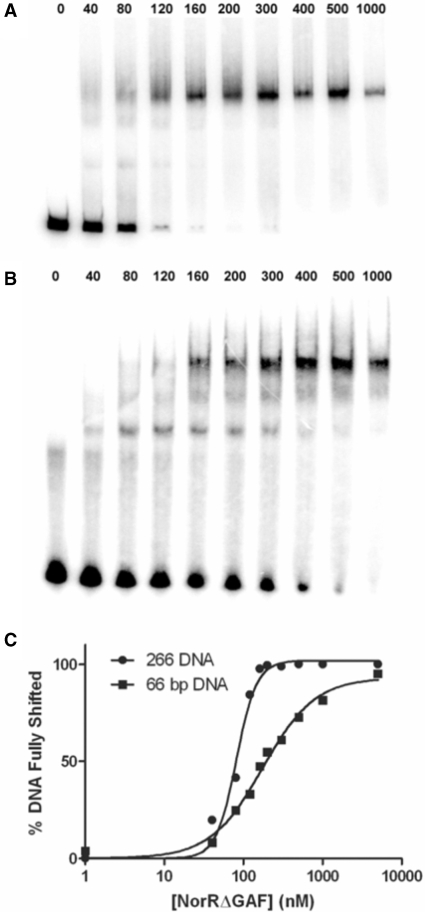
In order to further assess the differences between the 66 and 266 bp DNA, we used a gel retardation assay to compare the affinity of NorRΔGAF for these DNA fragments. Whereas only a single shifted species was observed on the 266 bp DNA (Figure 7A), partial occupancy of the enhancer sites was evident at low protein concentrations with the 66 bp DNA (Figure 7B). Quantitation of the fully shifted species showed that NorRΔGAF has a 2-fold higher affinity for the 266 bp DNA fragment (KD, 81 nM) compared to the 66 bp DNA (KD, 174 nM). Moreover, the NorRΔGAF-DNA-binding curve for the 266 bp DNA fragment exhibited increased positive cooperativity with a Hill coefficient of 3.4 compared with 1.3 for the 66 bp oligonucleotide (Figure 7C). This increase in affinity and cooperativity is consistent with a more stable hexameric ring formation, in agreement with the increased ATP turnover observed with the longer DNA fragment (Figure 6A). Although we cannot rule out the possibility that the increase in affinity is due to thermodynamic requirements for DNA recognition, it is highly likely that the affinity increase is due to additional interactions between protein and DNA, such as those encountered through the DNA wrapping around NorR. Nevertheless, under the conditions at which the ATPase activity and gel filtration were measured (micromolar concentrations), the DNA should be fully saturated with protein, thus the differences in gel filtration profile and ATPase activity (Figures 5 and 6) are likely due to the different nature of the nucleoprotein complexes formed rather than the different affinities for the protein–DNA interaction.
Figure 7.
NorRΔGAF has a higher affinity for the 266 bp DNA fragment than for the 66 bp DNA fragment NorR123. NorRΔGAF concentrations between 0 and 1000 nM (as indicated above the gels) were incubated with either the 266 bp DNA fragment (A) or the 66 bp DNA fragment (B). The percentage of fully shifted DNA was quantified using a Fujix BAS 1000 phosphorimager (C).
EM studies of NorRΔGAF bound to 266 bp NorR123 DNA
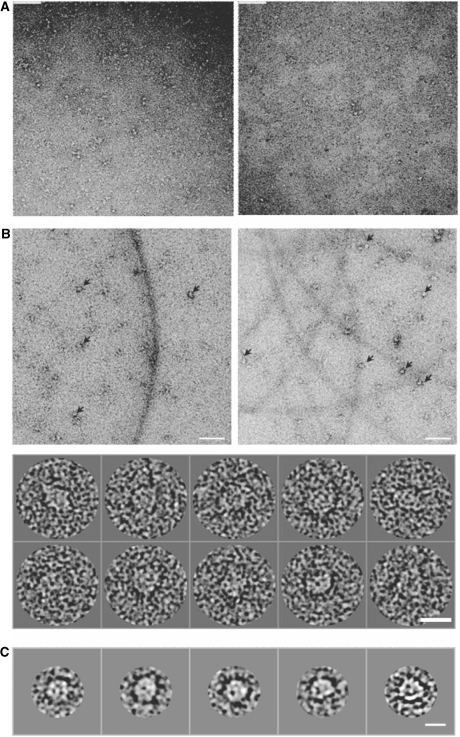
To investigate the mechanism behind the increased ATPase activity stimulated by the longer 266 bp DNA fragment, we analysed these protein–DNA complexes, prepared either in situ or after purification by gel filtration chromatography, using negatively-stained electron microscopy. As shown in Figure 8B, NorRΔGAF does in fact form high order oligomers in the presence of 266 bp DNA. Image analysis and classification of 5000 particles into class averages of 7–10 particles per class (Figure 8C) allowed visualisation of ring-shaped particles with dimensions of 126 Å in diameter, consistent with a hexameric ring observed in cryo-EM studies of other bEBPs such as PspF1–275 and NtrC (20,38). These ring shaped particles were not observed in the presence of the 21 or 66 bp oligonucleotides (Figure 8A), in agreement with the observations from size exclusion chromatography that the stable higher order oligomer is only observed upon binding of NorRΔGAF to the longer 266 bp DNA fragment (Figure 5B and 6B).
Figure 8.
Negative-stain electron microscopy of NorRΔGAF in complex with 266 bp dsDNA, containing all three enhancer sites. (A) Raw micrographs of NorRΔGAF in complex with 21 bp NorR1 (right panel) or 66 bp NorR123 (left panel) DNA fragments. Scale bar 90 nm. (B) Top, raw micrograph of NorRΔGAF in complex with 266 bp dsDNA, with arrows showing some of the higher order oligomers. Scale bar 90 nm. Bottom, gallery of picked particles. Scale bar 15 nm. (C) Selected class averages of 7–10 particles per class generated from 5000 particles show ring-shaped particles with a diameter of 126 Å. Scale bar 11 nm.
DISCUSSION
It has now been experimentally confirmed that all three NorR-binding sites are required for transcriptional activation of the E. coli norV promoter. Similar conclusions have been derived from deletion analysis of this promoter (39) and also the Ralstonia eutropha norB promoter (40). Our data extend these observations and characterise the requirement for the three enhancers in vitro. The presumed physiological role of bacterial enhancers is to tether activators at high local concentration close to the promoter and to facilitate the formation of higher oligomeric forms that are active for transcriptional activation. While multiple enhancers are common in σ54-dependent promoters, an absolute dependency on more than one target site is unusual. For example, two enhancers are sufficient to assemble higher order oligomers of NtrC, in which some protomers are bound by protein–protein interactions and apparently do not contact DNA (41). In contrast, our data for NorR indicate that all three enhancer sites are necessary for the formation of an active oligomeric species. When present at high concentration, bEBPs can activate transcription from solution in the absence of enhancer DNA in vitro and some σ54-dependent activators naturally lack a DNA-binding domain (42). Moreover, in many cases the DNA-binding domain does not appear to be essential for transcriptional activation. For example, PspF lacking the enhancer-binding domain (PspF1–275) can form high-order oligomers and activate transcription in vitro.
The increased ATPase activity of NorRΔGAF observed upon binding to the three enhancer sites could be due to one or all of the following three reasons: (i) DNA binding induces conformational changes that promote self-association and therefore increases ATPase activity, (ii) DNA binding stimulates ATPase activity per se, (iii) since there are three NorR-binding sites, binding to DNA increases the local protein concentration and thus promotes self-association, which increases the ATPase activity. Our data suggest that the significant increase in ATPase activity upon binding to enhancer sites is not likely to be a consequence of self-association resulting from an increase in local protein concentration. It is more likely due to the conformational changes induced by DNA binding that promote hexameric ring formation and ATP hydrolysis per se, therefore increasing the ATPase activity. Hence in the case of NorR, in addition to GAF domain activation, the enhancer DNA sites provide an important ligand to promote assembly of the active oligomeric form of the activator. This is in stark contrast to the activation mechanism of other bEBPs such as NtrC, where activation of the receiver domain is key to hexamer formation and ATPase activity (20). Our results are consistent with the observation that all three NorR-binding sites are required for catalytic activity. Mutating any one of the sites abolishes binding of one NorR dimer to the DNA, therefore preventing proper hexameric ring formation. However, in the absence of DNA, ATPase activity increases with protein concentration, in agreement with the observation that open complex formation can be achieved in the absence of enhancer DNA at relatively high concentrations of NorR. This demonstrates that although the native conformation in the absence of DNA is not optimal for formation of higher order oligomers (as shown in gel filtration experiments), at higher protein concentrations and in the presence of ATP, oligomerisation can occur to enable catalytic activity. However, the higher order complexes formed under these conditions are likely to be far less stable than the nucleoprotein complexes formed in the presence of the three enhancer sites. In the absence of the DNA-binding domain, the isolated AAA+ domain of NorR fails to form higher order oligomers and the ATPase activity is further reduced compared to NorRΔGAF, consistent with the hypothesis that the DNA-binding domain can promote oligomerisation, and hence ATPase activity. DNA binding appears to shift the conformation of NorRΔGAF to a form that favours hexameric ring formation at physiological protein concentrations, thus providing the necessary ATPase activity required for open complex formation.
The requirement for three consecutive enhancer sites is reminiscent of the EBP-related protein TyrR, which is proposed to bind as a dimer to three TyrR boxes, forming a hexameric species that is active in transcriptional repression (43). Another unusual feature of the NorR–DNA interaction is the heparin resistance of the protein–DNA complexes, which implies that NorR makes extensive DNA contacts, possibly forming a topologically distinct nucleoprotein complex. Our EM data show that in the presence of the 266 bp DNA fragment carrying the three enhancer sites, NorRΔGAF forms oligomeric rings, similar to other bEBPs in their active functional states. The precise mechanism whereby the enhancer DNA stabilises ring formation is currently unclear. One possible model is that the DNA wraps around the hexameric ring, making extensive contacts that help to stabilise it. Assuming that the diameter of the hexameric ring assembly of the AAA+ domain is ∼120 Å (38), a minimum of 450 Å (the total diameter to the centre of duplex DNA is 120 + 25 Å) or ∼130 bp DNA is required to wrap around the ring. The 66 bp NorR123 oligonucleotide would therefore be insufficient, consistent with our finding that the stimulatory effect of this DNA fragment, which contains all three UAS sites, is similar that of the 21 bp oligonucleotide, which carries only a single UAS site.
The assembly of bEBPs into at least a hexamer is necessary for activation of the ATPase activity required to drive the transition of the σ54-RNA polymerase promoter complex from the closed to the open DNA-melted state. Although the experiments described here have been performed with a constitutive form of NorR lacking the regulatory GAF domain, the ATPase activity of wild-type NorR is also enhancer dependent, and in addition requires the binding of NO to the ferrous iron centre to activate the catalytic activity of the AAA+ domain. Given that the NorR apoprotein is fully competent for DNA binding, the three enhancers clearly provide a scaffold for the assembly of a stable heparin-resistant NorR nucleoprotein complex that is poised at the promoter, ready to perceive the NO signal. In contrast to other EBPs such as NtrC and DctD which are dimeric in their inactive forms and are regulated through control of the oligomerisation state, the activity of wild-type NorR is apparently regulated when bound to DNA as a higher order oligomer.
In summary, our data support a unique activation mechanism for NorR. Three NorR dimers readily bind to the three consecutive UAS sites. DNA binding by NorR induces conformational changes that stimulate hexameric ring formation. NorR then forms a hexameric ring with extensive DNA interactions (possibly through DNA wrapping) for increased stability. In the presence of the NO signal, intramolecular repression of the AAA+ domain by the GAF domain is released, activating ATPase activity and allowing the NorR hexamer to interact with RNAP-σ54 and activate transcription.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biotechnology and Biological Sciences Research Council (to R.D.) (grant number BB/D009588/1); Wellcome Trust (to X.Z.) (076909/Z/05). Funding for open access charge: Biotechnology and Biological Sciences Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dan Bose for his advice and help with data collection and image processing and Nicolas Joly for his help with the ATPase assays. The authors also thank Richard Little for advice and assistance with protein purification and Stephen Spiro for his input during the early stages of this project.
REFERENCES
- 1.Zumft W. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacMicking J, Xie Qw, Nathan C. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 3.Poole RK, Hughes MN. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. MicroReview. Mol. Microbiol. 2000;36:775–783. doi: 10.1046/j.1365-2958.2000.01889.x. [DOI] [PubMed] [Google Scholar]
- 4.Gardner AM, Helmick RA, Gardner PR. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J. Biol. Chem. 2002;277:8172–8177. doi: 10.1074/jbc.M110471200. [DOI] [PubMed] [Google Scholar]
- 5.Poock SR, Leach ER, Moir JWB, Cole JA, Richardson DJ. Respiratory detoxification of nitric oxide by the cytochrome c nitrite reductase of Escherichia coli. J. Biol. Chem. 2002;277:23664–23669. doi: 10.1074/jbc.M200731200. [DOI] [PubMed] [Google Scholar]
- 6.Gomes CM, Giuffre A, Forte E, Vicente JB, Saraiva LM, Brunori M, Teixeira M. A novel type of nitric-oxide reductase. Escherichia coli flavorubredoxin. J. Biol. Chem. 2002;277:25273–25276. doi: 10.1074/jbc.M203886200. [DOI] [PubMed] [Google Scholar]
- 7.Hutchings MI, Mandhana N, Spiro S. The NorR protein of Escherichia coli activates expression of the flavorubredoxin gene norV in response to reactive nitrogen species. J. Bacteriol. 2002;184:4640–4643. doi: 10.1128/JB.184.16.4640-4643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner AM, Gessner CR, Gardner PR. Regulation of the nitric oxide reduction operon (norRVW) in Escherichia coli. Role of NorR and sigma54 in the nitric oxide stress response. J. Biol. Chem. 2003;278:10081–10086. doi: 10.1074/jbc.M212462200. [DOI] [PubMed] [Google Scholar]
- 9.Pohlmann A, Cramm R, Schmelz K, Friedrich B. A novel NO-responding regulator controls the reduction of nitric oxide in Ralstonia eutropha. Mol. Microbiol. 2000;38:626–638. doi: 10.1046/j.1365-2958.2000.02157.x. [DOI] [PubMed] [Google Scholar]
- 10.Studholme DJ, Dixon R. Domain architectures of sigma54-dependent transcriptional activators. J. Bacteriol. 2003;185:1757–1767. doi: 10.1128/JB.185.6.1757-1767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'A;utreaux B, Tucker NP, Dixon R, Spiro S. A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature. 2005;437:769–772. doi: 10.1038/nature03953. [DOI] [PubMed] [Google Scholar]
- 12.Tucker NP, D'A;utreaux B, Yousafzai FK, Fairhurst SA, Spiro S, Dixon R. Analysis of the nitric oxide-sensing non-heme iron center in the NorR regulatory protein. J. Biol. Chem. 2008;283:908–918. doi: 10.1074/jbc.M705850200. [DOI] [PubMed] [Google Scholar]
- 13.Tucker NP, D'A;utreaux B, Studholme DJ, Spiro S, Dixon R. DNA binding activity of the Escherichia coli nitric oxide sensor NorR suggests a conserved target sequence in diverse proteobacteria. J. Bacteriol. 2004;186:6656–6660. doi: 10.1128/JB.186.19.6656-6660.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoover TR, Santero E, Porter S, Kustu S. The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell. 1990;63:11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Martin J, De Lorenzo V. Integration host factor suppresses promiscuous activation of the sigma 54-dependent promoter Pu of Pseudomonas putida. Proc. Natl Acad. Sci. USA. 1995;92:7277–7281. doi: 10.1073/pnas.92.16.7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss DS, Batut J, Klose KE, Keener J, Kustu S. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- 17.Cannon WV, Gallegos MT, Buck M. Isomerization of a binary sigma-promoter DNA complex by transcription activators. Nat. Struct. Biol. 2000;7:594–601. doi: 10.1038/76830. [DOI] [PubMed] [Google Scholar]
- 18.Schumacher J, Zhang X, Jones S, Bordes P, Buck M. ATP-dependent Transcriptional Activation by Bacterial PspF AAA+Protein. J. Mol. Biol. 2004;338:863–875. doi: 10.1016/j.jmb.2004.02.071. [DOI] [PubMed] [Google Scholar]
- 19.Sallai L, Tucker PA. Crystal structure of the central and C-terminal domain of the sigma54-activator ZraR. J. Struct. Biol. 2005;151:160–170. doi: 10.1016/j.jsb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 20.De Carlo S, Chen B, Hoover TR, Kondrashkina E, Nogales E, Nixon BT. The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev. 2006;20:1485–1495. doi: 10.1101/gad.1418306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rappas M, Schumacher, Niwa H, Buck M, Zhang X. Structural basis of the nucleotide driven conformational changes in the AAA+domain of transcription activator PspF. J. Mol. Biol. 2006;357:481–492. doi: 10.1016/j.jmb.2005.12.052. [DOI] [PubMed] [Google Scholar]
- 22.Rombel I, Peters-Wendisch P, Mesecar A, Thorgeirsson T, Shin YK, Kustu S. MgATP binding and hydrolysis determinants of NtrC, a bacterial enhancer-binding protein. J. Bacteriol. 1999;181:4628–4638. doi: 10.1128/jb.181.15.4628-4638.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Austin S, Dixon R. The prokaryotic enhancer binding protein NTRC has an ATPase activity which is phosphorylation and DNA dependent. EMBO J. 1992;11:2219–2228. doi: 10.1002/j.1460-2075.1992.tb05281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter S, North A, Wedel A, Kustu S. Oligomerization of NTRC at the glnA enhancer is required for transcriptional activation. Gen. Dev. 1993;7:2258–2273. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- 25.Perez-Martin J, de Lorenzo V. In vitro activities of an N-terminal truncated form of XylR, a sigma 54-dependent transcriptional activator of Pseudomonas putida. J. Mol. Biol. 1996;258:575–587. doi: 10.1006/jmbi.1996.0270. [DOI] [PubMed] [Google Scholar]
- 26.Lowry OH, Rosenberg NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J.Biol.Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 27.Ito W, Ishiguro H, Kurosawa Y. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene. 1991;102:67–70. doi: 10.1016/0378-1119(91)90539-n. [DOI] [PubMed] [Google Scholar]
- 28.Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 29.Norby JG. Coupled assay of Na+,K+-ATPase activity. Methods Enzymol. 1988;156:116–119. doi: 10.1016/0076-6879(88)56014-7. [DOI] [PubMed] [Google Scholar]
- 30.D'A;utreaux B, Tucker N, Spiro S, Dixon R. Characterization of the nitric oxide-reactive transcriptional activator NorR. Methods Enzymol. 2008;437:235–251. doi: 10.1016/S0076-6879(07)37013-4. [DOI] [PubMed] [Google Scholar]
- 31.van Heel M, Gowen B, Matadeen R, Orlova EV, Finn R, Pape T, Cohen D, Stark H, Schmidt R, Schatz M, et al. Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- 32.Bose D, Pape T, Burrows PC, Rappas M, Wigneshweraraj SR, Buck M, Zhang X. Organization of an activator-bound RNA Polymerase holoenzyme. Mol. Cell. 2008;32:337–346. doi: 10.1016/j.molcel.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grant T. Advances in Single Particle Electron Microscopy. London: Imperial College London; 2007. [Google Scholar]
- 34.Mukhopadhyay P, Zheng M, Bedzyk LA, LaRossa RA, Storz G. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc. Natl Acad. Sci. USA. 2004;101:745–750. doi: 10.1073/pnas.0307741100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eydmann T, Söderbäck E, Jones T, Hill S, Austin S, Dixon R. Transcriptional activation of the nitrogenase promoter in vitro: adenosine nucleosides are required for inhibition of NIFA activity by NIFL. J. Bacteriol. 1995;177:1186–1195. doi: 10.1128/jb.177.5.1186-1195.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.North AK, Kustu S. Mutant forms of the enhancer-binding protein NtrC can activate transcription from solution. J. Mol. Biol. 1997;267:17–36. doi: 10.1006/jmbi.1996.0838. [DOI] [PubMed] [Google Scholar]
- 37.Joly N, Schumacher J, Buck M. Heterogeneous nucleotide occupancy stimulates functionality of phage shock protein F, an AAA+ transcriptional activator. J. Biol. Chem. 2006;281:34997–35007. doi: 10.1074/jbc.M606628200. [DOI] [PubMed] [Google Scholar]
- 38.Rappas M, Schumacher J, Beuron F, Niwa H, Bordes P, Wigneshweraraj S, Keetch CA, Robinson CV, Buck M, Zhang X. Structural insights into the activity of enhancer-binding proteins. Science. 2005;307:1972–1975. doi: 10.1126/science.1105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Justino MC, Gonçalves VMM, Saraiva LM. Binding of NorR to three DNA sites is essential for promoter activation of the flavorubredoxin gene, the nitric oxide reductase of Escherichia coli. Biochem. Biophys. Res. Commun. 2005;328:540–544. doi: 10.1016/j.bbrc.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 40.Busch A, Pohlmann A, Friedrich B, Cramm R. A DNA region recognized by the nitric oxide-responsive transcriptional activator NorR is conserved in beta- and gamma-proteobacteria. J. Bacteriol. 2004;186:7980–7987. doi: 10.1128/JB.186.23.7980-7987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyman C, Rombel I, North AK, Bustamante C, Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 42.Beck LL, Smith TG, Hoover TR. Look, no hands! Unconventional transcriptional activators in bacteria. Trends Microbiol. 2007;15:530–537. doi: 10.1016/j.tim.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 43.Wilson TJ, Maroudas P, Howlett GJ, Davidson BE. Ligand-induced self-association of the Escherichia coli regulatory protein TyrR. J. Mol. Biol. 1994;238:309–318. doi: 10.1006/jmbi.1994.1294. [DOI] [PubMed] [Google Scholar]
- 44.Casadaban MJ, Chou J, Cohen SN. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translation initiation signals. J. Bacteriol. 1980;143:971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.