Abstract
Ionising radiation induces clustered DNA damage sites which pose a severe challenge to the cell’s repair machinery, particularly base excision repair. To date, most studies have focussed on two-lesion clusters. We have designed synthetic oligonucleotides to give a variety of three-lesion clusters containing abasic sites and 8-oxo-7, 8-dihydroguanine to investigate if the hierarchy of lesion processing dictates whether the cluster is cytotoxic or mutagenic. Clusters containing two tandem 8-oxoG lesions opposing an AP site showed retardation of repair of the AP site with nuclear extract and an elevated mutation frequency after transformation into wild-type or mutY Escherichia coli. Clusters containing bistranded AP sites with a vicinal 8-oxoG form DSBs with nuclear extract, as confirmed in vivo by transformation into wild-type E. coli. Using ung1 E. coli, we propose that DSBs arise via lesion processing rather than stalled replication in cycling cells. This study provides evidence that it is not only the prompt formation of DSBs that has implications on cell survival but also the conversion of non-DSB clusters into DSBs during processing and attempted repair. The inaccurate repair of such clusters has biological significance due to the ultimate risk of tumourigenesis or as potential cytotoxic lesions in tumour cells.
INTRODUCTION
Ionising radiation (IR) induces clustered DNA damage, defined as two or more lesions within one or two helical turns of the DNA double-helix by a single radiation track. Clustered damage is thought to be a unique feature of IR and some radiomimetic drugs. Clustered damage sites induced in cells by radiation are predicted to consist of modified bases, with 8-oxo-7,8-dihydroguanine (8-oxoG) and thymine glycol being two of the major oxidatively generated lesions induced by radiation in mammalian cells (1,2), along with apurinic/apyrimidinic (AP) sites, 2-deoxyribonolactone, single-strand breaks (SSBs) or double-strand breaks (DSBs) (3–6). AP sites are thought to arise from scission of a weakened glycosidic bond resulting from radical induced nucleobase modifications (5,6). Damage complexity increases with an increase in the linear energy transfer (LET) of the radiation (7). Biophysical modelling studies have shown that with low LET radiation, such as γ-irradiation, 30% of DSBs formed are complex, where at least one lesion is found in close proximity to the DSB terminus. High LET, however, such as α-radiation, results in a yield of complex DSBs of over 90% (8–10). Several groups have confirmed that exposure of mammalian cells to IR results in the formation of non-DSB clustered damage, where a mixture of base modifications and SSBs are found in close proximity to each other, with yields that are 4–8 times greater than that of DSBs (11–14). It was hypothesised (8) that clustered DNA damage sites, including complex DSB, are more difficult to repair than endogenously induced damage formed as isolated lesions, and as such are the major determinant of the biological effects of IR. The likelihood of clustered damage sites arising endogenously is low (15,16).
The cell predominantly uses the base excision repair (BER) pathway to repair base lesions, AP sites and SSBs which are induced in cells either endogenously or by IR [reviewed in refs. (17–19)]. Several studies using models of clustered DNA damage sites have verified experimentally that the repair of lesions within a cluster occurs by BER (20–22), although depending on the orientation of the lesions to each other, the efficiency of BER may be reduced. Using cell extracts it has been shown that the repair of lesions within a bistranded clustered damage site depends upon the orientation of the lesions and whether repair occurs mainly by short patch BER or if there is a significant contribution from the long patch BER pathway (23–29).
Using purified proteins, the repair of lesions found within clustered damage sites not only depends upon the orientation of the lesions to the other vicinal lesions, but also upon the types of lesions present in the cluster and the interplay between the different repair activities in the processing of the various lesions in the cluster (20–22,27,30–35). A hierarchy of repair was initially noted (31,32,35) with both mammalian cell extracts and purified bacterial proteins with an AP site being more readily cleaved than base lesions; the resulting SSB retards excision of any base modifications within ∼5 base pairs when present within the cluster until the SSB has been repaired (24–37). With the exception of clusters containing bistranded AP sites (25,29,30,35,36,38,39) this hierarchy limits the formation of DSBs which may lead to chromosome rearrangements and/or be lethal. However, when two AP sites are present in a bistranded cluster (25,30,35,36,38,39) both AP sites are efficiently cleaved by AP endonuclease to form a DSB unless they are positioned within three base pairs 5′ to each other. In this orientation the SSB resulting from incision of one of the AP sites then retards the incision of the other AP site (25,30,35).
The existence of a hierarchy of repair has the advantage of preventing DSBs but due to the retardation of excision of other types of lesions it could lead to mutations by extending the lifetime of the lesions in the cluster so that they may persist through to replication in cycling cells. A hierarchy of processing has been confirmed in both prokaryotic (24,40–46) and eukaryotic cells (36,47,48), where it was found that, depending on the lesions present within a cluster, attempted repair leads to either mutation frequencies that are greater than if the lesions are present in isolation or to DSB formation. The mutation frequency of base lesions in clusters decreases with increasing interlesion distance approaching that of the isolated lesion, suggesting the lesions are conferring less retardation of repair on the other as they become further apart. Additionally, the relative abundance and efficiency of base glycosylases in Escherichia coli also play a decisive role in the initial stages of processing some clusters (28,41). MutY, the bacterial mismatch repair protein that removes adenine misincorporated opposite 8-oxoG, is the most important of the glycosylases in preventing mutations arising from 8-oxoG. When bistranded clusters containing uracil residues or AP sites are transformed into E. coli, DSB formation occurs if lesions are <7 base pairs apart, irrespective of their orientation (45), in contrast to the lack of DSB formation for a bistranded cluster containing 8-oxoG and an AP site.
The majority of previous studies have focussed on the effect of bistranded clusters. However, lesions can also occur in tandem to each other, when they are located on the same strand of DNA. In the few studies with tandem lesions containing either 8-oxoG or 5,6-dihydrouracil (DHU), retardation of incision of the second lesion occurs after incision of the first (49,50). In tandem clusters containing an AP site and a base modification, repair of the AP site is attempted prior to excision of the base which is retarded by the presence of the resulting SSB (24,51,52).
To date few studies (33,34,36,38) have investigated clusters containing more than two lesions. Clustered damage sites induced by IR, particularly those formed by more densely IR, could be more complex containing three or more lesions. Therefore, we designed synthetic oligonucleotides containing three lesions made up of AP sites and 8-oxoG at specified sites (Table 1). We have previously shown that when 8-oxoG is in tandem with an AP site that the mutation frequency of 8-oxoG is reduced (24). Based upon these findings (24) and the hierarchy of processing seen with bistranded clusters containing two lesions, we aim to gain more insight into the co-ordinated process of more complex clusters and identify which intermediates give rise to mutations or DSBs. Using cell extracts, wild-type and glycosylase-deficient strains of E. coli, we present evidence that when 8-oxoG is present in a cluster containing bistranded AP sites, DSB formation is not prevented. However, when two tandem 8-oxoG lesions oppose an AP site, DSBs are not formed but we see an elevated incidence of mutation, predominantly at the site of the more 3′ 8-oxoG.
Table 1.
Oligonucleotide sequences
| Cluster | Sequence | Strand |
|---|---|---|
| U/8-oxoG+1+5 | 5′ CTCTTAGTCAGGAATAAXTATXAATGCTGGGAGCGCAGGC | 1 |
| 3′ GAGAATCAGTCCTTATYCATACTTACGACCCTCGCGTCCG | 2 | |
| U/8-oxoG+2−2 | 5′ CTCTTAGTCAGGAATAAXTATXAATGCTGGGAGCGCAGGC | 1 |
| 3′ GAGAATCAGTCCTTATTCAYACTTACGACCCTCGCGTCCG | 2 | |
| U/8-oxoG−1−5 | 5′ CTCTTAGTCAGGAATAAXTATXAATGCTGGGAGCGCAGGC | 1 |
| 3′ GAGAATCAGTCCTTATTCATACYTACGACCCTCGCGTCCG | 2 | |
| U/U+1 8-oxoG+5 | 5′ CTCTTAGTCAGGAATAAYTATXAATGCTGGGAGCGCAGGC | 1 |
| 3′ GAGAATCAGTCCTTATYCATACTTACGACCCTCGCGTCCG | 2 | |
| U/U+2 8-oxoG−2 | 5′ CTCTTAGTCAGGAATAAXTATYAATGCTGGGAGCGCAGGC | 1 |
| 3′ GAGAATCAGTCCTTATTCAYACTTACGACCCTCGCGTCCG | 2 | |
| U/U+5 8-oxoG+1 | 5′ CTCTTAGTCAGGAATAAXTATYAATGCTGGGAGCGCAGGC | 1 |
| 3′ GAGAATCAGTCCTTATYCATACTTACGACCCTCGCGTCCG | 2 | |
| U/U-1 8-oxoG−5 | 5′ CTCTTAGTCAGGAATAAXTATYAATGCTGGGAGCGCAGGC | 1 |
| 3′ GAGAATCAGTCCTTATTCATACYTACGACCCTCGCGTCCG | 2 | |
| U/U-2 8-oxoG+2 | 5′ CTCTTAGTCAGGAATAAYTATXAATGCTGGGAGCGCAGGC | 1 |
| 3′ GAGAATCAGTCCTTATTCAYACTTACGACCCTCGCGTCCG | 2 | |
| U/U-5 8-oxoG−1 | 5′ CTCTTAGTCAGGAATAAYTATXAATGCTGGGAGCGCAGGC | 1 |
| 3′ GAGAATCAGTCCTTATTCATACYTACGACCCTCGCGTCCG | 2 | |
| Tandem 8-oxoG | 5′ CTCTTAGTCAGGAATAAXTATXAATGCTGGGAGCGCAGGC | 1 |
| 3′ GAGAATCAGTCCTTATTCATACTTACGACCCTCGCGTCCG | 2 | |
| U control | 5′ CTCTTAGTCAGGAATAAGTATGAATGCTGGGAGCGCAGGC | 1 |
| 3′ GAGAATCAGTCCTTATTCAYACTTACGACCCTCGCGTCCG | 2 | |
| No lesion control | 5′ CTCTTAGTCAGGAATAAGTATGAATGCTGGGAGCGCAGGC | 1 |
| 3′ GAGAATCAGTCCTTATTCATACTTACGACCCTCGCGTCCG | 2 |
Y represents a uracil residue and X represents an 8-oxoG lesion. The lesions on strand 1 have been given a number relating their positions to the uracil residue on the complementary strand 2. The number denotes the base separation. A positive number is given if the lesion on strand 1 is in the 5′ direction to that on strand 2 and a negative number is given if the lesion on strand 1 is in the 3′ direction to that on strand 2. The U control oligonucleotide consists of a single uracil residue on strand 2.
MATERIALS AND METHODS
Substrate oligonucleotides
The oligonucleotide sequences, as depicted in Table 1, were purchased PAGE purified from Eurogentec. Strand 1 contains two lesions: uracil (Y), 8-oxoG (X), or a mix of both at fixed positions. Strand 2 contains a single uracil at variable positions. The uracil control oligonucleotide contains a single uracil (Y) present in strand 2 and no lesion in strand 1. The no lesion control oligonucleotide does not contain any modified bases. Based on the previous nomenclature (31) lesions situated on strand 1 3′ to the single uracil found on strand 2 within the cluster would be given a negative number and 5′ lesions a positive number, the number relating to the distance, in base pairs, between the lesions.
Preparation of 5′-end-labelled oligonucleotides
Oligonucleotides (0.2 μg) were 32P 5′-end-labelled by incubating 10 U T4 polynucleotide kinase (Invitrogen) with 15 μCi of [γ-32P] ATP (6000 Ci/mmol, 10 mCi/ml, Perkin Elmer LAS) in 20 μl of buffer (70 mM Tris–HCl pH 7.6, 10 mM MgCl2, 100 mM KCl, 1 mM 2-mercaptoethanol) for 30 min at 37°C. Following purification through a G-25 column (GE Healthcare) the labelled oligonucleotide was hybridised to its complementary strand, present in a 2-fold excess, by heating at 90°C for 5 min and leaving to cool slowly over 2–3 h. Hybridisation of the oligonucleotides was verified on a 12% native polyacrylamide gel.
Preparation of 3′-end-labelled oligonucleotides
Oligonucleotides (0.2 μg) were 32P 3′-end-labelled using 30 U recombinant terminal deoxynucleotidyl transferase (Invitrogen) with 30 μCi of [α-32P] dATP (5000 Ci/mmol, 10 mCi/ml, Perkin Elmer LAS) in 40 μl buffer (100 mM potassium cacodylate pH 7.2, 2 mM CoCl2, 200 mM DTT) for 1 h at 37°C. The reaction was stopped by incubating at 70°C for 10 min. Following purification through a G-25 column (GE Healthcare), the labelled oligonucleotide was hybridised to its complementary strand, present in a 2-fold excess, by heating at 90°C for 5 min and leaving to cool slowly over 2–3 h. Hybridisation of the oligonucleotides was verified on a 12% native polyacrylamide gel.
Conversion of a uracil residue into an AP site
The double-stranded oligonucleotides containing uracil were incubated with 1 U uracil DNA glycosylase (Invitrogen) in 50 μl buffer (10 mM Tris–HCl pH 7.5, 50 mM NaCl, 1 mM EDTA) for 30 min at 37°C to convert the uracil residue into an AP site. The percentage conversion into AP sites was verified by treatment with APE1 as previously described (30). These AP site-containing oligonucleotides were freshly prepared after each 32P-labelling and found to be stable for 2 weeks stored at 4°C in Tris–EDTA buffer pH 8.0.
Preparation of nuclear extract
The nuclear extracts were prepared as previously described (31) from CHO-K1 cells. In summary, cells were harvested in exponential phase and the pelleted cells were resuspended in an equal volume of buffer (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT). To lyse the cytoplasmic membranes the cells were drawn through a 0.5 μm diameter needle 10 times. Following a brief centrifugation at 13 000 r.p.m. at 4°C the nuclei were collected and resuspended in two-third volume of high salt buffer (20 mM HEPES pH 7.9, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF) for 30 min with agitation, on ice. Following centrifugation at 13 000 r.p.m. for 10 min at 4°C, the supernatant was dialysed for 16 h against 1 l of buffer (20 mM HEPES pH 7.9, 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF). The protein concentration was determined using the Bradford colorimetric technique and was found to be between 6.6 and 11.1 mg/ml. Aliquots of nuclear extracts were stored at −80°C.
Repair assay
The double-stranded oligonucleotides (5000 c.p.m.) were incubated with 0.9 μg CHO-K1 nuclear extract in 5 μl repair buffer (70 mM Tris–HCl pH 7.5, 10 mM MgCl2, 10 mM DTT, 4 mM ATP, 40 mM phosphocreatine, 1.6 μg/ml phosphocreatine kinase, 0.1 mM each of dATP, dTTP, dCTP, dGTP) at 37°C for 0, 1, 5, 15, 30 or 60 min. To stop the reactions, 5 μl of denaturing stop solution (98% formamide, 2 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol) was added. The samples were then subjected to electrophoresis on a 12% denaturing polyacrylamide gel containing 8 M urea in 1× TBE (89 mM Tris–HCl, 89 mM boric acid, 2 mM EDTA pH 8.3) for 1 h at a constant power of 90 W. The dried gel was exposed to a Bio-Rad Phosphorimager screen for visualisation of repair products using phosphorimaging technology (Bio-Rad, Molecular Imager FX) and the image was quantified with Quantity One software (Bio-Rad, Hercules, CA). To follow the time dependence of the repair of the AP site, the intensity of the bands representing either single-stranded DNA, single-stranded DNA with one or more bases added (before ligation), or rejoined DNA (ligation of the AP site) was expressed as a percentage of the total intensities for all bands. The efficiency of repair of AP sites within a clustered damage site was compared to that of the single AP site. The error bars represent standard deviations from at least three experiments.
Assessment of DSB induction during repair
The repair assay was performed as described above but native stop solution was substituted for denaturing stop solution (40% sucrose, 5 mM EDTA, 0.025% bromophenol, 0.025% xylene cyanol). The samples were then subjected to electrophoresis on a 12% native polyacrylamide gel in 1× TBE (89 mM Tris–HCl, 89 mM boric acid, 2 mM EDTA pH 8.3) for 2 h at a constant voltage of 300 V. The dried gel was exposed to a Bio-Rad Phosphorimager screen for visualisation of repair products using phosphorimaging technology (Bio-Rad, Molecular Imager FX) and the image was quantified with Quantity One software (Bio-Rad, Hercules, CA). The intensity of the band representing the DSB was expressed as a percentage of the total intensities for the bands resulting from the DSB and intact strands. The error bars represent standard deviations from at least three experiments.
Processing of APE1-SSB by purified polymerase β
The 3′-end-labelled double-strand oligonucleotides (10 000 cpm) were incubated at 37°C for 30 min in the absence or presence of purified BER enzymes (amounts determined by titration); HAP1 alone at 25 ng, HAP1 at 25 ng plus polβ at 2 ng or HAP1 at 25 ng plus polβ at 2 ng and ligase III at 15 ng, in a 10 μl reaction containing purified protein buffer (80 mM HEPES pH 7.9, 10 mM MgCl2, 2 mM DTT, 200 mM EDTA, 4 mM ATP, 800 μg/ml bovine serum albumin, 40 mM each of dATP, dTTP, dCTP and dGTP). All samples were then treated with 2 μl 0.5 M sodium borohydride for 10 min on ice and the reaction was stopped by the addition of 10 μl denaturing stop solution (98% formamide, 2 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol). The addition of sodium borohydride stabilises the end groups of the 5′ terminus during electrophoresis. The samples were incubated at 90°C for 3 min and then subjected to electrophoresis on a 20% denaturing polyacrylamide gel containing 8 M urea in 1× TBE (89 mM Tris–HCl, 89 mM boric acid, 2 mM EDTA pH 8.3) for 90 min at a constant power of 90 W. The gel was exposed to a Bio-Rad Phosphorimager screen for visualisation and quantified as above.
E. coli strains
Isogenic strains CC104 (wild-type) (53) and BH980 (mutY::KanR) were a kind gift from Dr S. Boiteux (CEA, France). Uracil DNA glycosylase-deficient strain BW310 (54) was obtained from the E. coli Genetic Stock Center.
Preparation of oligonucleotides for E. coli reporter system
Oligonucleotides (20 pmol, see Table 1) were hybridised in Tris–EDTA buffer pH 8.0 by heating at 90°C for 5 min and left to cool slowly over 2–3 h. Oligonucleotides were then phosphorylated on the 5′ termini by incubating 10 U T4 polynucleotide kinase (Invitrogen) in 50 µl buffer (70 mM Tris–HCl pH 7.6, 10 mM MgCl2, 100 mM KCl, 1 mM 2-mercaptoethanol) and 2 mM ATP for 30 min at 37°C. Samples were purified by passage through a Qiagen nucleotide removal column as per the manufacturer’s instructions and the concentration of each double-stranded oligonucleotide was determined spectrophotometrically.
Plasmid preparation and ligation
pUC18 plasmid DNA was linearised by incubating 20 µg with 300 U SmaI (NEB) in 150 µl buffer (20 mM Tris–acetate, 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM DTT) at 25°C for 3 h. To minimise the linear plasmid rejoining, the 5′ termini were dephosphorylated by incubating the linear pUC18 with 20 U Antarctic phosphatase (NEB) in 150 µl buffer (50 mM Bis–Tris–Propane–HCl, 1 mM MgCl2, 0.1 mM ZnCl2) for 1 h at 37°C followed by heat inactivation at 65°C for 10 min. DNA was subjected to electrophoresis on a 1% agarose gel in 1× TBE (89 mM Tris–HCl, 89 mM boric acid, 2 mM EDTA pH 8.3) for 1 h at a constant 80 V. Linearised DNA was recovered using a Qiagen MinElute gel extraction kit and the concentration was determined spectrophotometrically. Aliquots of 200 fmol of linearised pUC18 plasmid DNA were ligated to 5 pmol of hybridised oligonucleotide by incubating with 5 U T4 DNA ligase (Roche) in 20 µl buffer (66 mM Tris–HCl, 5 mM MgCl2, 1 mM DTT, 1 mM ATP) at 16°C for 16 h with agitation. The control plasmids contained the undamaged oligonucleotide (no lesion control) as shown in Table 1. Samples were purified by dialysis against H2O using 0.025 µm Millipore nitrocellulose filters at room temperature for 1 h.
Transformation of electrocompetent E. coli
Aliquots of 60 ng of ligated pUC18/oligonucleotide were incubated with 60 µl of electrocompetent E. coli on ice for 10 min. The bacteria were transformed by electroporation at 1.8 kV using a Bio-Rad E. coli pulser, followed by immediate addition of 500 µl SOB before incubating for 1 h at 37°C. Transformants were selected for by plating on LB agar containing ampicillin (100 µg/ml) and incubating at 37°C for 16 h.
Sequence analysis
From the above transformations 30 colonies were used to inoculate 30 individual aliquots of 5 ml LB broth containing ampicillin (100 µg/ml) and incubated at 37°C until an OD600nm of 0.5 of the bacterial culture was reached. Sequencing was performed by GeneService on 200 µl aliquots using the following primers: forward 5′-CTTCGCTATTACGCCAGCTGGCGA-3′ and reverse 5′-GCTTTCCAGTCGGGAAACCTGTCGTGCCAGC-3′. The sequencing data was analysed using FinchTV software, available online for free download. Of the plasmids sent for sequencing, a small percentage did not contain an inserted oligonucleotide (0–6.7%) as a result of either re-ligation of the pUC18 plasmid or the insert had been removed during processing. These sequenced plasmids containing no insert were not included in the calculations of the mutation frequency.
RESULTS
Effect of two tandem 8-oxoG lesions on the repair of an opposing AP site
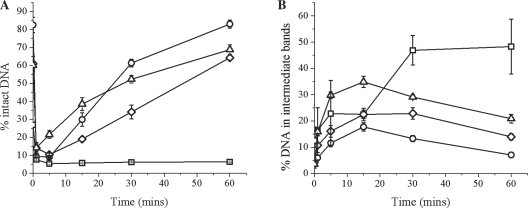
We first investigated the efficiency of repair of an AP site by CHO-K1 nuclear extract over 60 min when present in a cluster opposing two tandem 8-oxoG residues, shown in Figure 1A (for representative scan of a denaturing polyacrylamide gel see Supplementary Figure 1). For the AP control, rapid cleavage of the AP site by APE1 occurs within the first minute, seen by a sharp decrease of intact DNA followed by restoration of the full length 40 bp oligonucleotide. When two tandem 8-oxoG lesions are inserted into the opposing strand, the efficiency of cleavage of the AP site is unaffected. However, the efficiency of repair of the resulting SSB is retarded, with little or no repair seen with the AP/8-oxoG+1+5 cluster. The reduced efficiency of repair of the other two clusters leads to a lower level of rejoining. For instance, at 30 min the extent of repair of the AP site for AP/8-oxoG+2−2 and AP/8-oxoG-1−5 is 1.8 and 1.2-fold lower, respectively, than that for the AP control.
Figure 1.
Repair of an AP site when present in a cluster with two tandem 8-oxoG lesions on the opposing strand. (A) Repair of the AP site after incubation with CHO-K1 nuclear extract. (B) Accumulation of DNA in the intermediate repair bands (SSB+1 and SSB+2): (circle) AP control, (square) AP/8-oxoG+1+5, (diamond) AP/8-oxoG+2−2, (triangle) AP/8-oxoG−1−5. Error bars represent the standard error of the mean from at least three independent experiments.
Despite reduced repair of the three-lesion clusters compared to that of the AP site control, addition of bases into the break site occurs. Figure 1B shows the percentage accumulation of DNA in the intermediate repair bands (total SSB + 1 and SSB + 2). As can be seen for the control AP oligonucleotide, the amount of the SSB +1 base initially increases then declines over the time course as the full length 40mer is restored. AP/8-oxoG+2−2 and AP/8-oxoG–1−5 follow the same trend as that of the AP control but the level of accumulation is slightly increased. Both of these clusters also show a small contribution from the long patch repair pathway, as indicated by the SSB+2 band (see Supplementary Figure 1). In contrast to the other two clustered damage sites containing tandem 8-oxoG lesions, the amount of SSB+1 band with the AP/8-oxoG+1+5 cluster is 48%, compared with 7% for the AP control after 60 min. With the AP/8-oxoG+2−2 and AP/8-oxoG−1−5 clusters and the control, the amount of DNA present within the reaction as SSB+1 (and +2) decreases after the first 15 min as the intact oligonucleotide is restored (Figure 1B). In contrast, a decrease of the SSB+1 band was not seen with AP/8-oxoG+1+5, with a fraction of the SSB band persisting to 60 min.
During repair of an AP site via BER, polβ has to remove the 5′-dRP residue of the SSB created from cleavage of an AP site by APE1 to form a ligatable 5′-phosphate residue. If the dRPase activity of polβ is reduced when an 8-oxoG lesion is in the vicinity of the break, the efficiency of rejoining of the SSB would be reduced. To address this, substrates were 3′-32P-end labelled and using purified proteins it was confirmed that the dRP residue is in fact removed following incision of the AP site by APE1 in the AP site/8-oxoG clusters as efficiently as the removal of the dRP residue from the incised AP control when polβ is present (indicated by the band shift from SSB to SSB-dRP in Supplementary Figure 2). Consistent with the retarded repair (Figure 1A), little or no ligation was seen with AP/8-oxoG+1+5 when ligase III was included in the repair reaction, compared with the level of ligation seen with the other clusters. Therefore the retardation in repair of the AP site in the clustered damage is mainly due to retardation of ligation and not due to a compromise in the dRPase activity of polβ.
DSB formation from two opposing AP sites in the presence of 8-oxoG
Since the two-tandem 8-oxoG/AP site clusters do not induce DSBs in the extracts, we substituted one of the 8-oxoG lesions located on strand 1 (Table 1) for an AP site to see if these clusters give DSBs as they now contain bistranded AP sites with a vicinal 8-oxoG residue. We found that these clusters fell into two defined groups; those that promptly formed DSBs from rapid incision of both AP sites and the other subset that showed a ‘two population’ effect where the AP site in tandem with 8-oxoG is preferentially incised, and as a consequence incision of the other AP site on strand 2 is retarded.
Prompt DSB formation from rapid incision of both opposing AP sites
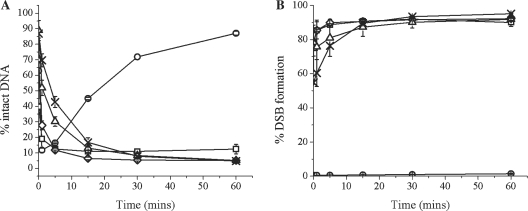
When two bistranded AP sites are five base pairs in the 5′ orientation or at any position studied in the 3′ orientation, a DSB is formed within 10-15 min due to incision of both AP sites when incubated with nuclear extract, as shown in Figure 2B (for a representative scan of a polyacrylamide gel see Supplementary Figure S3). With the clusters containing AP/AP+5 8-oxoG+1, AP/AP−1 8-oxoG−5, AP/AP−2 8-oxoG+2 and AP/AP−5 8-oxoG−1, a DSB is formed irrespective of the position of 8-oxoG within the clustered damage site.
Figure 2.
Prompt formation of DSBs where incision in clusters containing bistranded AP sites, of either of the bistranded AP sites is not inhibited. (A) Repair of the AP site after incubation with CHO-K1 nuclear extract. (B) Formation of DSBs: (circle) AP control, (square) AP/AP+5 8-oxoG+1, (diamond) AP/AP−5 8-oxoG−1, (triangle) AP/AP−2 8-oxoG+2, (cross) AP/AP−1 8-oxoG−5. Error bars represent the standard error of the mean from at least three independent experiments.
To investigate if any repair of the AP sites within these clusters occurs, either strand 1 or 2 was 32P 5′-end-labelled dependent upon which lesion was being investigated (Table 1), and incubated with CHO-K1 nuclear extract for 0–60 min. Data has only been shown for strand 2 containing the single AP site as processing of the AP site on either strand 1 or 2 is identical. A single AP site control has been included for comparison. For oligonucleotides AP/AP+5 8-oxoG+1 and AP/AP−5 8-oxoG−1 APE1 cleaves both of the AP sites within the first minute of incubation with the extract, as seen by the sharp decrease in the amount of intact DNA represented in Figure 2A, consistent with neither AP site conferring significant retardation of incision of the other AP site. With oligonucleotides AP/AP–1 8-oxoG–5 and AP/AP–2 8-oxoG+2 a slight retardation of incision of the AP site can be seen in the initial stages up to ∼20 min, shown by a 2.6- and 1.9-fold reduction, respectively, in the amount of DNA present as a SSB in the first 5 min of the assay, compared with that of the AP control. Once cleavage of the first AP site has occurred, a DSB is formed rapidly due to incision of the second AP site.
Retardation of incision of the single AP site when opposite an AP site and 8-oxoG in tandem, giving rise to ‘two populations’ of lesion
When the opposing AP sites are up to two bases apart in the 5′ orientation and an 8-oxoG lesion is present as the third lesion, namely AP/AP+1 8-oxoG+5 and AP/AP+2 8-oxoG–2 clusters, a preferential incision of the AP site found in tandem with the 8-oxoG lesion was seen, together with a reduced efficiency of incision of the single AP site on strand 2 (Table 1). This differential efficiency of incision of the AP sites gives rise to what we have termed a ‘two population’ effect. However, previous studies (25) with bistranded AP sites in the +1 orientation showed that cleavage of the AP sites is random, inferring that either of the sites could be initially cleaved.
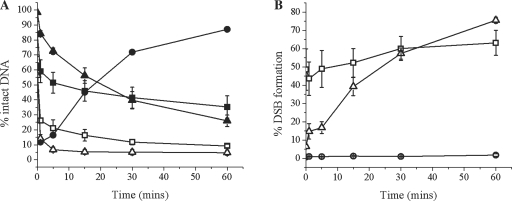
After incubation of AP/AP+1 8-oxoG+5 with nuclear extract we quantified the repair levels of each AP site. Within 1 min APE1 cleaves 70% of the AP sites found in tandem with 8-oxoG (Figure 3A and Supplementary Figure S4) and by 60 min only a residual 10% remains as 40 bp intact oligonucleotide. This is in stark contrast with the single AP site on the opposing strand where 40% of the AP sites are cleaved within 1 min and increasing by a further 25% by 60 min, leaving 35% of the single AP sites intact at 60 min (Figure 3A). As a result of the retardation of incision of the single AP site, only 63% of the clusters form a DSB within 60 min (Figure 3B). This is consistent with the level of cleavage of the single AP site leading us to conclude that the DSB forms immediately once the single AP site has been incised. The SSB formed through incision of either AP site persists as repair intermediates reflecting attempted repair of the SSB are not apparent (Supplementary Figure S4).
Figure 3.
Slow DSB formation in clusters containing bistranded AP sites, when the incision of the AP site in tandem with 8-oxoG confers retardation of incision on the opposing, single AP site. (A) Repair of the AP site after incubation with CHO-K1 nuclear extract: (filled circle) AP control, (filled square) *AP/AP+1 8-oxoG+5, (filled triangle) *AP/AP+2 8-oxoG−2, (open square) AP/*AP+1 8-oxoG+5, (open triangle) AP/*AP+2 8-oxoG−2. The 32P-labelled strand is denoted by asterisk in each case. (B) Formation of DSBs: (circle) AP control, (square) AP/AP+1 8-oxoG+5, (triangle) AP/AP+2 8-oxoG−2. Error bars represent the standard error of the mean from at least three independent experiments.
This ‘two population’ effect is initially much more pronounced in the AP/AP+2 8-oxoG–2 cluster. The cleavage of the AP site in tandem with 8-oxoG is unaffected by the presence of the AP site on strand 2 as shown by the level of incision of the AP site being 84% after 1 min (Figure 3A), comparable with cleavage of the control AP site. However, with the single AP site on the other strand, we see only 14% of the substrate cleaved within 1 min, in comparison with 84% of the AP site in tandem with 8-oxoG on strand 1. A titration with purified APE1 for 30 min revealed that 14 pmol of the endonuclease is insufficient for cleavage of the single AP site when placed in this cluster, whereas 0.3 pmol is adequate for efficient incision of a control AP site (data not shown). Therefore, the cleavage seen within 60 min is probably the result of another endonuclease, possibly NEIL-1, as this has been shown to act as an AP endonuclease independent back up pathway in some cases (55–57) and does indeed cleave the substrate AP site on strand 2 in the AP/AP+2 8-oxoG–2 cluster, after a 30 min incubation with 4.5 pmol of the enzyme (data not shown). Despite the initial higher level of retardation of cleavage of the single AP site (located on strand 2) within this cluster than seen with AP/AP+1 8-oxoG+5, the level of incision at 60 min is 74% so that a further 60% cleavage occurs. Due to the retardation of cleavage of the single AP site on strand 2 some attempted repair of the SSBs formed after cleavage of the AP site in tandem with 8-oxoG occurs, as demonstrated by the addition of a base into the break site (Supplementary Figure S4). However, no further repair intermediates were seen within 60 min. The level of DSB formation (Figure 3B) at 60 min reflects the level of cleavage of the single AP site on strand 2. As with AP/AP+1 8-oxoG+5, we deduce that DSBs are formed immediately upon cleavage of the single AP site on strand 2 since the SSB or the SSB + 1 base intermediate resulting from rapid incision of the AP site in tandem with 8-oxoG, persists up to 60 min.
The cytotoxic and mutagenic potential of the three lesion clusters when used in an E. coli reporter assay
To assess the cytotoxic and mutagenic potential of these clusters we used a plasmid-based E. coli reporter assay as previously developed in our laboratory (40). This system allows us to examine the mutation frequency of the lesions within surviving colonies by DNA sequencing through the site of the cluster. For this assay the uracil residues within the clusters were not converted to AP sites, in contrast to the in vitro repair assay where the oligonucleotides were used containing AP sites. Previous studies (40,44,45) have shown that uracil residues are rapidly converted into AP sites using this assay. Consistent with their reports we observed similar findings regardless of whether the clusters contained uracil residues or AP sites prior to transformation (data not shown). If DSB formation occurs causing the pUC18 to linearise which prevents the ampicillin resistance cassette being transcribed, a loss of colonies will be observed. Linear pUC18 used to insert the lesion-containing oligonucleotides was transformed into E. coli and the transformation efficiency was found to be extremely low (>100-fold reduction), relative to the transformation efficiency of the purified circular plasmids.
DSB formation with bistranded uracil clusters leads to colony loss
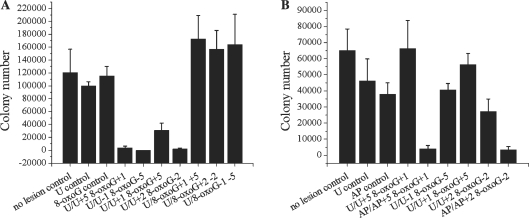
The results obtained from the repair assay showed that those clusters containing bistranded AP sites fell into two groups: those that form rapid DSBs and those that show a ‘two population’ effect. To test the E. coli response to such clusters we chose to transform both clustered damage sites that show the ‘two population’ effect, U/U+1 8-oxoG+5 and U/U+2 8-oxoG–2, and two representative clusters that rapidly form DSBs, U/U+5 8-oxoG+1 and U/U–1 8-oxoG–5. Figure 4A graphically expresses the survival of the transformants after overnight incubation. All of the tested bistranded uracil clusters show a dramatic loss of colonies, consistent with our biochemical data showing DSBs form from these clustered damaged sites. With the clusters which form DSBs (U/U–1 8-oxoG–5, U/U+2 8-oxoG–2 and U/U+1 8-oxoG+5), sequence analysis of the plasmids obtained from the few colonies surviving showed low frequencies of mutations with the most common mutations being small deletions, up to 4 base pairs in length, mainly at the AP sites as shown in Table 2. With U/U+1 8-oxoG+5, which biochemically showed a ‘two population’ effect, we observed a slightly higher colony survival than for the other three substrates which form DSBs. The increased survival may reflect some repair of the SSB prior to cleavage of the other AP site prior to replication.
Figure 4.
E. coli colony survival after transformation with clustered damage sites. (A) Colony survival after transformation of clustered DNA damage into wild-type E. coli. Mean colony numbers: no lesion control (120 650), U control (100 050), 8-oxoG control (115 650), U/U+5 8-oxoG+1 (4000), U/U−1 8-oxoG−5 (0), U/U+1 8-oxoG+5 (31 125), U/U+2 8-oxoG−2 (2000), U/8-oxoG+1 +5 (173 000), U/8-oxoG+2 −2 (157 000), U/8-oxoG−1 −5 (164 312.5). (B) Colony survival after transformation of clustered DNA damage into ung1 E. coli. Mean colony numbers: no lesion control (65 000), U control (46 333.3), AP control (38 000), U/U+5 8-oxoG+1 (66 250), AP/AP+5 8-oxoG+1 (4000), U/U-1 8-oxoG−5 (40666.7), U/U+1 8-oxoG+5 (56 333.3), U/U+2 8-oxoG−2 (27 250), AP/AP+2 8-oxoG−2 (3333.3). Clustered damage sites transformed are shown along the horizontal axes. No lesion control refers to an oligonucleotide containing no damage inserted into the plasmid before transformation. Error bars represent the standard error of the mean from at least three independent experiments.
Table 2.
Mutation frequencies of clustered damage sites in wild-type E. coli
| Oligonucleotide | Mutation | Frequency % |
|---|---|---|
| U/U-1 8-oxoG−5 | 1 bp deletion at single Ua | 3.7 |
| U/U+1 8-oxoG+5 | GC:TA transversion at U+1 | 16.0 |
| U/U+2 8-oxoG−2 | TA:GC transversion at single Ua | 4.8 |
| 2 bp deletion at single Ua | 4.8 | |
| U/U+5 8-oxoG+1 | 4 bp deletion at U+5 | 5.9 |
| U/8-oxoG+1 +5 | GC:TA transversion at 8-oxoG+1 | 3.4 |
| A:T point mutation at U | 3.4 | |
| 4 bp deletion at 8-oxoG+1 | 3.4 | |
| U/8-oxoG+2 −2 | GC:TA transversion at 8-oxoG+2 | 3.7 |
| 1 bp deletion at 8-oxoG+2 | 11.1 |
The frequency for each type of mutation was determined from 30 colonies by dividing the number of mutant clones by the total number of sequenced clones.
aSingle U, the uracil residue found alone on strand 2.
To assess whether DSBs were formed via lesion processing rather than during replication, we used an ung1-deficient E. coli strain, BW310, which would be unable to remove the uracil residues from within the clusters, so lesion processing would not occur. Figure 4B shows the survival of the uracil-containing clusters for comparison with those containing AP sites, formed prior to transformation. Clusters containing bistranded uracil residues no longer form DSBs and their survival rates are comparable with those of the controls. In contrast, with the clusters that contained AP sites we see a marked loss of colonies, due to DSB formation.
Mutation frequency of lesions persisting within the clustered damage sites
Clusters containing two tandem 8-oxoG lesions did not lead to loss of colony formation in wild-type E. coli (Figure 4A), consistent with the in vitro repair assays. We therefore investigated whether these clusters lead to enhanced mutation frequencies and the types of mutations formed from the sequence analysis of the plasmids obtained for the colonies. For oligonucleotides containing no damage, a single uracil or a single 8-oxoG, we have previously shown the mutation frequencies to be ∼0.5, 3.5 and 2%, respectively (40), consistent with the lack of mutations seen from assaying only 30 colonies in the present study. The mutation frequency and the types of mutations are shown in Table 2 for the three-lesion clusters in wild-type E. coli. The most mutagenic cluster is U/8-oxoG+2−2 with a frequency of ∼15%. In contrast to the mutations at the site of the uracil residues for the clusters leading to loss of colony formation, the majority of the mutations now occur at the site of one or both of the 8-oxoG lesions.
Since the majority of the mutations occurred at the site of the 8-oxoG, the types and frequency of the mutations were determined in the mutY null strain. A marked increase in the mutation frequencies of these clusters (Table 3) was seen compared with those in wild-type E. coli (Table 2). A low level of mutations are formed for a single 8-oxoG and for two 8-oxoG lesions in tandem in mutY E. coli. The increase in mutation frequency for the clustered sites emphasises the importance of mutY in mutation prevention and also the lifetime extension of the 8-oxoG lesions due to the presence of uracil within the cluster. With the exception of a low (3.4%) incidence of deletions in the U/8-oxoG+2 −2 cluster, the majority of the mutations detected in the clusters containing tandem 8-oxoG are GC:TA transversions, predominantly at the site of the 8-oxoG lesion closer to the 3′-end of the oligonucleotide insert. Interestingly, GC:TA transversions are seen at both the 8-oxoG sites within several of the rescued plasmids.
Table 3.
Mutation frequencies of clustered damage sites in mutY E. coli
| Oligonucleotide | Mutation | Frequency % |
|---|---|---|
| Control 8-oxoG | GC:TA transversion | 3.4 |
| Tandem 8-oxoG | 1 bp deletion at the more 3′ 8-oxoG | 4.8 |
| U/8-oxoG+1 +5 | GC:TA transversion at 8-oxoG+5 | 22.2 |
| GC:TA transversion at 8-oxoG+1 | 7.4 | |
| GC:TA transversion at 8-oxoG+1 and +5 | 3.7 | |
| U/8-oxoG+2 −2 | GC:TA transversion at 8-oxoG+2 | 20.7 |
| GC:TA transversion at 8-oxoG−2 | 3.4 | |
| GC:TA transversion at 8-oxoG+2 and −2 | 10.3 | |
| 1 bp deletion at 8-oxoG+2 | 3.4 |
The frequency for each type of mutation was determined from 30 colonies by dividing the number of mutant clones by the total number of sequenced clones.
DISCUSSION
Since IR induces clustered damage sites containing two or more lesions formed within one or two helical turns of the DNA double helix by a single radiation track, and the majority of studies to date have focussed on clusters containing two lesions, synthetic oligonucleotides have been designed containing a variety of three-lesions within a clustered damage site. We have extended understanding of the hierarchy of repair of clustered damage based on the finding that the reparability of an AP site(s) in three-lesion clusters depends upon the composition of the lesions within the cluster and whether its incision leads to potentially cytotoxic DSBs or are highly mutagenic in E. coli. For instance, the presence of 8-oxoG when present in a cluster with bistranded AP sites does not prevent the formation of DSB whereas with clusters containing two 8-oxoG in tandem opposite to an AP site, DSBs are not formed. Although a proportion of the mutations for clusters containing tandem 8-oxoG opposite to an AP site contain GC to AT transversions at both 8-oxoG sites, surprisingly the mutations occur predominantly at the 8-oxoG located closer to the 3′ terminus of the inserted oligonucleotide.
We have confirmed from the dramatic loss of colonies in wild-type E. coli that DSBs form with clusters containing two AP sites, one on each strand. DSB formation was also confirmed when the clusters containing two bistranded AP sites were incubated with the nuclear extract, even in the case of those clusters which showed a ‘two population’ effect where some repair was also seen. These latter clusters showed a reduced efficiency for APE1 cleavage of the second AP site. With bistranded uracil clusters we see a rescue of colony survival in the ung1 E. coli strain, since the lack of ung1 prevents DSB formation, implying that in wild-type bacteria the uracils are converted into AP sites, resulting ultimately in a DSB. This finding is also consistent with the formation of DSBs in wild-type E. coli when the uracils had been converted to AP sites prior to transformation, and the findings with bistranded AP site clusters after transformation into yeast (29) and bacterial (44) strains deficient in various AP endonucleases. Evidence that the DSB arises through BER processing of the AP sites and not due to replication-induced DSB comes from the observation that when a single-strand nick (no loss of base) is present in close proximity to an 8-oxoG on the other strand, the nick is repaired before excision of 8-oxoG, minimising DSB formation (58). In contrast, when the 8-oxoG is replaced by an AP site, a DSB rapidly forms as there is insufficient time for the nick to be repaired before rapid incision of the AP site. From the few surviving colonies in wild-type E. coli with these clusters, the most prevalent mutations seen are small deletions at the site of one of the uracil residues. No mutations were seen at the site of 8-oxoG with these particular clusters.
In contrast, DSBs are not formed in E. coli with clusters containing two tandem 8-oxoG lesions opposite a uracil/AP site. In bacteria there are two main glycosylases involved in the prevention of mutation arising from 8-oxoG; fpg that excises 8-oxoG when present within the DNA double helix, and mutY, that removes adenine residues misincorporated opposite 8-oxoG lesions, with a smaller role for mutT which hydrolyses 8-oxoGTP to 8-oxoGMP in the nucleotide pool to prevent its integration into the DNA sequence. The mutation frequency for single 8-oxoG is low in wild-type and MutY E. coli, whereas a dramatic increase was seen following transformation of clustered damage containing tandem 8-oxoG lesions opposite a uracil/AP site into mutY bacteria, consistent with mutY being the main glycosylase to protect against mutations at 8-oxoG (28,40,41). The overall mutation frequency in mutY E. coli is similar to that determined with the bistranded AP/8-oxoG containing clusters (40). Surprisingly, the vast majority of the mutations with the clusters containing tandem 8-oxoG are GC:AT transversions, occurring at the 8-oxoG closer to the 3′-end of the inserted oligonucleotide. This preferential site for mutations reinforces the concept of a hierarchy of lesion excision within clustered damage sites previously shown biochemically (49) with two tandem 8-oxoG lesions using cell extracts and purified proteins. The increased incidence of mutation at the more 3′ lesion implies that the other 8-oxoG, in our case the 8-oxoG at −2 within U/8-oxoG+2−2 and at +1 in U/8-oxoG+1+5, is excised preferentially. When two 8-oxoG residues are placed in tandem to each other and treated with purified Fpg (0.65 pmol) it was verified that the lesion located closer to the 5′ terminus of the oligonucleotide is preferentially incised (data not shown). Therefore, in our system we propose the following mechanisms of repair; the uracil residue is rapidly excised and the resulting AP site rapidly incised to leave a cluster containing a SSB and two tandem 8-oxoG lesions. The resultant SSB is slowly repaired as shown in cell extracts so that excision of 8-oxoG is retarded (23). Once the SSB is rejoined one of the two 8-oxoG residues may then be excised, with the more 5′ lesion being excised preferentially. The resultant SSB from excision of one of the 8-oxoG residues would be repaired with reduced efficiency, prior to potential excision of the remaining 8-oxoG, which if still present at replication results in mutations. If the SSB resulting from excision of uracil is still present at replication, this would lead to loss off the SSB-containing strand (40,41) and mutations at both 8-oxoG sites, as is indeed seen (Table 3). Clusters containing a SSB and two tandem 8-oxoG lesions did not show induction of DSBs as no loss of colonies was observed (Figure 4A) so it is confirmed that excision of 8-oxoG occurs once the SSB on the opposing strand has been rejoined.
In support of this mechanism, it was confirmed using nuclear extracts that the rejoining of the SSB, arising from incision of the AP site in clusters containing two tandem 8-oxoG lesions opposing a single AP site (AP/8-oxoG+1+5, AP/8-oxoG+2−2, AP/8-oxoG−1−5), is indeed retarded. We had previously shown that the AP site is incised with an efficiency that is 7.5x that of excision of 8-oxoG (31) so under the conditions used 8-oxoG would not be expected to be excised to any great extent. Further, the resulting strand break from incision of the AP site would additionally retard excision of 8-oxoG by glycosylases. Using purified proteins, the ligation step involving ligase III was shown to be retarded and not polymerase β which efficiently adds a base into the break site of these clusters and removes the dRP residue on the 5′ terminus of the SSB, leaving a 5′-phosphate ready for ligation. With the cluster AP/8-oxoG+1+5, the subsequent ligation was not observed after incubation with nuclear extract. This observation was unexpected since with bistranded AP site/8-oxoG clusters (23), ligation products were seen even when the AP site is at +1 to the 8-oxoG, suggesting that the presence of the second 8-oxoG lesion interferes greatly with the efficiency of ligase III. The footprint of ligase III is four base pairs 5′ and 14 base pairs 3′ to the SSB, on the break-containing strand (59) and ligase I fully encircles the DNA double-helix (60). The greater retarded ligation seen may reflect enzyme–DNA contacts on the opposing strand containing the tandem 8-oxoG. Further, ligase I may compensate for a loss of ligase III activity for all orientations with bistranded AP site/8-oxoG clusters, except when in the +1 position to each other (26). The zinc finger and DNA binding domain of ligase III combine to form a novel intrinsic DNA sensing module which differs to ligase I which does not contain a zinc finger motif (61).
Several studies have shown that cleavage of two bistranded AP sites is retarded when in the +1 to +3 orientation to each other (25,30,35). This may in part explain the ‘two population’ effect seen with the clusters AP/AP+2 8-oxoG−2 and AP/AP+1 8-oxoG+5, when the two AP sites are in this orientation. Additionally, AP endonuclease is a structure specific binding enzyme that recognises the kink introduced into the DNA by the formation of an AP site and has a footprint on the lesion-containing strand of six base pairs 5′ and five base pairs 3′ to the AP site (62). It also makes contact with a base on the opposing strand to the AP site to ensure that the active site of the enzyme is correctly positioned for the lesion to be repaired. If an AP site at either position +1 or +2 interferes with the contact made on the opposing strand it may cause cleavage retardation. Due to the lack of retardation of incision of the AP site found on strand 1, the 8-oxoG in tandem with it has no effect. However, incision of the remaining AP site is retarded possibly reflecting some interference of the APE1 contact due to the 8-oxoG and the SSB on the opposite strand. As a consequence the SSB may persist through to replication leading to a replication-induced DSB or to incorporation of adenine as the preferred nucleotide across from the non-coding AP site during both replication and transcription (63,64).
In summary, novel insights are presented into how more complex clustered damage sites formed as a result of exposure to IR, provide a serious challenge to the repair machinery of the cell. It is not only the prompt formation of DSBs that have implications on cell survival but also the conversion of non-DSB clusters to DSBs during processing and attempted repair. With the three-lesion clusters we have enhanced understanding on the hierarchy of processing of tandem lesions when opposed to an AP site/SSB. Due to the complexity of these clustered damage sites, BER is compromised and as a result the mutation frequency is increased. The inefficient repair of such clustered damage sites has great biological significance due to the ultimate risk of tumourogenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Studentship from Medical Research Council to L.J.E. Funding for open access charge: Medical Research Council, UK.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Grigory Dianov (GI-ROB, University of Oxford, UK) for providing the purified proteins used in this study. They would also like to thank Siobhan Cunniffe for technical assistance.
REFERENCES
- 1.Frelon S, Douki T, Cadet J. Radical oxidation of the adenine moiety of nucleoside and DNA: 2-hydroxy-2′-deoxyadenosine is a minor decomposition product. Free Radic. Res. 2002;36:499–508. doi: 10.1080/10715760290025889. [DOI] [PubMed] [Google Scholar]
- 2.Pouget JP, Frelon S, Ravanat JL, Testard I, Odin F, Cadet J. Formation of modified DNA bases in cells exposed either to gamma radiation or to high-LET particles. Radiat. Res. 2002;157:589–595. doi: 10.1667/0033-7587(2002)157[0589:fomdbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 3.Regulus P, Duroux B, Bayle PA, Favier A, Cadet J, Ravanat JL. Oxidation of the sugar moiety of DNA by ionizing radiation or bleomycin could induce the formation of a cluster DNA lesion. Proc. Natl Acad. Sci. USA. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog. Nucleic Acid Res. Mol. Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 5.von Sonntag C. Free-Radical-Induced DNA Damage and Its Repair. Springer, Verlag: Heidelberg; 2006. [Google Scholar]
- 6.Xue L, Greenberg MM. Use of fluorescence sensors to determine that 2-deoxyribonolactone is the major alkali-labile deoxyribose lesion produced in oxidatively damaged DNA. Angew. Chem. Int. Ed. Engl. 2007;46:561–564. doi: 10.1002/anie.200603454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodgkins PS, O'N;eill P, Stevens D, Fairman MP. The severity of alpha-particle-induced DNA damage is revealed by exposure to cell-free extracts. Radiat. Res. 1996;146:660–667. [PubMed] [Google Scholar]
- 8.Goodhead DT. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int. J. Radiat. Biol. 1994;65:7–17. doi: 10.1080/09553009414550021. [DOI] [PubMed] [Google Scholar]
- 9.Nikjoo H, O'N;eill P, Terrissol M, Goodhead DT. Quantitative modelling of DNA damage using Monte Carlo track structure method. Radiat. Environ. Biophys. 1999;38:31–38. doi: 10.1007/s004110050135. [DOI] [PubMed] [Google Scholar]
- 10.Nikjoo H, O'N;eill P, Wilson WE, Goodhead DT. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat. Res. 2001;156:577–583. doi: 10.1667/0033-7587(2001)156[0577:cafdts]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 11.Gulston M, Fulford J, Jenner T, de Lara C, O'N;eill P. Clustered DNA damage induced by gamma radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and Nth sensitive sites. Nucleic Acids Res. 2002;30:3464–3472. doi: 10.1093/nar/gkf467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutherland BM, Bennett PV, Sidorkina O, Laval J. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl Acad. Sci. USA. 2000;97:103–108. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutherland BM, Bennett PV, Sutherland JC, Laval J. Clustered DNA damages induced by x rays in human cells. Radiat. Res. 2002;157:611–616. doi: 10.1667/0033-7587(2002)157[0611:cddibx]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 14.Georgakilas AG. Processing of DNA damage clusters in human cells: current status of knowledge. Mol. Biosyst. 2008;4:30–35. doi: 10.1039/b713178j. [DOI] [PubMed] [Google Scholar]
- 15.Bennett PV, Cintron NS, Gros L, Laval J, Sutherland BM. Are endogenous clustered DNA damages induced in human cells? Free Radic. Biol. Med. 2004;37:488–499. doi: 10.1016/j.freeradbiomed.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Sutherland BM, Bennett PV, Cintron NS, Guida P, Laval J. Low levels of endogenous oxidative damage cluster levels in unirradiated viral and human DNAs. Free Radic. Biol. Med. 2003;35:495–503. doi: 10.1016/s0891-5849(03)00327-7. [DOI] [PubMed] [Google Scholar]
- 17.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair. 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zharkov DO. Base excision DNA repair. Cell Mol. Life Sci. 2008;65:1544–1565. doi: 10.1007/s00018-008-7543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic. Biol. Med. 2002;33:1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 21.Lomax ME, Gulston MK, O'N;eill P. Chemical aspects of clustered DNA damage induction by ionising radiation. Radiat. Prot. Dosimetry. 2002;99:63–68. doi: 10.1093/oxfordjournals.rpd.a006840. [DOI] [PubMed] [Google Scholar]
- 22.Weinfeld M, Rasouli-Nia A, Chaudhry MA, Britten RA. Response of base excision repair enzymes to complex DNA lesions. Radiat. Res. 2001;156:584–589. doi: 10.1667/0033-7587(2001)156[0584:robere]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 23.Lomax ME, Cunniffe S, O'N;eill P. 8-OxoG retards the activity of the ligase III/XRCC1 complex during the repair of a single-strand break, when present within a clustered DNA damage site. DNA Repair. 2004;3:289–299. doi: 10.1016/j.dnarep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Cunniffe SM, Lomax ME, O'N;eill P. An AP site can protect against the mutagenic potential of 8-oxoG when present within a tandem clustered site in E. coli. DNA Repair. 2007;6:1839–1849. doi: 10.1016/j.dnarep.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Lomax ME, Cunniffe S, O'N;eill P. Efficiency of repair of an abasic site within DNA clustered damage sites by mammalian cell nuclear extracts. Biochemistry. 2004;43:11017–11026. doi: 10.1021/bi049560r. [DOI] [PubMed] [Google Scholar]
- 26.Mourgues S, Lomax ME, O'N;eill P. Base excision repair processing of abasic site/single-strand break lesions within clustered damage sites associated with XRCC1 deficiency. Nucleic Acids Res. 2007;35:7676–7687. doi: 10.1093/nar/gkm947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Budworth H, Dianova, Podust VN, Dianov GL. Repair of clustered DNA lesions. Sequence-specific inhibition of long-patch base excision repair be 8-oxoguanine. J. Biol. Chem. 2002;277:21300–21305. doi: 10.1074/jbc.M201918200. [DOI] [PubMed] [Google Scholar]
- 28.Bellon S, Shikazono N, Cunniffe S, Lomax M, O'N;eill P. Processing of thymine glycol in a clustered DNA damage site: mutagenic or cytotoxic. Nucleic Acids Res. 2009;37:4430–4440. doi: 10.1093/nar/gkp422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozmin SG, Sedletska Y, Reynaud-Angelin A, Gasparutto D, Sage E. The formation of double-strand breaks at multiply damaged sites is driven by the kinetics of excision/incision at base damage in eukaryotic cells. Nucleic Acids Res. 2009;37:1767–1777. doi: 10.1093/nar/gkp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.David-Cordonnier MH, Cunniffe SM, Hickson ID, O'N;eill P. Efficiency of incision of an AP site within clustered DNA damage by the major human AP endonuclease. Biochemistry. 2002;41:634–642. doi: 10.1021/bi011682l. [DOI] [PubMed] [Google Scholar]
- 31.David-Cordonnier MH, Laval J, O'N;eill P. Clustered DNA damage, influence on damage excision by XRS5 nuclear extracts and Escherichia coli Nth and Fpg proteins. J. Biol. Chem. 2000;275:11865–11873. doi: 10.1074/jbc.275.16.11865. [DOI] [PubMed] [Google Scholar]
- 32.Harrison L, Hatahet Z, Wallace SS. In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J. Mol. Biol. 1999;290:667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- 33.Eot-Houllier G, Eon-Marchais S, Gasparutto D, Sage E. Processing of a complex multiply damaged DNA site by human cell extracts and purified repair proteins. Nucleic Acids Res. 2005;33:260–271. doi: 10.1093/nar/gki165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eot-Houllier G, Gonera M, Gasparutto D, Giustranti C, Sage E. Interplay between DNA N-glycosylases/AP lyases at multiply damaged sites and biological consequences. Nucleic Acids Res. 2007;35:3355–3366. doi: 10.1093/nar/gkm190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaudhry MA, Weinfeld M. Reactivity of human apurinic/apyrimidinic endonuclease and Escherichia coli exonuclease III with bistranded abasic sites in DNA. J. Biol. Chem. 1997;272:15650–15655. doi: 10.1074/jbc.272.25.15650. [DOI] [PubMed] [Google Scholar]
- 36.Kozmin SG, Sedletska Y, Reynaud-Angelin A, Gasparutto D, Sage E. The formation of double-strand breaks at multiply damaged sites is driven by the kinetics of excision/incision at base damage in eukaryotic cells. Nucleic Acids Res. 2009;37:1767–1777. doi: 10.1093/nar/gkp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pascucci B, Stucki M, Jonsson ZO, Dogliotti E, Hubscher U. Long patch base excision repair with purified human proteins. DNA ligase I as patch size mediator for DNA polymerases delta and epsilon. J. Biol. Chem. 1999;274:33696–33702. doi: 10.1074/jbc.274.47.33696. [DOI] [PubMed] [Google Scholar]
- 38.Paap B, Wilson DM, 3rd, Sutherland BM. Human abasic endonuclease action on multilesion abasic clusters: implications for radiation-induced biological damage. Nucleic Acids Res. 2008;36:2717–2727. doi: 10.1093/nar/gkn118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Z, de los Santos C. NMR characterization of clustered bistrand abasic site lesions: effect of orientation on their solution structure. J. Mol. Biol. 2001;308:341–352. doi: 10.1006/jmbi.2001.4587. [DOI] [PubMed] [Google Scholar]
- 40.Pearson CG, Shikazono N, Thacker J, O'N;eill P. Enhanced mutagenic potential of 8-oxo-7,8-dihydroguanine when present within a clustered DNA damage site. Nucleic Acids Res. 2004;32:263–270. doi: 10.1093/nar/gkh150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shikazono N, Pearson C, O'N;eill P, Thacker J. The roles of specific glycosylases in determining the mutagenic consequences of clustered DNA base damage. Nucleic Acids Res. 2006;34:3722–3730. doi: 10.1093/nar/gkl503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malyarchuk S, Brame KL, Youngblood R, Shi R, Harrison L. Two clustered 8-oxo-7,8-dihydroguanine (8-oxodG) lesions increase the point mutation frequency of 8-oxodG, but do not result in double strand breaks or deletions in Escherichia coli. Nucleic Acids Res. 2004;32:5721–5731. doi: 10.1093/nar/gkh911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malyarchuk S, Youngblood R, Landry AM, Quillin E, Harrison L. The mutation frequency of 8-oxo-7,8-dihydroguanine (8-oxodG) situated in a multiply damaged site: comparison of a single and two closely opposed 8-oxodG in Escherichia coli. DNA Repair. 2003;2:695–705. doi: 10.1016/s1568-7864(03)00040-5. [DOI] [PubMed] [Google Scholar]
- 44.D'S;ouza DI, Harrison L. Repair of clustered uracil DNA damages in Escherichia coli. Nucleic Acids Res. 2003;31:4573–4581. doi: 10.1093/nar/gkg493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harrison L, Brame KL, Geltz LE, Landry AM. Closely opposed apurinic/apyrimidinic sites are converted to double strand breaks in Escherichia coli even in the absence of exonuclease III, endonuclease IV, nucleotide excision repair and AP lyase cleavage. DNA Repair. 2006;5:324–335. doi: 10.1016/j.dnarep.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blaisdell JO, Wallace SS. Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli. Proc. Natl Acad. Sci. USA. 2001;98:7426–7430. doi: 10.1073/pnas.131077798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malyarchuk S, Harrison L. DNA repair of clustered uracils in HeLa cells. J. Mol. Biol. 2005;345:731–743. doi: 10.1016/j.jmb.2004.10.079. [DOI] [PubMed] [Google Scholar]
- 48.Malyarchuk S, Castore R, Harrison L. DNA repair of clustered lesions in mammalian cells: involvement of non-homologous end-joining. Nucleic Acids Res. 2008;36:4872–4882. doi: 10.1093/nar/gkn450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Budworth H, Matthewman G, O'N;eill P, Dianov GL. Repair of tandem base lesions in DNA by human cell extracts generates persisting single-strand breaks. J. Mol. Biol. 2005;351:1020–1029. doi: 10.1016/j.jmb.2005.06.069. [DOI] [PubMed] [Google Scholar]
- 50.Venkhataraman R, Donald CD, Roy R, You HJ, Doetsch PW, Kow YW. Enzymatic processing of DNA containing tandem dihydrouracil by endonucleases III and VIII. Nucleic Acids Res. 2001;29:407–414. doi: 10.1093/nar/29.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lomax ME, Salje H, Cunniffe S, O'N;eill P. 8-OxoA inhibits the incision of an AP site by the DNA glycosylases Fpg, Nth and the AP endonuclease HAP1. Radiat. Res. 2005;163:79–84. doi: 10.1667/rr3284. [DOI] [PubMed] [Google Scholar]
- 52.Imoto S, Bransfield LA, Croteau DL, Van Houten B, Greenberg MM. DNA tandem lesion repair by strand displacement synthesis and nucleotide excision repair. Biochemistry. 2008;47:4306–4316. doi: 10.1021/bi7021427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cupples CG, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl Acad. Sci. USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duncan BK, Weiss B. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J. Bacteriol. 1982;151:750–755. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 56.Rosenquist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair. 2003;2:581–591. doi: 10.1016/s1568-7864(03)00025-9. [DOI] [PubMed] [Google Scholar]
- 57.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl Acad. Sci. USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shikazono N, O'N;eill P. Biological consequences of potential repair intermediates of clustered base damage site in Escherichia coli. Mutat. Res. 2009;669:162–168. doi: 10.1016/j.mrfmmm.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 59.Leppard JB, Dong Z, Mackey ZB, Tomkinson AE. Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair. Mol. Cell Biol. 2003;23:5919–5927. doi: 10.1128/MCB.23.16.5919-5927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pascal JM, O'B;rien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 61.Cotner-Gohara E, Kim IK, Tomkinson AE, Ellenberger T. Two DNA-binding and nick recognition modules in human DNA ligase III. J. Biol. Chem. 2008;283:10764–10772. doi: 10.1074/jbc.M708175200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat. Res. 2000;460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 63.Sagher D, Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983;22:4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- 64.Zhou W, Doetsch PW. Effects of abasic sites and DNA single-strand breaks on prokaryotic RNA polymerases. Proc. Natl Acad. Sci. USA. 1993;90:6601–6605. doi: 10.1073/pnas.90.14.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.