Abstract
Lin28 acts as a repressor of microRNA processing and as a post-transcriptional regulatory factor for a subset of mRNAs. Here we report that in human embryonic stem cells Lin28 facilitates the expression of the pivotal pluripotency factor Oct4 at the post-transcriptional level. We provide evidence that Lin28 binds Oct4 mRNA directly through high affinity sites within its coding region and that an interaction between Lin28 and RNA helicase A (RHA) may play a part in the observed regulation. We further demonstrate that decreasing RHA levels impairs Lin28-dependent stimulation of translation in a reporter system. Taken together with previous studies showing that RHA is required for efficient translation of a specific class of mRNAs, these findings suggest a novel mechanism by which Lin28 may affect target mRNA expression and represent the first evidence of post-transcriptional regulation of Oct4 expression by Lin28 in human embryonic stem cells.
INTRODUCTION
Lin28 is an evolutionarily conserved RNA-binding protein that plays a critical role in the developmental timing in Caenorhabditis elegans (1). In the mouse, Lin28 is widely expressed in early stage embryos, with expression declining and becoming restricted to a limited number of tissues as embryonic development proceeds (2). In human tissues, Lin28 expression has been detected in normal ovarian surface epithelium and in mature oocytes (3,4). Lin28 expression is high in human and mouse embryonic stem (ES) cells, and decreases dramatically during ES cell differentiation (5,6). The biological importance of Lin28 is further underscored by its ability to facilitate the reprogramming of human somatic cells to induced pluripotent stem (iPS) cells (7).
Predominantly cytoplasmic, Lin28 plays pleiotropic roles in the regulation of gene expression. Lin28 has been shown to block let-7 microRNA processing in embryonic cells (8–12). Two recent studies, however, have demonstrated that the let-7 family microRNAs are not the only targets of Lin28 regulation—a handful of other microRNAs are also substrates of Lin28 inhibition (13,14). Lin28 has also been reported to bind a specific subset of mRNAs and to modulate their translation. For example, in differentiating skeletal muscle cells, Polesskaya et al.(15) demonstrated that Lin28 binds IGF-2 mRNA and stimulates its translation. Similarly, our studies using mouse ES cells identified mRNAs of a subset of cell cycle-related genes as putative Lin28 targets (16,17). In an attempt to further understand Lin28-mediated translational regulation in muscle cells, Polesskaya et al.(15) performed affinity chromatography and proteomics analysis and found that the translation initiation factor eIF3beta interacts specifically with Lin28, suggesting a role for Lin28 in the modulation of translation initiation. However, this interaction has not yet been reported in other cell types.
Oct4, known also as Oct3 and Oct3/4, is a POU-domain transcription factor that regulates transcription via binding to an octamer motif located in the promoter and enhancer regions of target genes (18). The mouse and human Oct4 orthologs exhibit a high conservation of nucleotide sequence and genomic organization. Essential for regulating cell fate during early development, Oct4 is also critical for maintaining self-renewal and pluripotency of ES cells. A less than two-fold increase in Oct4 level results in ES cell differentiation into primitive endoderm and mesoderm, while reduction of Oct4 expression induces differentiation towards trophectoderm, suggesting that a precise level of Oct4 is required for maintaining pluripotency (19). To further highlight its key role in pluripotency, Oct4 was recently demonstrated to be both necessary and sufficient to reprogram mouse adult neural stem cells to pluripotency (20). These results also suggest that ES cells must possess a sophisticated regulatory network to maintain Oct4 expression at an optimal level. In line with this view, multiple factors have been identified that regulate Oct4 expression at the transcriptional level. Among the well-studied regulators are Nanog, Sox2, FoxD3 and Oct4 itself, which partner with one another to activate the transcription of Oct4. In fact, Oct4 represses its own transcription when over-expressed (18). Thus, these factors apparently work in concert to establish regulatory circuitry composed of autoregulatory and feed-forward loops that contribute to the maintenance of pluripotency and self-renewal.
In this report, we reveal a novel mechanism of regulation of Oct4 expression: regulation at the post-transcriptional level by Lin28. Importantly, we identify RNA helicase A (RHA) as a novel interacting partner of Lin28 and provide evidence that this interaction may play a role in the Lin28-mediated post-transcriptional regulation of gene expression.
MATERIALS AND METHODS
Antibodies, plasmids and siRNAs
The anti-Lin28 antibody (Abcam, ab46020), anti-RHA (Abcam, ab54593), anti-beta-actin (Sigma, A2228), anti-beta-tubulin (Abcam, ab6046), anti-gapdh (Abcam, ab9484), anti-Oct4 (Chemicon, AB3209), anti-Flag (Sigma, F3165), anti-PABP (Santa Cruz, sc-32318), anti-eIF3beta (Santa Cruz, sc-30251) and rabbit pre-immune serum (SouthernBiotech, 0040-01) were purchased. siLin28 (ON-TARGETplus SMARTpool, L-018411-01), siLin28-2 (an equal molar mixture of two siRNAs, J-018411-09 and J-018411-11), siRHA (ON-TARGETplus SMARTpool, L-009950-00-0005) and consiRNA (D-001810-10-05) were purchased from Dharmacon.
Cell culture and siRNA transfection
Human ES line H1 (listed in the NIH hES registry under the name WA01, WiCell) was maintained as undifferentiated state by culturing on Matrigel-coated plates (B&D) in feeder-free and components defined conditions to avoid any feeder-related contamination to the experiment (21). Briefly, cells were cultured in DMEM/F12 media (Invitrogen, Carlsbad, CA, USA) supplemented with 1% MEM-nonessential amino acids (Invitrogen), 1 mM l-glutamine, 1% penicillin-streptomycin (P/S), 50 ng/ml basic fibroblast growth factor (Millipore), 1X N2 supplements (Invitrogen), and 1X B27 supplements (Invitrogen), and with daily media change. Cells were passaged weekly by dissociation with 1 mg/mL Dispase (StemCell technology). The H1 cells used were between passages 30 and 70 with normal karyotype and expressed common hES cell markers. PA-1 and HEK293 cells were cultured using standard protocols provided by ATCC. Cell transfections were carried out as described (22).
Protein extraction and Western blot analyses
These were done as described by Xu et al. (16).
IP, RNA extraction and RT-qPCR
These were carried out based on protocols as previously described (16). The RT primers specific for the human genes are: beta-actin forward: 5′-ATCAAGATCATTGCTCCTCCTGAG; beta-actin reverse: 5′-CTGCTTGCTGATCCACATCTG; tubulin forward: 5′-CGTGTTCGGCCAGAGTGGTGC, tubulin reverse: 5′-GGGTGAGGGCATGACGCTGAA; Lin28 forward: 5′-CGGGCATCTGTAAGTGGTTC, Lin28 reverse: 5′-CAGACCCTTGGCTGACTTCT; Oct4 forward: 5′- GCCGGTTACAGAACCACACT, Oct4 reverse: 5′-GTGGAGGAAGCTGACAACAA. Firefly luciferase forward: 5′-GCTGGGCGTTAATCAGAGAG, Firefly luciferase reverse: 5′-GTGTTCGTCTTCGTCCCAGT; Renilla forward: 5′-GCAAATCAGGCAAATCTGGT, Renilla reverse: 5′-GGCCGACAAAAATGATCTTC. Nanog forward: 5′-TGCCTCACACGGAGACTGTC, Nanog reverse: 5′-TGCTATTCTTCGGCCAGTTG. Sox2 forward: 5′-ACACCAATCCCATCCACACT, Sox2 reverse: 5′-GCAAACTTCCTGCAAAGCTC.
Sucrose gradient polysome fractionation
2 × 107 H1 cells were collected, washed with PBS, and homogenized in 0.5 mL of MCB buffer [100 mM KOAc, 0.1% Triton, 50 mM HEPES, pH 7.4, 2 mM Mg (OAc)2, 10% glycerol, 1 mM DTT, 20U/mL RNase out (invitrogen), 1X complete mini EDTA-free protease inhibitor cocktail (Roche)]. The lysate was centrifuged at 1300g at 4°C for 10 min. The supernatant was applied onto the top of a 15–55% (W/W) linear sucrose gradient made by Density Gradient Fractionation System (Teledyne ISCO Inc.). The gradient was centrifuged at 150 000g for 3 h (Beckman, CA, USA). Fractions (0.3 ml) were collected, and polysome fractions pooled and used for IP. IP were carried out essentially as described above with some modifications. Briefly, pooled polysome fractions in a total of ∼4 ml were divided into two tubes and incubated with protein A sepharose beads pre-bound with either anti-Lin28 antibody or pre-immune IgG at 4°C overnight. The following day, beads were collected and washed with MCB buffer supplemented with 250 mM NaCl. Bound RNAs were extracted and used in RT-qPCR analysis.
Firefly reporter constructs and Luciferase activity assays
The various firefly reporter constructs were created by inserting the indicated fragments generated by PCR using the human Oct4 gene (NM_203289) as a template. The fragments were inserted into the 3′ UTR of the parent firefly reporter opened at NotI and XhoI. The resulting clones were confirmed by sequencing. The B1U1 luciferase construct was described previously (16). The constructs were transfected into HEK293 cells, together with increasing amounts of Flag-Lin28 DNA. In addition, a Renilla reporter was included in all transfections for normalization purposes. Transfection was carried out in a 48-well plate scale. The amount of total plasmid DNA per well was 400 ng that included 100 ng of firefly reporter DNA, 2 ng of Renilla DNA, and the indicated amounts of Flag-Lin28. Luciferase activities were determined using a TD 20/20n (Turner BioSystems) and the Dual Luciferase Assay System (Promega) according to the manufacturer’s instructions. Luciferase mRNA levels were determined by RT-qPCR and levels plotted after normalization against beta-tubulin and Renilla mRNAs. In the case of the combined RHA siRNA knockdown and luciferase assays, HEK293 cells were transfected with siRHA or control siRNA as usual. Seventy-two hours later, cells were transfected with the indicated plasmid DNAs which include 100 ng of firefly reporter DNA containing the Oct4 R2, 2 ng of Renilla DNA, and 0 or 20 ng of Flag-Lin28. Luciferase activities were assayed 24 h later.
In vitro transcription and UV-XL assays
These were performed as previously described (17). Briefly, 5′ T7 promoter-containing transcription templates were created by PCR using the firefly reporter constructs described above as templates. The resulting PCR fragments were gel purified and used to generate R2, R3, R4 and B1U1 RNAs using MEGAscript T7 (Ambion, AM1334) according to the manufacturer’s instructions. The resulting RNA fragments were used in the XL and competition assays. To prepare cell extract for XL, Flag-Lin28 was transfected into HEK293 cells and cell extracts prepared 24 h later by incubating cells in 10 cell volumes of lysis buffer [0.5% Triton X-100, 10 mM NaCl, 10 mM Tris–HCl, pH 7.5, 10 mM EDTA, 0.5 mM PMSF, 1 mM DTT, 1X protease inhibitor cocktail (Calbiochem)] on ice for 20 min, followed by centrifugation to remove insoluble material. In a 40 µl of XL reaction, there were 2 µl of extract, 20 nM of in vitro transcribed and 32P-UTP labeled F2 or F3 RNA, 6.5 µl of XL buffer (1 mM MgCl2, 280 mM KCl, 1 mg/ml of yeast total RNA, 35 mg/ml of heparin, 20 mM HEPES, pH 7.9, 3% glycerol, and 1 mM DTT), and 140, 420, and 1260 nM of the indicated unlabeled competitor RNA. The mixture was incubated at 30°C for 5 min and then exposed to UV light (254 nm) on ice for 5 min. After RNase A (working concentration 1 mg/ml) treatment at 37°C for 30 min, cross-linked product was immunoprecipitated in 300 µl of IP buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 10 mM EDTA, 0.5% Triton X-100, 0.5 mM PMSF, 1 mM DTT, 1X protease inhibitor cocktail) containing 10 µl of protein A Sepharose pre-bound to 20 µg of anti-Flag antibody. IP was carried out at 4°C overnight. After washing 5 times with 1 ml of cold IP buffer, bound proteins were eluted using 3× SDS sample buffer and resolved on 12% SDS–PAGE, followed by autoradiography.
Immunofluorescence
These were carried out as described previously (22). The polyclonal anti-Lin28 and anti-Oct4, and the monoclonal anti-RHA were used at 1 : 2000, 1 : 1000 and 1 : 500 dilutions, respectively.
RESULTS
Lin28 specifically associates with Oct4 mRNA in embryonic cells
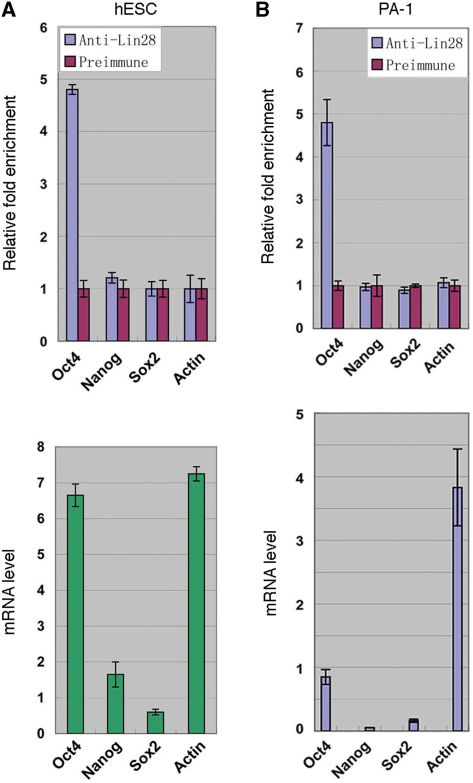
In our previous work with mouse ES cells, we noticed a specific enrichment of Lin28 in ribonucleoprotein (RNP) complexes containing Oct4 mRNA (17). To explore the possibility of Oct4 mRNA being a regulatory target of Lin28, we performed immunoprecipitation (IP) experiments. We used an antibody specific for Lin28 (11,17) to isolate RNPs from human embryonic stem (hES) cell line H1 and embryonic carcinoma (EC) PA-1 cells. Both cells express Lin28 and Oct4, which are localized predominantly to the cytoplasm and nucleus, respectively (Supplementary Figure S1). Following IP, RNAs were extracted from the IP complexes and used to generate cDNAs that were subsequently subjected to quantitative real-time PCR (RT-qPCR) analysis. In Figure 1 (upper panels), the amounts of mRNAs present in the Lin28-containing complexes (blue bars) relative to those in the pre-immune IgG IP complexes (which were arbitrarily set as 1, red bars) are shown. Beta-actin mRNA was used as a control for non-specific RNA binding. While Oct4 mRNA exhibited ∼5-fold enrichment in the Lin28-containing RNPs in both H1 and PA-1 cells, the mRNAs for Nanog and Sox2, two other pluripotency factors, did not. The preferential enrichment of Oct4 mRNA in the Lin28-containing complexes likely reflects its high affinity for Lin28, which is suggested by the lack of enrichment of the actin mRNA that is present at a comparable level in the extract (Figure 1, bottom panels). Therefore, these results together suggest that Oct4 mRNA could be a target for Lin28 regulation at the post-transcriptional level.
Figure 1.
Lin28 is specifically enriched in Oct4 mRNA-containing RNPs. RNPs were isolated from H1 (A) or PA-1 cells (B) using anti-Lin28 antibody or pre-immune rabbit serum, followed by RNA extraction and RT-qPCR analysis. Upper panels: relative abundance of the indicated mRNAs associated with anti-Lin28 versus pre-immune IP complexes plotted as relative fold enrichment. Bottom panels: relative mRNA levels after normalization against beta-tubulin mRNA levels in the cell extracts. Error bars are mean ± SD (n = 3).
Down-regulation of Lin28 expression reduces Oct4 protein levels
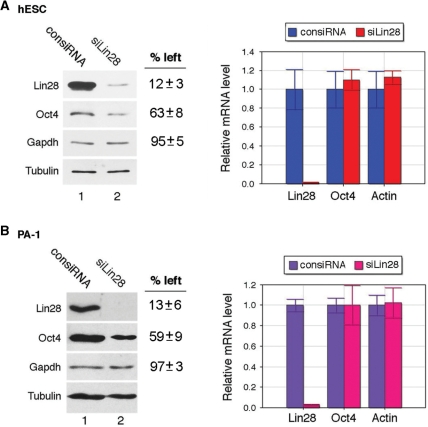
Evidence exists that Lin28 acts to stimulate the translation of target mRNAs (15–17). We therefore asked whether Lin28 affects Oct4 translation. First, we performed siRNA knockdown experiments to assess the effect of Lin28 depletion on Oct4 expression. Lin28-specific siRNA (6) or a control siRNA was transfected into H1 or PA-1 cells. Seventy-two hours later, RNAs and proteins were extracted from the transfected cells and subjected to RT-qPCR and Western blot analysis, respectively. We observed an average of 87–88% reduction of Lin28 at the protein level in Lin28 siRNA transfected compared to control siRNA transfected H1 and PA-1 cells, respectively (Figure 2, left panels, top blots, compare lanes 2 to 1). Importantly, we also observed an average of 37–41% reduction in Oct4 protein levels in H1 and PA-1 cells, respectively, that were transfected with Lin28 siRNA versus control siRNA (Figure 2, left panels, second blots from the top, compare lanes 2 to 1), while the housekeeping Gapdh protein levels were not significantly affected (Figure 2, left panels, third blots from the top, compare lanes 2 to 1). The efficacy and specificity of the Lin28 siRNA was confirmed by RT-qPCR. As shown in Figure 2, right panels, transfection of Lin28-specific siRNA leads to a reduction in Lin28 mRNA level by at least 95% in both H1 and PA-1 cells, compared to that of control siRNA. Meanwhile, the levels of the untargeted Oct4 and beta-actin mRNAs were not affected, suggesting that the Lin28 siRNA effect was specific. We also tested Lin28-specific siRNAs targeted to different regions of Lin28 mRNA and obtained similar knockdown effects (Supplementary Figure S2). While this appears to be in contrast to our previous work with mouse ES cells where we did not observe a decrease in Oct4 protein level following Lin28 siRNA treatment (16), it is likely that the discrepancy is due to insufficient Lin28 knockdown in the previous experiments. Insufficient Lin28 knockdown may also account for the failure to observe Oct4 level reduction in hES cells, which was indicated by the estimated siRNA knockdown efficiency of <70% (6). Consistent with this, we have been unable to observe significant Oct4 protein level decreases in experiments where Lin28 knockdown efficiencies were <80% (data not shown). Taken together with the fact that Oct4 mRNA levels were not affected by Lin28 siRNA knockdown (Figure 2, right panels), our results suggest that Lin28 may regulate Oct4 at the translational level.
Figure 2.
Lin28 depletion leads to decreased levels of Oct4 in hES cells and EC cells. H1 (A) or PA-1 cells (B) were transfected with control siRNA (consiRNA) or Lin28-specific siRNA (siLin28). Seventy-two hours later, RNAs and proteins were extracted from transfected cells, and the levels measured by RT-qPCR and Western blot, respectively. In the left panels, representative results of two Western blot analyses are shown. In lanes 1 and 2, cells were transfected with consiRNA and siLin28, respectively. The antibodies used in the Western blot analyses are labeled on the left. The Lin28, Oct4 and Gapdh protein levels in siLin28-transfected cells relative to those in consiRNA-transfected cells (which were set as 100%) are shown on the right. Protein levels on Western gels were determined using Bio-Rad Quantity One software, and calculated after normalization against beta-tubulin loading control. Average numbers of two (A) or three (B) independent experiments are presented. In the right panels, relative mRNA levels are shown after normalization against beta-tubulin control. mRNA levels in cells transfected with consiRNA were arbitrarily set as 1. Numbers are mean ± SD (n = 3).
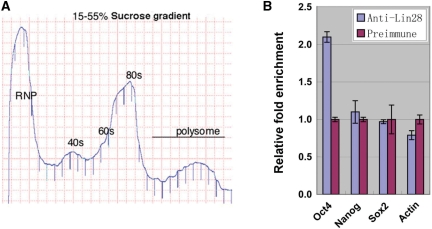
Based on the fact that mRNAs actively being translated are associated with polysomes, we asked whether Lin28 is associated with Oct4 mRNA in polysomes. Thus, we carried out IP experiments using polysome fractions prepared from H1 cells. We reasoned that if stimulation of translation by Lin28 is target mRNA-specific, we would expect to see an enrichment of a target mRNA in Lin28-containing polysomes, while non-target mRNAs would not be enriched. Consistent with this prediction, we observed a reproducible 2-fold enrichment of Oct4 mRNA in Lin28-associated polysomes, while no enrichment was seen with mRNAs for Nanog, Sox2 and beta-actin (Figure 3). The observed difference in the fold of enrichment between IP RNP (5-fold enrichment, Figure 1A) and polysomes (2-fold enrichment, Figure 3B) is likely due to the different conditions used for the two types of IP experiments. Taken together, these results suggest that Lin28 may play a role in Oct4 mRNA translation.
Figure 3.
Lin28 preferentially associates with Oct4 mRNA in polysomes. (A) Polysome profile of H1 cells. Cell lysate was fractionated through a 15–55% linear sucrose gradient and the polysome profile recorded. (B) Polysome fractions were pooled and subjected to IP using anti-Lin28 or preimmune serum as a negative control. RNAs were extracted from the IP complexes, followed by RT-qPCR. Relative abundance of the indicated mRNAs associated with anti-Lin28 (blue bars) versus pre-immune IP complexes (red bars) were plotted as relative fold enrichment after normalization against beta-tubulin mRNA levels. Error bars are mean ± SD (n = 3).
Oct4 mRNA coding region contains sequence elements capable of translational stimulation
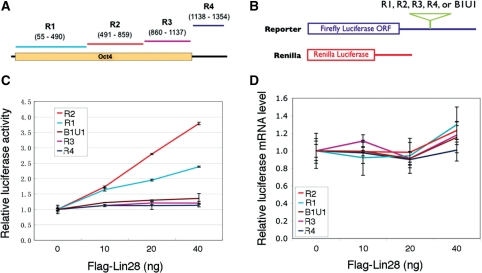
To provide further evidence supporting Lin28’s role in the regulation of Oct4 mRNA translation, we utilized a luciferase reporter system (16,17,23). To ask whether Oct4 mRNA contains sequence elements that stimulate translation in a Lin28-dependent fashion, we used a previously described strategy (16,17) to initiate mapping the sequences. Thus, we inserted fragments derived from various regions of the Oct4 mRNA (Figure 4A) into the 3′ untranslated region (3′ UTR) of a firefly luciferase reporter construct (Figure 4B, upper panel). The ability of the individual sequences to stimulate the translation of the reporter transcript was measured. As shown in Figure 4C, the Oct4 R2 sequence exhibited the strongest Lin28-dependent stimulatory effect on firefly luciferase activity, followed by R1. R3 and R4 did not have effects, as was the case for the negative control B1U1 sequence, which was derived from the 3′UTR of mouse cyclin B1 mRNA (16). Importantly, our RT-qPCR analysis confirmed that the alterations in the luciferase activity did not result from changes in the luciferase mRNA levels (Figure 4D). These results suggest that the Oct4 mRNA coding region contains cis-acting sequence elements capable of translational stimulation in a Lin28-dependent fashion.
Figure 4.
Luciferase reporter assays. (A) A schematic drawing of human Oct4 mRNA. The orange box represents the open reading frame (ORF), while the black line represents the 3′UTR. The various fragments used to create the firefly luciferase reporter constructs are marked above. The numbers in nucleotides depict positions of the fragments relative to the Oct4 transcriptional start site. (B) Schematic structures of reporter constructs. The fragments cloned into the firefly luciferase reporter construct at the 3′UTR (blue line) are indicated above. The sizes in nucleotides for the R1, R2, R3, R4 and B1U1 fragments are 436, 369, 278, 217 and 297, respectively. (C) Luciferase activity results. The reporter constructs were transfected into HEK293 cells that do not express endogenous Lin28, together with increasing amounts of Flag-Lin28. In addition, a Renilla reporter (B) that produces red fluorescence proteins was included in all transfection for normalization purposes. Luciferase activities and mRNA levels were measured 24 h post-transfection. Firefly luciferase activities (after normalization against Renilla luciferase) (C) and firefly mRNA levels (D) from cells without Flag-Lin28 transfection were arbitrarily set as 1. Numbers are mean ± SD (n = 3). B1U1 was used as a negative control for the stimulatory activity (16).
The coding region of Oct4 mRNA contains recognition sites for Lin28
To ask whether R2 preferentially binds Lin28, we performed in vitro UV-crosslinking (XL) experiments similar to those described previously (17). XL reveals direct contact between RNA and protein based on the natural photoreactivity of nucleic acids and amino acids upon UV irradiation. Given that the specific affinity of Lin28 for let-7 microRNA precursors is relatively low (10), it is not surprising that Lin28 can be crosslinked nonspecifically to many RNAs (17; Supplementary Figure S3). This is not unprecedented, as other sequence-specific RNA-binding proteins such as the fragile X mental retardation protein (FMRP) have been reported to do so (24).
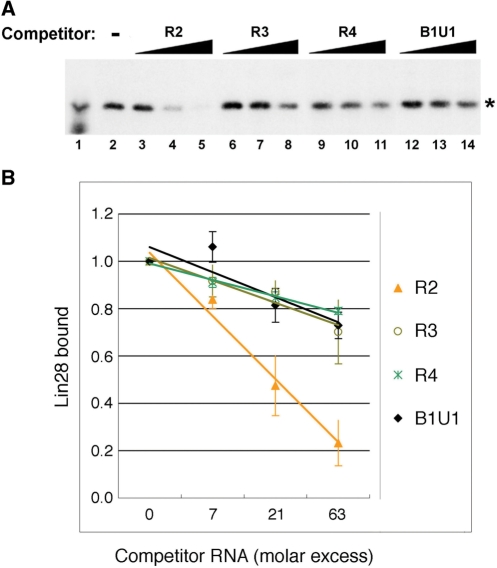
To characterize the interaction between R2 and Lin28, a fixed amount of radioactively labeled R2 RNA was incubated with an extract of HEK293 cells (which do not express endogenous Lin28) transfected with Flag-Lin28, in the presence of increasing amounts of unlabeled R2, R3, R4 and B1U1 RNA fragments (Figure 5A). The reactions were UV-irradiated, followed by RNase digestion. Cross-linked Lin28 was captured by IP using an anti-Flag antibody and visualized by autoradiography. Figure 5A shows a representative gel image of three independent XL experiments. Lin28 binding to the radioactively labeled R2 RNA was competed far more efficiently by the unlabeled R2 RNA itself than by any of the others (compare lanes 3–5 with lanes 6–14). Under the conditions used, the affinity of R2 RNA for Lin28 was estimated as ∼4-fold higher than that of R3 and R4 RNAs (Figure 5B). When labeled R3 was used, more efficient competition by R2 RNA compared to R3, R4 and B1U1 RNAs was also observed (Supplementary Figure S3). These results suggest that Lin28 preferentially binds to R2 in vivo.
Figure 5.
XL and competition assays. (A) Flag-Lin28 was transfected into HEK293 cells and cell extract prepared. XL were carried out using radioactively labeled R2 RNA in the absence (lane 2) or presence of increasing amounts of unlabeled competitor R2 (lanes 3–5), R3 (lanes 6–8), R4 (lanes 9–11), and B1U1 (lanes 12–14), followed by IP using anti-Flag antibody. The unlabeled RNAs were at 7 (lanes 3, 6, 9, 12), 21 (lanes 4, 7, 10, 13) and 63 (lanes 5, 8, 11, 14) molar excess relative to the labeled R2 RNA. Five percent of total cross-linked product prior to IP (lane 1) and anti-Flag IP products were resolved by SDS–PAGE, followed by autoradiography. The bands marked with * indicate Flag-Lin28. (B) Amounts of cross-linked Flag-Lin28 plotted against unlabeled competitor RNA in molar excess. Each point represents three independent experiments. Numbers are mean ± SD.
Lin28 interacts with RHA
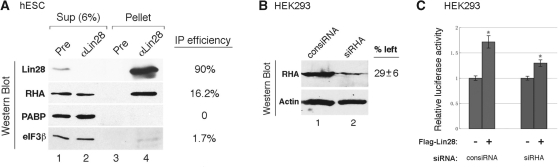
The above results together provide compelling evidence that Lin28 binds Oct4 mRNA through specific recognition of high affinity sequences within the coding region and acts to facilitate Oct4 translation. To further dissect the molecular mechanism by which Lin28 may modulate Oct4 translation, we set out to identify Lin28-interacting proteins in hES cells. We hypothesized that Lin28 stimulates Oct4 mRNA translation by actively recruiting components of translational machinery to the mRNA. Thus, we performed co-IP experiments using the anti-Lin28 antibody to isolate Lin28-containing protein complexes from H1 cells. Co-IP was carried out in the presence of an excess amount of RNase A to disrupt any RNA-bridged interactions that are formed through two RNA-binding proteins simultaneously binding to the same RNA molecule. Protein components in the Lin28-containing complexes were resolved by SDS–PAGE, followed by mass spectrometry analysis of protein bands that were present in the anti-Lin28 IP complexes and not in the pre-immune IP complexes. We identified RHA as a putative Lin28-interacting protein (Supplementary Figure S4). This was subsequently confirmed by co-IP and Western blot analyses. As shown in Figure 6, while ∼90% of Lin28 was immunoprecipitated by the anti-Lin28 antibody (lane 4, top panel), ∼16% of RHA was also brought down by the anti-Lin28 co-IP (Lane 4, second panel from the top). Importantly, the interaction between Lin28 and RHA is direct since poly(A)-binding protein (PABP), which binds to all polyadenylated mRNAs, was not present in the anti-Lin28 co-IP complexes (Lane 4, third panel from the top). Interestingly, we also observed an interaction between Lin28 and the translation initiation factor eIF3beta that was identified by Polesskaya et al.(15) as a Lin28-interacting partner in muscle cells, although under our assay conditions the interaction with Lin28 was much weaker compared to RHA.
Figure 6.
Lin28 interacts with RHA. (A) Results from co-IP experiments. Lin28-containing protein complexes were immunoprecipitated in the presence of excess amounts of RNAse A from H1 cells using anti-Lin28 or pre-immune IgG. Six percent IP supernatants (lanes 1 and 2) and co-IP complexes (lanes 3 and 4) were resolved by SDS–PAGE, followed by Western blot analysis using the indicated antibodies shown on the left. The estimated IP efficiencies were marked on the right. Bands on the Western gels were quantitated using Bio-Rad Quantity One software. (B) Results of RHA siRNA knockdown. HEK293 cells were transfected with control siRNA (lane 1) or a siRNA specific for RHA (lane 2). Seventy-two hours later, proteins were extracted from the transfected cells, followed by Western blot analysis. The specific RHA knockdown at the protein level was quantitated as 71% (an average of three experiments), using beta-actin as a loading control. (C) Luciferase reporter assays. HEK293 cells were transfected with firefly luciferase reporter DNA containing Lin28-binding sequences, with or without co-transfection of Flag-Lin28. The transfections were performed 72 h after the transfection of control siRNA or siRHA. Renilla luciferase was included in the plasmid DNA transfection for normalization purposes. Luciferase activities were measured 24 h following plasmid DNA transfection. Firefly luciferase activities in the absence of Flag-Lin28 expression were set as 1. Numbers are mean ± SD (n = 3), *P < 0.01.
To determine whether the observed interaction is biologically significant, we performed RHA siRNA knockdown experiments. Transfection of a RHA-specific siRNA (siRHA) led to an average of 71% reduction of RHA at the protein level (Figure 6B, compare lane 2 to 1, top panel). Importantly, in the cells where RHA was down-regulated, the ability of Lin28 to stimulate the translation of a firefly luciferase reporter gene was also decreased. As shown in Figure 6C, in control siRNA-transfected cells, luciferase activity was ∼75% higher in cells transfected with Flag-Lin28 than in cells transfected with an empty vector (compare the blue bars with and without the red asterisk). In siRHA-transfected cells, the same level of Flag-Lin28 expression stimulated firefly luciferase activity by only ∼30% (compare the red bars with and without the red asterisk), a 2.5-fold reduction in the stimulation of translation. These results thus indicate that RHA may be an important partner of Lin28 in the regulation of translation of target mRNAs.
DISCUSSION
We show here that Lin28 associates with Oct4 mRNA in RNPs and polysomes in human ES cells and EC cells. The functional significance of these associations is underscored by the fact that inhibition of Lin28 leads to reduced Oct4 protein levels and that Lin28 stimulates luciferase activity from reporter transcripts containing putative Lin28 recognition sites from the Oct4 mRNA. Taken together, these observations suggest a role for Lin28 in the regulation of Oct4 translation. We have further identified RHA as a novel Lin28-interacting protein and provide evidence that this interaction may be important for Lin28’s function in regulating target mRNA translation.
In humans, two isoforms of Oct4, Oct4A and Oct4B, are produced via alternative splicing (25). Oct4A has been demonstrated to be essential for ‘stemness’ by acting as a transcription factor with a pivotal role in the regulation of expression of genes important for the maintenance of pluripotency and the self-renewal ability of ES cells. On the other hand, the function of Oct4B is currently unknown, although multiple Oct4B isoforms resulting from alternative translation initiation have been reported (26). Owing to its clear role in pluripotency, it is not surprising that cells have evolved complex mechanisms to control Oct4 expression to ensure appropriate levels of its expression (18). Until now, however, all pathways involved in the regulation of Oct4 expression have been reported to be at the transcriptional level (18). Our studies demonstrating that Oct4 expression is also modulated at the post-transcriptional level by another pluripotency factor, Lin28, further highlight the complexity of this important regulatory circuitry.
Our studies in mouse ES cells have shown that Lin28 plays a role in ES cell proliferation partly by modulating the expression of cell cycle-related genes including those for cyclin A, cyclin B, cdk4 and the replication-dependent histone H2a at the post-transcriptional level (16,17). Here, we reveal yet another function of Lin28—affecting the expression of Oct4 which is essential for the pluripotency and self-renewal of ES cells. Taken together with the notion that Lin28 inhibits the production of let-7 microRNAs (8–12) involved in cell growth and differentiation, we speculate that all three functions of Lin28 may contribute collaboratively in the process of reprogramming human somatic cells to iPS cells (7).
Related to this work, Darr and Benvenisty (6) have reported that overexpression of Lin28 in human ES cells triggers ES cell differentiation preferentially towards the primitive endoderm lineage, a phenotype similar to that seen when Oct4 expression was slightly elevated (<2-fold) (19). Based on our findings that Lin28 stimulates Oct4 production, Lin28 overexpression might well lead to an increased level of Oct4 [which was not examined in the work of Darr and Benvenisty (6)], which subsequently permits lineage-specific differentiation.
Despite accumulating evidence pointing to the role of Lin28 as a target-specific translational regulator (15–17), the precise mechanism underlying the process is unclear. The finding that Lin28 interacts with the translation initiation factor eIF3beta (this report and ref. 15) is intriguing, given that translation initiation is the rate-limiting step of translation. During initiation, ribosomes are recruited to the mRNA by eukaryotic translation initiation factors (eIFs) that include eIF4E, eIF4A, eIF4G and eIF3. eIF3beta is one of the 13 subunits of the mammalian eIF3 complex (27 and references therein). The interaction between Lin28 and eIF3beta may increase the efficiency of assembly of translation initiation complexes on an mRNA, thus facilitating ribosome loading.
In addition to eIF3beta, we have identified RHA as another Lin28-interacting partner in hES cells. RHA (also called DHX9 and NDHII) is a highly conserved DEAD-box protein that functions in multiple cellular processes, including transcription, splicing, nuclear export, translation, and RNAi, by catalyzing RNA–RNA and RNA–protein rearrangements in RNP complexes (28). In particular, RHA has been shown to promote translation of a specific class of mRNAs that contain highly structured 5′ UTRs (29,30). These include the cellular JUND growth-control gene and viral transcripts that contain a post-transcriptional control element (PCE) in their 5′ UTRs. It has been proposed that complex 5′ UTR structures create a barrier for efficient ribosomal scanning of the transcripts and that direct binding of RHA to the PCE stimulates RNA–RNA and RNA–protein rearrangements necessary for efficient translation (29,30).
Most RNA helicases characterized in vitro lack RNA substrate specificity, yet in vivo perform very specific functions with no redundancy, supporting a view that these RNA helicases require cofactors to convey specificity in vivo (28). We speculate that RHA may function through interaction with Lin28 as a cofactor to specify its RNA substrates. This is supported by the following observations. First, Lin28 interacts with RHA in an RNA-independent manner (Figure 6A). Second, down-regulation of RHA impedes Lin28-dependent stimulation of translation from transcripts containing Lin28-binding sites (Figure 6B and C). Third, Lin28-binding sequences that confer translational stimulatory effects have been mapped to 3′ UTRs (16) as well as to the coding regions of target mRNAs (17; Figures 4 and 5), instead of 5′UTRs. Fourth, we find no obvious sequence and/or structural similarity between the Lin28-binding sequences and the post-transcriptional control element PCE (29,30). We therefore postulate that Lin28 may selectively bind RNA substrates by recognition of high affinity binding sites and subsequently recruit eIF3beta (to enhance translation initiation) as well as RHA (to facilitate RNP remodeling during translation) to enhance translation efficiency.
Finally, RHA has recently been reported to be associated with RNA-induced silencing complexes (RISC), and to promote their formation (31). In addition, Lin28 has been shown to inhibit Dicer (a component of the RISC) cleavage of let-7 pre-microRNAs in vitro (9). Taken together, these observations raise the possibility that RHA-Lin28 interaction provides a molecular link between these processes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Connecticut Stem Cell Research Grants (06SCA02 and 09SCAYALE14) to Y.H.; Connecticut Stem Cell Research Grants Program Core Award (06SCD01) and Hybrid Award (06SCE01) Project 2 to Haifan Lin. Funding for open access charge: Connecticut Innovations (grants 09SCAYALE14 and 06SCD01).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
This material is based upon work supported by the State of Connecticut under the Connecticut Stem Cell Research Grants Program. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the State of Connecticut, the Department of Public Health of the State of Connecticut or Connecticut Innovations, Inc.
REFERENCES
- 1.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 2.Yang DH, Moss EG. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr. Patterns. 2003;3:719–726. doi: 10.1016/s1567-133x(03)00140-6. [DOI] [PubMed] [Google Scholar]
- 3.Assou S, Cerecedo D, Tondeur S, Pantesco V, Hovatta O, Klein B, Hamamah S, De Voset J. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics. 2009 doi: 10.1186/1471-2164-10-10. doi:10.1186/1471-2164-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, PhillipS LA, Lockhart VL, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richard M, Tan SP, Tan JH, Chan WK, Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- 6.Darr H, Benvenisty N. Genetic analysis of the role of the reprogramming gene LIN-28 in human embryonic stem cells. Stem Cells. 2009;27:352–362. doi: 10.1634/stemcells.2008-0720. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 8.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin-28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 10.Piskounova E, Viswanathan SR, Janas M, LaPierreR J, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 11.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA Uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Trabucchi M, Briata P, Carcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15:357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu B, Huang Y. Histone H2a mRNA interacts with Lin28 and contains a Lin28-dependent posttranscriptional regulatory element. Nucleic Acids Res. 2009;37:4256–4263. doi: 10.1093/nar/gkp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pei D. Regulation of pluripotency and reprogramming by transcription factors. J. Biol. Chem. 2009;284:3365–3369. doi: 10.1074/jbc.R800063200. [DOI] [PubMed] [Google Scholar]
- 19.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 20.Kim JB, Sebastiano V, Wu G, Araúzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 21.Yao S, Chen S, Clark J, Hao E, Beattie GM, Hayek A, Ding S. Long-term self-renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc. Natl Acad. Sci. USA. 2006;103:6907–6912. doi: 10.1073/pnas.0602280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M, Guller S, Huang Y. Method to enhance transfection efficiency of cell lines and placental fibroblasts. Placenta. 2007;28:779–782. doi: 10.1016/j.placenta.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 23.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeda J, Seino S, Bell GI. Human Oct3 gene family cDNA sequences, alternative splicing, gene organization, chromosomal location, and expression at low levels in adult tissues. Nucleic Acids Res. 1992;20:4613–4620. doi: 10.1093/nar/20.17.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Zhao Y, Xiao Z, Chen B, Wei Z, Wang B, Zhang J, Han J, Gao Y, Li L, et al. Alternative translation of OCT4 by an internal ribosome entry site and its novel function in stress response. Stem Cells. 2009;27:1265–1275. doi: 10.1002/stem.58. [DOI] [PubMed] [Google Scholar]
- 27.Martineua Y, Derry MC, Wang X, Yanagiya A, Berlanga JJ, Shyu AB, Imataka H, Gehring K, Sonenberget N. The poly(A)-binding protein-interacting protein 1 binds to eIF3 to stimulate translation. Mol. Cell Biol. 2008;28:6658–6667. doi: 10.1128/MCB.00738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol. Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 29.Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 30.Bolinger C, Yilmaz A, Hartman TR, Kovacic MB, Fernandez S, Ye J, Forget M, Green PL, Boris-Lawrie K. RNA helicase A interacts with divergent lymphotropic retroviruses and promotes translation of human T-cell leukemia virus type 1. Nucleic Acids Res. 2007;35:2629–2642. doi: 10.1093/nar/gkm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.