Abstract
In bacterial 16S rRNAs, methylated nucleosides are clustered within the decoding center, and these nucleoside modifications are thought to modulate translational fidelity. The N4, 2′-O-dimethylcytidine (m4Cm) at position 1402 of the Escherichia coli 16S rRNA directly interacts with the P-site codon of the mRNA. The biogenesis and function of this modification remain unclear. We have identified two previously uncharacterized genes in E. coli that are required for m4Cm formation. mraW (renamed rsmH) and yraL (renamed rsmI) encode methyltransferases responsible for the N4 and 2′-O-methylations of C1402, respectively. Recombinant RsmH and RsmI proteins employed the 30S subunit (not the 16S rRNA) as a substrate to reconstitute m4Cm1402 in the presence of S-adenosylmethionine (Ado-Met) as the methyl donor, suggesting that m4Cm1402 is formed at a late step during 30S assembly in the cell. A luciferase reporter assay indicated that the lack of N4 methylation of C1402 increased the efficiency of non-AUG initiation and decreased the rate of UGA read-through. These results suggest that m4Cm1402 plays a role in fine-tuning the shape and function of the P-site, thus increasing decoding fidelity.
INTRODUCTION
The ribosome is the translator of genetic information on mRNAs that results in protein synthesis. The crystal structures of ribosomal subunits and their functional complexes with various substrates have helped us to elucidate the mechanisms of protein synthesis. The small (30S) subunit of the ribosome, containing the 16S RNA, plays a critical role in deciphering the codons on mRNAs (1–4). The neck region of the 30S subunit provides a path for mRNA along with the three tRNA binding sites, the A-, P- and E-sites. At the P-site, precise codon–anticodon pairing occurs between the mRNA and a peptidyl–tRNA during the elongation cycle to maintain the reading frame (5–7). In the initiation step, the AUG codon at the P-site is recognized by formyl-methionyl (fMet)-tRNAiMet and this recognition is assisted by initiation factors (8,9). This step is kinetically controlled by the formation and stability of the 30S initiation complex (30S IC), which is followed by the formation of the 70S initiation complex (70S IC) (10,11). The flexibility of the 16S rRNA during these conformational rearrangements is believed to affect the fidelity of initiation (12), suggesting that the local structure of the P-site affects the fidelity of AUG codon selection. The P-site in the small subunit is formed by several conserved residues (positions 1400–1405 and 1496–1502) in the top part of helix 44 in the 16S rRNA (Figure 1A) (7). Certain point mutations in the P-site of Escherichia coli 16S rRNA resulted in growth reduction (13,14). In particular, a single base deletion of C1400 resulted in a lethal phenotype (15). In an in vitro study, a mutant ribosome in which G1401–C1501 was flipped to C1401–G1501 showed full tRNA-binding activity at the P-site and mostly normal peptide-bond formation; however, formation of the first peptide bond was largely inhibited in the mutant ribosome (16). A U1498G mutation also affected the formation of the first peptide bond (17,18). These data suggest that certain conserved residues in the P-site of the 16S rRNA are required for the initiation of translation by the ribosome.
Figure 1.
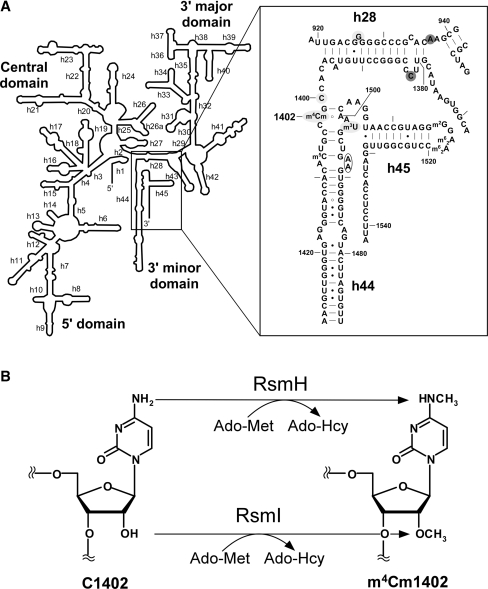
The secondary structure of the decoding center in the E. coli 16S rRNA and the enzymatic formation of m4Cm1402. (A) The secondary structure of the E. coli 16S rRNA is shown on the left. Helix 44 and surrounding regions are shown on the right, with helix and base numbers indicated. A number of modified nucleosides including 5-methylcytidine (m5C), N4, 2′-O-dimethylcytidine (m4Cm) and 3-methyluridine (m3U) are also shown. The bases annotated as A-site (blank circle), P-site (shaded with light gray) and E-site (shaded with dark gray) are indicated (4,6). (B) Enzymatic formation of m4Cm1402. N4-methylation and 2′-O-methylation of C1402 are catalyzed by RsmH and RsmI, respectively, using Ado-Met as a methyl group donor. After the reaction, Ado-Met is converted to Ado-Hcy.
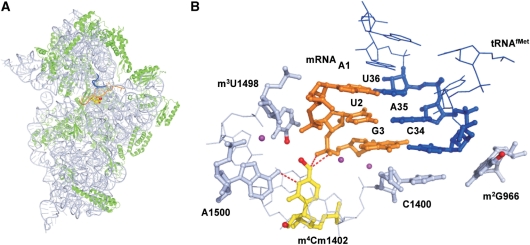
Ribosomal RNAs are decorated with various post-transcriptional modifications (19). Seven species of RNA modifications are found at 11 positions in the 16S rRNA, and some are clustered in the decoding center (20). They are required for the fine-tuning of local rRNA structure (21), RNA–RNA interactions (21), 30S subunit assembly (22,23) and antibiotic resistance (24). Recently, Umesh and his colleagues showed that methyl-modifications in the 16S rRNA are required for the stringent selection of the initiator tRNA (25), suggesting that the translation initiation step is affected and modulated by certain methyl-modifications. However, it is still unclear which modifications are dominantly associated with the translation initiation. At the P-site of the 16S rRNA there is a unique dimethyl modification, N4, 2′-O-dimethylcytidine (m4Cm) at position 1402 (Figure 1A and B), which was discovered in 1966 (26). C1402 is universally conserved (27), and the dimethyl modification is thought to be conserved in bacteria (28–30). A C to U replacement at position 1402 resulted in a slow growth phenotype (13), and m4Cm1402 is thought to be important for ribosome function. However, the enzymes and biogenesis for this modification have not been characterized. In the crystal structure of the E. coli 30S subunit complexed with mRNA and tRNAiMet, the codon–anticodon helix at the P-site interacts with the major groove on the top part of helix 44 (5–7). The backbone of the P-site codon is recognized by m4Cm1402 through hydrogen bonds between the N4 of m4Cm1402 and a phosphate between the 2nd and 3rd letters of the P-site codon (Figure 5B) (5–7). These observations suggest that m4Cm1402 is involved in the fine-tuning of the local structure of the P-site, and the correct recognition of the initiation codon.
Figure 5.
Structural insights into the P-site function of m4Cm1402. (A) Crystal structure of the 30S subunit with the P-site tRNAfMet and mRNA (5). The 16S rRNA, the ribosomal proteins, the anticodon stem–loop of the P-site tRNA, and the mRNA are depicted in light blue, green, blue, and orange, respectively. The m4Cm1402 is shown as a set of yellow spheres, and its methyl groups are depicted as red spheres. The coordinates of the methyl groups of m4Cm1402 are calculated based on a previously published structure (6). (B) The tertiary structure of the P-site in the 30S subunit, including the AUG codon of the mRNA, the CAU anticodon of tRNAiMet and m2G966, C1400, m4Cm1402, m3U1498 and A1500 in the 16S rRNA. The same color code was used as described for (A). Mg2+ ions are depicted as magenta spheres. The methyl group of m3U1498 is also depicted as red. The hydrogen bonds formed by m4Cm1402 are shown as red dash.
To investigate the function and biogenesis of m4Cm1402, it is necessary to identify the genes that encode enzymes responsible for m4Cm1402 formation. We have performed a genome-wide screen for genes responsible for RNA modifications using a reverse genetics approach combined with mass spectrometry (ribonucleome analysis) (31). By this approach we have identified many genes responsible for tRNA modifications (32–34). For this study, we modified the approach in order to screen for genes responsible for rRNA modifications. We successfully identified two methyltransferases responsible for m4Cm1402 formation in the E. coli 16S rRNA (Figure 1B), and further characterized the biogenesis and function of m4Cm1402.
MATERIALS AND METHODS
Strains and mediums
A series of single deletion strains each bearing a kanamycin resistance marker (Kmr) was obtained from the Genetic Stock Research Center, National Institute of Genetics, Japan (35). A series of knockout strains with transposon insertions was kindly provided by Dr T. Miki (36). The E. coli K-12 strain BW25113 (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) was used for the ‘one-step inactivation of chromosomal genes’ procedure (37). The following primers were used to amplify the chloramphenicol acetyltransferase gene (Cmr) with 40-nt extensions at both ends that were homologous to sequences of the yraL gene: ΔyraL-F (Cmr) (5′-atcggcaatctggcggatatcacccagcgtgcgttagaggactaaatcagtaagttggcagc-3′) and ΔyraL-R (Cmr) (5′-tgcggccagcgccgccgcttttttcagcggcagttctgccaaccagcaatagacataagcggc-3′). The pBT vector (Stratagene) was used as a template for Cmr. The amplified sequence was used to construct BW25113 ΔyraL::Cm. To construct ΔmraW/ΔyraL, ΔyraL::Cm was transferred to ΔmraW (Kmr) by P1-transduction. The E. coli strains were grown in 1 ml of Luria–Bertani (LB) medium in 96-well plates at 37°C overnight. Doubling time was determined using OD600 measurements in a plate reader (Molecular Device, Inc.) every 15 min.
Ribonucleome analysis
The mutant E. coli strains were grown in 1 ml of LB medium in 96-well plates at 37°C overnight. Total RNA was prepared from the harvested cells using an RNA preparation kit (RNeasy, Qiagen) and the rRNAs were isolated by electrophoresis in a 4% polyacrylamide gel containing 7 M urea. About 100 fmol of rRNAs were digested with RNase T1 or RNase A, and the digests were analyzed by capillary liquid chromatography (LC) nano electrospray ionization/mass spectrometry as described previously (31,38) with the following modifications. The concentration of RNase T1 was adjusted to 5 U/µl. We employed a linear ion trap-orbitrap hybrid mass spectrometer (LTQ Orbitrap XL, Thermo Fisher Scientific) equipped with a custom-made nanospray ion source, a Nanovolume Valve (Valco Instruments), and a splitless nano HPLC system (DiNa, KYA Technologies). Each digest was mixed with TEAA and loaded onto a nano-LC trap column (C18, Φ0.5 × 1.0 mm), desalted, and then concentrated with 0.1 M TEAA (pH 7.0). RNA fragments were eluted from the trap column and directly injected into a C18 capillary column (HiQ Sil; 3 µm C18, 100-Å pore size; Φ0.1 × 100 mm, KYA Technologies). The solvent system consisted of 0.4 M 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP; pH 7.0, adjusted with triethylamine; solvent A) and 0.4 M HFIP in 50% methanol (solvent B), and the samples were chromatographed at a flow rate of 300 nl/min using a linear gradient of 10–90% solvent B over 35 min. The chromatographic eluent was sprayed from a sprayer tip attached to the capillary column. The ionization voltage was set to –1.9 kV and ions were scanned in the negative polarity mode. For nucleoside analysis, RNAs were digested into nucleosides and analyzed by LC/MS using ion-trap mass spectrometry as described previously (31).
Preparation of rRNA fragments
Escherichia coli ribosomes were obtained basically as described previously (39,40). rRNAs were prepared from the ribosomes by phenol/chloroform extraction and ethanol precipitation (41). The 49-mer fragment covering positions 1378–1426 was carved out from the 16S rRNA using a complementary oligodeoxynucleotides procedure (42). The 20 A260 unit rRNA was mixed with 4 nmol of deoxyoligonucleotide (5′-caacccactcccatggtgtgacgggcggtgtgtacaaggcccgggaacg-3′), complementary to positions 1378–1426 in a buffer consisted of 58 mM HEPES-KOH pH 7.6 and 115 mM KCl. The mixture was incubated at 90°C for 5 min then cooled to 37°C. One-ninth volume of 500 mM NH4OAc (pH 5.3), 0.5 µg RNase A and 500 U RNase T1 was added, and the mixture was incubated at 37°C for 1 h to digest the non-protected regions of the rRNA. The protected DNA/RNA heteroduplex was recovered by phenol/chloroform extraction and ethanol precipitation, and then purified by the electrophoresis in a 15% polyacrylamide gel containing 7 M Urea.
Expression and purification of recombinant RsmH and RsmI
The strains carrying pCA24N plasmids for expression of RsmH and RsmI were obtained from the ASKA clone collection [NBRP (NIG, Japan): E. coli] (43). The recombinant RsmH and RsmI proteins, with N-terminal 6 × His-tags, were expressed in soluble form by induction with 0.1 mM IPTG, and purified by using the AKTA chromatography system with the His-trap chelating HP column (Amersham Biosciences). The pooled proteins were dialyzed against a buffer consisting of 50 mM Tris–HCl (pH 7.5), 1 mM dithiothreitol and 100 mM KCl. To remove endogenous Ado-Met, the recombinant proteins were incubated with 2 mM S-adenosylhomocysteine (Ado-Hcy) in a buffer consisted of 50 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 100 mM NH4Cl for 1 h at 37°C,and then dialyzed (44). The protein concentration was determined using the Bio-Rad protein assay kit with bovine serum albumin as the standard. Glycerol was added to the protein solution to a final concentration of 30%.
In vitro reconstitution of m4Cm1402
The 30S subunit and the tightly-coupled 70S ribosomes were isolated from the ΔrsmH/ΔrsmI strain by sucrose density gradient centrifugation as described previously (45). The 16S rRNA was prepared from the 30S fraction using Isogen (Nippon gene). In vitro methylations were performed at 37°C for 1 hour in 100-µl reaction mixtures containing 100 mM NH4Cl, 20 mM HEPES-KOH (pH 7.6), 10 mM Mg(OAc)2, 6 mM β-mercaptoethanol, 1 mM Ado-Met, 2.5 pmol 16S rRNA (or 5 pmol of 30S subunit or tightly-coupled 70S) and 10 pmol of recombinant proteins (RsmH and/or RsmI). rRNA was recovered from an aliquot of the reaction mixture using Isogen (Nippon Gene). This was digested with RNase T1 and then subjected to LC/MS analysis as described earlier.
Luciferase reporter constructs and assays
The ORFs of Rluc and Fluc were PCR-amplified from pJD375, which was kindly provided by Dr. Jonathan Dinman (Univ. of Maryland). A series of dual luciferase reporter constructs was made for this study. The sequences and names of DNA primers for vector construction were shown in Supplementary Table S1. The Firefly and Renilla luciferase fusion ORF was amplified from pJD375 using the primers LucNcoI-f and LucHindIII-r. The PCR product was cloned into the NcoI and HindIII sites of pQE60 (Qiagen), and resulting plasmid was named pQE60-FRLuc. For the series of reporter vectors designed to examine non-AUG initiation, the UAA stop codon was inserted at the end of the Rluc ORF, and the SD sequence of pQE60 was inserted upstream of the Fluc ORF by QuikChangeTM Site-Directed Mutagenesis (Stratagene), using the primers SD-f and SD-r. Next, the AAA start codon was inserted at the initiation position of Fluc ORF using the primers AAA-f and AAA-r, and the resulting plasmid was named pQE-Luc (AAA). Various codons were inserted at the initiation position of Fluc in pQE-Luc (AAA) using primers named by their corresponding start codons: AUG, AUU, UUG and GUG. Each construct was confirmed by DNA sequencing.
To construct the series of reporters designed to evaluate translational fidelity, window sequences were introduced between Fluc and Rluc using pQE60-FRluc as a template. The window sequences were designed based on constructs for a β-galactosidase assay (40,46) and are shown in Supplementary Table S2. For site-directed mutagenesis we employed the primers UGA-f and UGA-r for UGA read-through, UAG-f and UAG-r for UAG read-through, +1-f and +1-r for the +1 frameshift and −1-f and −1-r for the –1 frameshift, to yield pQE-Luc(UGA), pQE-Luc(UAG), pQE-Luc(+1) and pQE-Luc(-1), respectively. Each plasmid was confirmed by DNA sequencing.
The E. coli wild-type (BW25113), ΔrsmH, ΔrsmI and ΔrsmH/ΔrsmI strains were transformed with each of the constructs. Each transformant was cultivated at 37 degree-C in 2 ml LB liquid medium containing 100 µg/ml ampiciline. When the culture reached an A600 of 0.5, 0.5 ml of the cells were harvested and resuspended in 100 µl lysis buffer [50 mM HEPES-KOH (pH 7.6), 100 mM KCl, 10 mM MgCl2, 7 mM β-mercaptoethanol, 400 µM lysozyme]. Cell lysates were prepared by the lysozyme freeze–thaw method (47) and cleared by centrifugation for 15 min at 15 000 r.p.m. at 4°C. The cleared lysates (5 µl) were analyzed in a GLOMAX96 Microplate Luminometer (Promega) using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. The efficiency of non-AUG initiation, stop codon read-through and reading frame maintenance was measured by dividing Fluc chemiluminescence by that of Rluc (the F/R value) from the relevant constructs.
RESULTS
Identification of rsmH and rsmI, which are responsible for m4Cm1402 formation
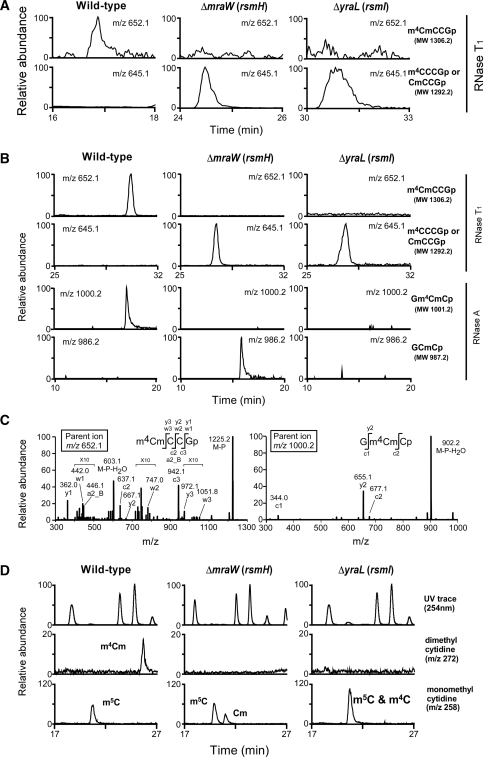
We employed a reverse genetics approach combined with MS (ribonucleome analysis) to identify the genes responsible for m4Cm1402 formation in E. coli. To specifically detect m4Cm at position 1402 in the 16S rRNA, we set up conditions LC/MS to analyze complex mixtures of rRNA fragments digested with base-specific ribonucleases (RNases). Total RNA was prepared from a wild-type strain of E. coli and digested with RNase T1, and then subjected to LC/MS. In the complex mixture of RNA fragments we clearly detected the doubly charged negative ion (m/z 652.1) in the tetramer fragment containing m4Cm (m4CmCCGp, MW 1306.2), which originated from the 16S rRNA (Figure 2A).
Figure 2.
Identification of rsmH and rsmI. (A) LC/MS analysis of the total RNA from E. coli wild-type (left panels), ΔmraW (rsmH) (middle panels) and ΔyraL (rsmI) (right panels), digested by RNase T1. The top and second panels show mass chromatograms detecting the tetramer fragments containing m4Cm1402 (m/z 652.1, top panels), or Cm1402 or m4C1402 (m/z 645.1, second panels). (B) LC/MS analysis of the 49-mer fragment (1378–1426) in the 16S rRNA from E. coli wild-type (left panels), ΔmraW (rsmH) (middle panels) and ΔyraL (rsmI) (right panels), after digested with RNase T1 (upper panels) and RNase A (lower panels). Top panels: mass chromatograms detecting the tetramer fragments containing m4Cm1402 (m/z 652.1). Second panels: detection of the tetramers containing Cm1402 or m4C1402 (m/z 645.1). Third panels: detection of trimer fragments containing m4Cm1402 (m/z 1000.2). Fourth panels: detection of trimer fragments containing Cm1402 (m/z 986.2). Although the dimer Gm4Cp must have been produced in this treatment, we could not detect it because of chemical noise in the early eluents. (C) Collision-induced dissociation (CID) spectra of the tetramer (left) and trimer (right) fragments bearing m4Cm1402. The doubly charged ion (m/z 652.1) of the tetramer and singly charged ion (m/z 1000.2) of the trimer were employed as parent ions for CID. The sequence of each fragment with modifications was confirmed by assignment of the product ions. The nomenclatures for product ions of nucleic acids are the same as those used in the literature (64). The regions with 10-fold magnification are indicated with ‘×10’. (D) Nucleoside analysis of the 49-mer fragment from wild-type (left panels), ΔmraW (rsmH) (middle panels) and ΔyraL (rsmI) (right panels). The top panels show the UV trace at 254 nm, the second and bottom panels show base peak presentations of the mass chromatograms to detect dimethylcytidine (m/z 272) and monomethylcytidine (m/z 258), respectively.
Since m4Cm is a dimethyl modification, we picked up 24 uncharacterized genes in E. coli that were annotated as methyltransferases in the Clusters of Orthologous Groups (COG) database (48), as candidates for enzymes involved in m4Cm1402 formation. Total RNAs were prepared from E. coli strains, each knocked out at one of the 24 candidate genes, and subjected them to the LC/MS analysis described earlier. As a result we identified two strains, ΔmraW and ΔyraL, which both produced mono-methylated tetramers (m4CCCGp or CmCCGp, MW 1292.2) instead of the dimethylated tetramer produced by wild-type E. coli (Figure 2A). Therefore, both genes encode methyltransferases for N4-methylation or 2′-O-methylation of C1402.
For a more detailed analysis, we used complementary oligodeoxynucleotides (42) to isolate an RNA fragment of 49 mer containing C1402 (positions 1378–1426), which was carved out from the 16S rRNA. We used LC/MS to analyze this fragment from wild-type and mutant E. coli strains. In the fragment isolated from wild-type E. coli we observed the dimethylated tetramer (m4CmCCGp, MW 1306.2) produced by RNase T1 digestion and the dimethylated trimer (Gm4CmCp, MW 1001.2) produced by RNase A digestion (Figure 2B). Each fragment was analyzed by collision-induced dissociation to confirm the exact positions of the dimethylation (Figure 2C). In the ΔmraW strain, a mono-methylated tetramer (m4CCCGp or CmCCGp, MW 1292.2) was observed in the RNase T1 digest and a mono-methylated trimer (GCmCp, MW 987.2) was observed in the RNase A digest. If mraW encodes a methyltransferase responsible for the 2′-O-methylation of m4Cm1402, the mono-methylated dimer (Gm4Cp, MW 682.1) would be produced by RNase A digestion. Therefore, this result demonstrated that mraW encodes a methyltransferase responsible for the N4-methylation of m4Cm1402. In the ΔyraL strain, a mono-methylated tetramer (m4CCCGp or CmCCGp, MW 1292.2) was observed in the RNase T1 digest, whereas the mono-methylated trimer (GCmCp, MW 987.2) was not detected in the RNase A digest. Although the dimer Gm4Cp must have been produced in this treatment, we could not detect it because of chemical noise in the early eluents. The data suggested that yraL encodes a methyltransferase responsible for the 2′-O-methylation of m4Cm1402.
To confirm these results we carried out nucleoside analyses of the 49-mer fragment from the 16S rRNA. In wild-type E. coli, we observed a positively charged proton-adduct of m4Cm (m/z 272) (Figure 2D). In the ΔmraW strain, no m4Cm could be found; instead Cm (m/z 258) appeared (Figure 2D). In ΔyraL strain, no m4Cm could be found, and we observed m4C (m/z 258) as an overlapping peak with that of m5C1407. This approximately doubled the peak intensity of the mono-methylated cytidines in the mass chromatogram (Figure 2D). Taken together, our results indicate that mraW and yraL encode methyltransferases responsible for the transfer of the N4-methyl and 2′-O-methyl groups, respectively, onto C1402. We therefore re-named mraW as rsmH, and yraL as rsmI, according to the established rules of nomenclature (49,50) (Figure 1B).
Growth properties of knockout strains of rsmH and/or rsmI
To evaluate the cellular functions of m4Cm1402 in E. coli, we measured the doubling times of single and double knockout strains of rsmH and rsmI (Table 1). When cultured at 37°C, the ΔrsmH and ΔrsmI strains showed 15% and 12% increases in doubling times, respectively, compared with wild-type E. coli. The ΔrsmH/ΔrsmI double knockout strain showed a 29% increase in doubling time. The knockout strains showed similar increases in doubling times when cultured at 33°C and 42°C. These data suggested that a clear growth phenotype was seen in the double knockout strain, whereas each of the single knockout strains showed similar but milder phenotypes. Thus, the defect in m4Cm modification resulted in a slight but significant change in cellular growth properties.
Table 1.
Doubling times of the ΔrsmH, ΔrsmI, and ΔrsmH/ΔrsmI strains
| Temperature (°C) | Doubling time (min) |
|||
|---|---|---|---|---|
| Wild-type | ΔrsmH | ΔrsmI | ΔrsmH/ΔrsmI | |
| 33 | 30.5 ± 0.4 | 34.5 ± 1.9 | 31.4 ± 0.2 | 36.5 ± 0.5 |
| 37 | 20.4 ± 0.3 | 23.5 ± 0.8 | 22.9 ± 0.6 | 26.3 ± 2.5 |
| 42 | 20.9 ± 0.8 | 22.7 ± 3.1 | 22.9 ± 0.3 | 26.8 ± 0.4 |
The doubling times shown here are the means and standard deviations of three independent experiments.
In vitro reconstitution of m4Cm1402
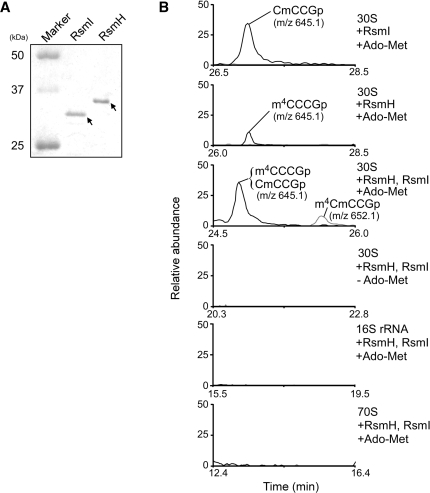
Next we tried to reconstitute m4Cm1402 in vitro using recombinant RsmH and RsmI. N-terminal His-tagged recombinant RsmH and RsmI were expressed and purified by Ni affinity chromatography (Figure 3A). We employed naked 16S rRNA, 30S subunits, and tightly-coupled 70S ribosomes without C1402 methylation, which were prepared from the ΔrsmH/ΔrsmI strain, as the substrates for the in vitro reconstitution. Recombinant proteins were incubated with the substrates in the presence of S-adenosylmethionine (Ado-Met) as a methyl group donor, and then the rRNAs were isolated for LC/MS analysis. As shown in Figure 3B, when the 30S subunit was used as substrate, m4C1402 and Cm1402 were formed by RsmH and RsmI, respectively, although the level of N4-methylation was quite low under these conditions. In addition, m4Cm1402 was successfully reconstituted when RsmH and RsmI were incubated together, but only in the presence of Ado-Met. Neither type of methylation was observed when the naked 16S rRNA or tightly-coupled 70S ribosomes were used as substrates (Figure 3B). These results confirmed that RsmH and RsmI are AdoMet-dependent methyltransferases responsible for the methylation of C1402. Our results also revealed that RsmH and RsmI recognize the 30S subunit as a substrate, but not the naked 16S rRNA or the tightly-coupled 70S ribosome.
Figure 3.
In vitro reconstitution of m4Cm1402 using recombinant RsmH and RsmI. (A) SDS-PAGE analysis of the purified recombinant RsmI and RsmH proteins (1 µg each). The proteins were visualized by CBB staining. (B) In vitro dimethylation of C1402 by RsmI and RsmH. Prior to this experiment, the recombinant proteins were treated with Ado-Hcy to remove endogenous Ado-Met. The 30S subunit, 16S rRNA and 70S complex from ΔrsmH/ΔrsmI were used as substrates. After the reaction, the rRNA was extracted, digested with RNase T1 and subjected to LC/MS analysis. All panels show mass chromatograms detecting the tetramer fragments produced by RNase T1, containing C1402 with methylation(s). When 30S subunit was used as substrate, Cm1402 (m/z 645.1, top panel) and m4C1402 (m/z 645.1, second panel) were reconstituted in vitro by RsmI and RsmH, respectively, in the presence of Ado-Met. m4Cm1402 (m/z 652.1, gray line in the third panel) was formed by RsmI and RsmH together in the presence of Ado-Met. In this reaction, tetramer fragments containing Cm1402 or m4C1402 were also detected (black line). No methylation by RsmI or RsmH took place without Ado-Met (fourth panel), or when the 16S rRNA (fifth panel) and the 70S complex (bottom panel) were used as substrates.
Dimethylation of C1402 is involved in the efficiency of non-AUG initiation and fidelity of decoding
Since m4Cm1402 directly interacts with the phosphate backbone of the P-site codon, the dimethyl modification may modulate P-site functions. We constructed a dual luciferase reporter system to evaluate the fidelity of initiation, reading frame maintenance, and stop codon read-through by ribosomes lacking the dimethyl-modification of C1402. First, to evaluate the efficiency of translation initiation at non-AUG codons, we employed reporter constructs encoding Renilla luciferase (Rluc) starting with the AUG codon, and Firefly luciferase (Fluc) starting with various codons (AUG, AUU, UUG, GUG and AAA) on a single transcript (Figure 4A). The GUG, UUG and AUU codons have been shown to serve as initiation codons in E. coli. The AAA codon was used as a negative control, and expression of Fluc starting with this codon was negligible in all strains, demonstrating that Fluc in this construct is translated only from its own initiation site. Each construct was introduced into wild-type, ΔrsmH, ΔrsmI and ΔrsmH/ΔrsmI. The efficiency of translation initiation in each strain was measured by dividing the chemiluminescence of Fluc by that of Rluc, and normalizing the ratio to that of wild-type E. coli (relative F/R value). As shown in Figure 4A, the ΔrsmH strain, lacking the N4-methylation of m4Cm1402, showed a 1.8-fold increase in the rate of AUU initiation (relative to AUG initiation) compared with those in wild-type and ΔrsmI strains. This result suggests that the N4-methylation of m4Cm1402 negatively regulates the efficiency of initiation at the AUU codon. In addition, ΔrsmH/ΔrsmI showed a 1.8-fold increase in relative AUU initiation, a 20% decrease in relative UUG initiation and a 10% decrease in relative GUG initiation compared with the wild type. These data indicate that both the 2′-O-methylation and N4-methylation of C1402 are needed for efficient initiation at the UUG and GUG codons.
Figure 4.
Non-AUG initiation, stop codon read-through and reading frame maintenance in the absence of rsmH and/or rsmI. (A) Efficiency of translation initiation at non-AUG codons in E. coli wild-type (WT) (white), ΔrsmI (gray), ΔrsmH (stripe) and ΔrsmI/ΔrsmH (black). The reporter constructs with various initiation codons are shown at the top. The reporter genes Rluc (Renilla), starting with AUG codon, and Fluc (Firefly), starting with various codons (AUG, AUU, UUG, GUG or AAA), are tandemly arranged in the same transcript. The efficiency of translation initiation was assessed by dividing the chemiluminescence of Fluc by that of Rluc (the F/R value). For each initiation codon, the relative F/R values for the knockout strains were normalized to that of the wild-type strain. SD: Shine–Dalgarno sequence. (B) Assessment of stop codon read-through and reading frame maintenance in E. coli wild-type (WT) (white), ΔrsmI (gray), ΔrsmH (stripe) and ΔrsmI/ΔrsmH (black). The reporter constructs shown at the top contain the Rluc (Renilla) and Fluc (Firefly) open reading frames with short windows to assess stop codon read-through (UGA or UAG) or reading frame maintenance (+1 or –1 frameshifts) inserted between them. The translational efficiency of each construct was measured by the F/R value. For each construct, the relative F/R values for the knockout strains were normalized to that of the wild-type strain.
Next, to measure the efficiency of stop codon read-through and reading frame maintenance in the absence of C1402 methylation, we employed fusion constructs containing the Rluc and Fluc open reading frames separated only by short windows containing a stop codon (UGA or UAG), or a frameshift site (+1 or –1) (Figure 4B). The translational efficiency of each construct was assessed by measuring the F/R values. As shown in Figure 4B, the ΔrsmH strain showed a decrease of 50% in UGA read-through. In contrast, the ΔrsmI strain showed a 1.3-fold increase in UGA read-through efficiency. However, the relative F/R value for the UGA construct in the ΔrsmH/ΔrsmI strain was similar to that of the ΔrsmH strain, indicating that rsmH plays a dominant role in the efficiency of UGA read-through. In addition, we observed that the ΔrsmI strain showed a 1.3-fold increase in +1 frameshift and a 1.2-fold increase in –1 frameshift. The ΔrsmH strain showed a 20% decrease in UAG read-through, a 10% decrease in +1 frameshift and a 1.2-fold increase in –1 frameshift. According to these data, the lack of each type of methylation affects translational efficiency to some extent, and the N4-methylation is especially important in UGA read-through.
DISCUSSION
We describe here the identification of two methyltransferases, RsmH and RsmI, which are responsible for the N4-methylation and 2′-O-methylation, respectively, of C1402 in the 16S rRNA of E. coli. According to the COG database, rsmH and rsmI are conserved in almost all species of bacteria (48), suggesting that m4Cm-modification at the P-site is a common structural feature of bacterial 16S rRNA. In contrast, neither gene has been found in archaeal species (48), suggesting that the corresponding site in archaeal species is unmodified or modified differently. In the eukaryotes, rsmH orthologs exist in several species of plants and vertebrates, including human (Supplementary Figure S1). However, the site corresponding to E. coli C1402 in the human 18S rRNA is modified to Cm (not to m4Cm). In addition, m4C was identified at position 841 in the hamster mitochondrial 12S rRNA (51), which is the position equivalent to C1402 in the E. coli 16S rRNA. These observations suggest that the human ortholog of rsmH appears to encode a mitochondrial N4-methyltransferase that forms m4C in the 12S rRNA. On the other hand, it seems that rsmI orthologs do not exist in most eukaryotes apart from some plants, even though 2′-O-methylation at the site corresponding to E. coli C1402 is found in Saccharomyces cerevisiae, humans and Arabidopsis thaliana. It is known that the C (UGAUGA) and D (CUGA) box (C/D box) snoRNA guides 2′-O-methylation at that site in eukaryotic cytoplasmic rRNA (52). These observations suggest that 2′-O-methylation at this site is conserved not only in bacteria but also in eukaryotes.
During the biogenesis of ribosomes, rRNA modifications are introduced at different stages of subunit formation to assist in the efficient assembly of ribosomal particles (53). Some modifications occur on naked rRNAs during transcription, while others are introduced at early or later stages of assembly. In an in vitro reconstitution of m4Cm1402, both RsmH and RsmI used the assembled 30S particle as a substrate for methylation, while neither enzyme recognized the naked 16S rRNA or the tightly coupled 70S ribosome. Considering that m4Cm1402 is localized at the P-site, RsmH and RsmI might access the 30S subunit from the inter-subunit side and enter the P-site to specifically recognize C1402. In this model, an association between the 50S and 30S subunits would disturb the interactions of both enzymes. This would explain why neither enzyme used the tightly coupled 70S ribosome as a substrate. Since the efficiency of N4-methylation by RsmH was quite low when the 30S subunit was used as a substrate, we speculate that RsmH (and RsmI) employs the intermediary particle of the 30S subunit as an actual substrate at the late stage of ribosomal assembly in the cell.
In the crystal structure of 70S ribosome complexed with mRNA, and tRNAs, the P-site in helix 44 of the 16S rRNA is shaped mainly by five conserved bases: m4Cm1402, C1403, m3U1498, A1499 and A1500 (Figures 1A and 5). m4Cm1402 confers the following interactions: the N4 nitrogen of the amino group forms a hydrogen bond with the oxygen atom of the phosphate backbone between the 2nd and 3rd letters of the P-site codon, the N3 nitrogen forms a hydrogen bond with the N6 amino group of A1500, and the base stacks on the cytosine ring of C1403. In addition, the N4-methyl group of m4Cm1402 is close to the N3-methyl group of m3U1498; this distance is calculated to be 3–4 Å (Figure 5B). Since the van der Waals diameter of a methyl group is 2.0 Å, these two methyl groups could interact via a van der Waals attraction. In addition, the N4-methyl group donates an electron to the base, to strengthen the stacking force to C1403 and the hydrogen bond between the N3 of m4Cm1402 and the N6 of A1500. It is known that 2′-O-methylation stabilizes the sugar pucker as a C3′-endo conformation (54,55). In fact, as seen in the crystal structure, m4Cm1402 adopts the C3′-endo form. If it adopted the C2′-endo form, m4Cm1402 could no longer associate with the P-site mRNA in the current structure. Taken together, the N4-methylation by RsmH and the 2′-O-methylation by RsmI are involved in stabilizing the local structure and interactions of the P-site to accommodate the codon–anticodon helix.
Using the dual luciferase reporter assay, we showed that the deletion of rsmH or rsmI affects the efficiency of non-AUG initiation and the fidelity of translation. A relative increase in initiation at the AUU codon was induced by a lack of N4-methylation. It is supposed that the formation of the 30S IC with a non-AUG initiation codon is kinetically excluded due to the unstable binding of initiator fMet-tRNAiMet and the lower rates of IF3 dissociation and 50S docking (10,11). As discussed earlier, the lack of N4-methylation at C1402 is thought to modulate the local structure of the P-site, and might allow the site to accept the non-canonical codon–anticodon helix. When the AUU codon is recognized by the initiator tRNA at the P-site, the 3rd letter of AUU codon is mismatched with the 1st letter of the anticodon. Considering that the N4 nitrogen of m4Cm1402 forms a hydrogen bond with the 5′ phosphate of the 3rd letter of the AUU codon, the lack of N4-methylation might directly influence this interaction to accommodate the AUU codon, leading to efficient formation of the 30S IC and then the 70S IC. In E. coli, AUU is used as the initiation codon in the infC gene, which encodes IF3. The expression level of IF3 is controlled by the efficacy of AUU initiation (56,57). Since IF3 is involved in the selection and formation of the 30S IC by accelerating the influx of initiator tRNA (10), it is thought that a low concentration of IF3 enhances the efficiency of AUU initiation and translation of infC, thus maintaining the cellular concentration of IF3. Based on our results, N4-methylation of m4Cm1402 by RsmH also plays a role in regulating AUU initiation. Furthermore, the dimethyl-modification of C1402 contributes slightly to the efficacy of translation initiation at GUG or UUG. When GUG or UUG is recognized by the initiator tRNA, the 1st letter of each codon is recognized by the 3rd letter of the anticodon as a G·U or U·U wobble pair, a situation that is different from the case in AUU initiation. The dimethyl-modification of C1402 might stabilize this non-canonical pairing at the P-site. Therefore we suggest that the dimethyl-modification of C1402 plays an important role in P-site function, especially in start codon selection.
The N3-methyl group of m3U1498 is located in the vicinity of the N4-methyl group of m4Cm1402 at the P-site. The gene rsmE was shown to encode a methyltransferase for m3U1498 (58). No growth phenotype could be observed when a ΔrsmE strain was grown alone, but it was defective when grown in competition with wild-type E. coli (58). In addition, when 16S rRNA methylation levels were low, the rsmE deletion apparently decreased the stringency of initiator tRNA selection (25). Taken together with our results, these observations indicate that methyl-modifications clustered at the P-site collectively work to maintain accurate translation initiation.
The dual luciferase assays also revealed that the relative rate of UGA read-through activity was increased in the absence of N4-methylation by RsmH. This result was unexpected because read-through efficiency is determined at the A-site (59,60). However, it is known that some genetic mutations at the P-site influence read-through activity (61). In addition, a recent report indicated that a mismatch introduced in the codon–anticodon pairing at the P-site elevated the affinity of the release factor for the A-site (62), suggesting that the geometry of the P-site codon–anticodon helix affects termination codon recognition at the A-site. It can be speculated that the lack of N4 methylation of C1402 influences the configuration of the P-site codon–anticodon helix, thus modulating the activity of RF2 and the UGA codon specificity at the A-site.
In this study, we showed that m4Cm1402 in the 16S rRNA modulates the accuracy of P-site function. In yeast, it has been reported that loss of rRNA modifications in the decoding center collectively affected biogenesis and function of the small subunit (63). It is possible that m4Cm1402 is also involved in other ribosomal functions and in the biogenesis of the 30S subunit. Further investigations are needed to clarify these issues.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan (MEXT) (to T.S.); the Global COE Program for Chemistry Innovation (to S.K.); and the New Energy and Industrial Technology Development Organization (NEDO) (to T.S.). Funding for open access charge: MEXT.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Drs J. Dinman, T. Miki and H. Mori for providing materials. We thank the Suzuki lab members including Drs Takeo Suzuki, Y. Ikeuchi, K. Kitahara, T. Yokoyama, and Y. Sakaguchi for technical support and for many fruitful discussions.
REFERENCES
- 1.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 2.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 3.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 4.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 5.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 6.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 7.Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl Acad. Sci. USA. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simonetti A, Marzi S, Jenner L, Myasnikov A, Romby P, Yusupova G, Klaholz BP, Yusupov M. A structural view of translation initiation in bacteria. Cell. Mol. Life Sci. 2009;66:423–436. doi: 10.1007/s00018-008-8416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson DN. Initiation of Protein Synthesis in Eubacteria. Weinheim: WILEY-VCH Verlag GmbH &KGaA; 2004. [Google Scholar]
- 10.Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol. Cell. 2006;23:183–193. doi: 10.1016/j.molcel.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 11.Milon P, Konevega AL, Gualerzi CO, Rodnina MV. Kinetic checkpoint at a late step in translation initiation. Mol. Cell. 2008;30:712–720. doi: 10.1016/j.molcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Qin D, Fredrick K. Control of translation initiation involves a factor-induced rearrangement of helix 44 of 16S ribosomal RNA. Mol. Microbiol. 2009;71:1239–1249. doi: 10.1111/j.1365-2958.2009.06598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jemiolo DK, Zwieb C, Dahlberg AE. Point mutations in the 3′ minor domain of 16S rRNA of E.coli. Nucleic Acids Res. 1985;13:8631–8643. doi: 10.1093/nar/13.23.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier N, Goringer HU, Kleuvers B, Scheibe U, Eberle J, Szymkowiak C, Zacharias M, Wagner R. The importance of individual nucleotides for the structure and function of rRNA molecules in E. coli. A mutagenesis study. FEBS Lett. 1986;204:89–95. doi: 10.1016/0014-5793(86)81392-8. [DOI] [PubMed] [Google Scholar]
- 15.Thomas CL, Gregory RJ, Winslow G, Muto A, Zimmermann RA. Mutations within the decoding site of Escherichia coli 16S rRNA: growth rate impairment, lethality and intragenic suppression. Nucleic Acids Res. 1988;16:8129–8146. doi: 10.1093/nar/16.16.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunningham PR, Nurse K, Bakin A, Weitzmann CJ, Pflumm M, Ofengand J. Interaction between the two conserved single-stranded regions at the decoding site of small subunit ribosomal RNA is essential for ribosome function. Biochemistry. 1992;31:12012–12022. doi: 10.1021/bi00163a008. [DOI] [PubMed] [Google Scholar]
- 17.Ringquist S, Cunningham P, Weitzmann C, Formenoy L, Pleij C, Ofengand J, Gold L. Translation initiation complex formation with 30 S ribosomal particles mutated at conserved positions in the 3′-minor domain of 16 S RNA. J. Mol. Biol. 1993;234:14–27. doi: 10.1006/jmbi.1993.1560. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham PR, Nurse K, Weitzmann CJ, Ofengand J. Functional effects of base changes which further define the decoding center of Escherichia coli 16S ribosomal RNA: mutation of C1404, G1405, C1496, G1497, and U1498. Biochemistry. 1993;32:7172–7180. doi: 10.1021/bi00079a014. [DOI] [PubMed] [Google Scholar]
- 19.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 20.Brimacombe R, Mitchell P, Osswald M, Stade K, Bochkariov D. Clustering of modified nucleotides at the functional center of bacterial ribosomal RNA. FASEB J. 1993;7:161–167. doi: 10.1096/fasebj.7.1.8422963. [DOI] [PubMed] [Google Scholar]
- 21.Grosjean H. Fine-Tuning of RNA Functions by Modification and Editing. New York: Springer; 2005. [Google Scholar]
- 22.Xu Z, O'F;arrell HC, Rife JP, Culver GM. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat. Struct. Mol. Biol. 2008;15:534–536. doi: 10.1038/nsmb.1408. [DOI] [PubMed] [Google Scholar]
- 23.Connolly K, Rife JP, Culver G. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol. Microbiol. 2008;70:1062–1075. doi: 10.1111/j.1365-2958.2008.06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douthwaite S, Fourmy D, Yoshizawa S. Fine-Tuning of RNA Functions by Modification and Editing. New York: Springer; 2005. [Google Scholar]
- 25.Das G, Thotala DK, Kapoor S, Karunanithi S, Thakur SS, Singh NS, Varshney U. Role of 16S ribosomal RNA methylations in translation initiation in Escherichia coli. EMBO J. 2008;27:840–851. doi: 10.1038/emboj.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nichols JL, Lane BG. N-4-methyl-2′-O-methyl cytidine and other methyl-substituted nucleoside constituents of Escherichia coli ribosomal and soluble RNA. Biochim. Biophys. Acta. 1966;119:649–651. doi: 10.1016/0005-2787(66)90147-x. [DOI] [PubMed] [Google Scholar]
- 27.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'S;ouza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinform. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guymon R, Pomerantz SC, Crain PF, McCloskey JA. Influence of phylogeny on posttranscriptional modification of rRNA in thermophilic prokaryotes: the complete modification map of 16S rRNA of Thermus thermophilus. Biochemistry. 2006;45:4888–4899. doi: 10.1021/bi052579p. [DOI] [PubMed] [Google Scholar]
- 29.de Crecy-Lagard V, Marck C, Brochier-Armanet C, Grosjean H. Comparative RNomics and modomics in Mollicutes: prediction of gene function and evolutionary implications. IUBMB Life. 2007;59:634–658. doi: 10.1080/15216540701604632. [DOI] [PubMed] [Google Scholar]
- 30.Emmerechts G, Barbe S, Herdewijn P, Anne J, Rozenski J. Post-transcriptional modification mapping in the Clostridium acetobutylicum 16S rRNA by mass spectrometry and reverse transcriptase assays. Nucleic Acids Res. 2007;35:3494–3503. doi: 10.1093/nar/gkm248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki T, Ikeuchi Y, Noma A, Suzuki T, Sakaguchi Y. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007;425:211–229. doi: 10.1016/S0076-6879(07)25009-8. [DOI] [PubMed] [Google Scholar]
- 32.Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell. 2006;21:97–108. doi: 10.1016/j.molcel.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Soma A, Ikeuchi Y, Kanemasa S, Kobayashi K, Ogasawara N, Ote T, Kato J, Watanabe K, Sekine Y, Suzuki T. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell. 2003;12:689–698. doi: 10.1016/s1097-2765(03)00346-0. [DOI] [PubMed] [Google Scholar]
- 35.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006–2008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miki T, Yamamoto Y, Matsuda H. A novel, simple, high-throughput method for isolation of genome-wide transposon insertion mutants of Escherichia coli K-12. Methods Mol. Biol. 2008;416:195–204. doi: 10.1007/978-1-59745-321-9_13. [DOI] [PubMed] [Google Scholar]
- 37.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spedding G. Isolation and Analysis of Ribosomes from Prokaryotes, Eukaryotes, and Organelles, Ribosomes and Protein Synthesis, A Practical Approach. New York: Oxford University Press; 1990. [Google Scholar]
- 40.Komoda T, Sato NS, Phelps SS, Namba N, Joseph S, Suzuki T. The A-site finger in 23 S rRNA acts as a functional attenuator for translocation. J. Biol. Chem. 2006;281:32303–32309. doi: 10.1074/jbc.M607058200. [DOI] [PubMed] [Google Scholar]
- 41.Moazed D, Stern S, Noller HF. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J. Mol. Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 42.Andersen TE, Porse BT, Kirpekar F. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA. 2004;10:907–913. doi: 10.1261/rna.5259404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 44.Lesnyak DV, Osipiuk J, Skarina T, Sergiev PV, Bogdanov AA, Edwards A, Savchenko A, Joachimiak A, Dontsova OA. Methyltransferase that modifies guanine 966 of the 16 S rRNA: functional identification and tertiary structure. J. Biol. Chem. 2007;282:5880–5887. doi: 10.1074/jbc.M608214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanada T, Suzuki T, Yokogawa T, Takemoto-Hori C, Sprinzl M, Watanabe K. Translation ability of mitochondrial tRNAsSer with unusual secondary structures in an in vitro translation system of bovine mitochondria. Genes Cells. 2001;6:1019–1030. doi: 10.1046/j.1365-2443.2001.00491.x. [DOI] [PubMed] [Google Scholar]
- 46.O'C;onnor M, Goringer HU, Dahlberg AE. A ribosomal ambiguity mutation in the 530 loop of E. coli 16S rRNA. Nucleic Acids Res. 1992;20:4221–4227. doi: 10.1093/nar/20.16.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirabayashi N, Sato NS, Suzuki T. Conserved loop sequence of helix 69 in Escherichia coli 23 S rRNA is involved in A-site tRNA binding and translational fidelity. J. Biol. Chem. 2006;281:17203–17211. doi: 10.1074/jbc.M511728200. [DOI] [PubMed] [Google Scholar]
- 48.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 49.Andersen NM, Douthwaite S. YebU is a m5C methyltransferase specific for 16 S rRNA nucleotide 1407. J. Mol. Biol. 2006;359:777–786. doi: 10.1016/j.jmb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 50.Ofengand J, Del Campo M. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Curtiss R, editor. Washington, DC: ASM Press; 2004. [Google Scholar]
- 51.Dubin DT, Taylor RH, Davenport LW. Methylation status of 13S ribosomal RNA from hamster mitochondria: the presence of a novel riboside, N4-methylcytidine. Nucleic Acids Res. 1978;5:4385–4397. doi: 10.1093/nar/5.11.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 2008;36:D178–D183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit. Rev. Biochem. Mol. Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 54.Davis DR. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, D.C.: ASM Press; 1998. pp. 85–102. [Google Scholar]
- 55.Mahto SK, Chow CS. Synthesis and solution conformation studies of the modified nucleoside N-4,2′-O-dimethylcytidine (m(4)Cm) and its analogues. Bioorg. Med. Chem. 2008;16:8795–8800. doi: 10.1016/j.bmc.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sussman JK, Simons EL, Simons RW. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol. 1996;21:347–360. doi: 10.1046/j.1365-2958.1996.6371354.x. [DOI] [PubMed] [Google Scholar]
- 57.Sacerdot C, Chiaruttini C, Engst K, Graffe M, Milet M, Mathy N, Dondon J, Springer M. The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol. Microbiol. 1996;21:331–346. doi: 10.1046/j.1365-2958.1996.6361359.x. [DOI] [PubMed] [Google Scholar]
- 58.Basturea GN, Rudd KE, Deutscher MP. Identification and characterization of RsmE, the founding member of a new RNA base methyltransferase family. RNA. 2006;12:426–434. doi: 10.1261/rna.2283106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 61.O'C;onnor M, Thomas CL, Zimmermann RA, Dahlberg AE. Decoding fidelity at the ribosomal A and P sites: influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res. 1997;25:1185–1193. doi: 10.1093/nar/25.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 64.Mcluckey SA, Vanberkel GJ, Glish GL. Tandem Mass-spectrometry of small, multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]