Abstract
In vitro compartmentalization (IVC) was employed for the first time to select for novel bacteriophage λ integrase variants displaying significantly enhanced recombination activity on a non-cognate target DNA sequence. These variants displayed up to 9-fold increased recombination activity over the parental enzyme, and one mutant recombined the chosen non-cognate substrate more efficiently than the parental enzyme recombined the wild-type DNA substrate. The in vitro specificity phenotype extended to the intracellular recombination of episomal vectors in HEK293 cells. Surprisingly, mutations conferring the strongest phenotype do not occur in the λ integrase core-binding domain, which is known to interact directly with cognate target sequences. Instead, they locate to the N-terminal domain which allosterically modulates integrase activity, highlighting a previously unknown role for this domain in directing integrase specificity. The method we describe provides a robust, completely in vitro platform for the development of novel integrase reagent tools for in vitro DNA manipulation and other biotechnological applications.
INTRODUCTION
Conservative site-specific DNA recombinases directing the manipulation of transgenes are essential tools for controlled genome modifications. Notably, the Cre and Flp recombinases have been developed into powerful tools facilitating excision, integration, inversion and translocations of DNA segments between their respective recombination target sites (also referred to as cognate sites) (1–4). However, the absence of endogenous cognate sites in mammalian genomes usually requires these to be stably introduced through either homologous recombination, e.g. in mouse embryonic stem cells, or by random integration (5). The ‘primed’, predetermined locus is then amenable to targeted manipulation by site-specific recombination reactions.
A potential strategy to overcome this limitation is to engineer recombinases with altered site specificities (6–8). To this end, Cre recombinase variants have been described that are able to specifically recombine novel target sites and excise HIV proviral genomic DNA in mammalian cells (9,10). Flp and bacteriophage phiC31 recombinase variants have also been described that utilize native genomic sequences as recombination target sites (11,12). Other approaches include chimeric enzymes comprising of a recombinase domain fused to zinc finger modules with defined DNA-binding specificities (13,14). Site-specific zinc finger nucleases that stimulate homologous recombination at the site of an induced genomic DNA double-strand break represent another strategy for achieving directed gene replacement inside eukaryotic cells (15,16).
Bacterial selection systems relying on identification of functional mutants through reporter gene activation (17–20) or substrate-linked protein evolution (10) are the predominant methodologies for engineering altered site-specificities in recombinases. A genetic selection system in yeast has also been described that yielded HIV-1 integrase variants displaying altered DNA-binding affinities (21). In vitro compartmentalization (IVC) is a cell-free directed evolution platform, wherein gene variants and the proteins they encode are clonally encapsulated in the aqueous compartments of an oil-in-water emulsion (22,23). It has been used to evolve several classes of nucleic-acid transacting proteins, including methylases, transcription factors and restriction enzymes (24–26). A related methodology utilizing compartmentalization of bacterial cells has also been used to evolve DNA polymerases with tailored properties (27,28).
In the present study, we demonstrate the use of IVC to evolve variants of bacteriophage λ integrase with altered site-specificity. λ integrase (Int) is the prototypical member of the large tyrosine-recombinase family that includes Cre and Flp. Int is central to the bacteriophage lifecycle, facilitating the controlled integration and excision of its genome into and out of the host bacterial chromosome, respectively (29,30). An Int variant, bearing two activating mutations (E174K/E218K) in the catalytic core domain, (Int-h/218) has been used in genome manipulation strategies in mammalian and plant cells, and thus represents an important tool for a variety of biotechnological applications (31–33).
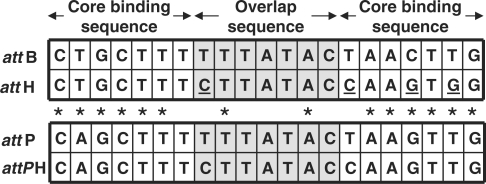
Int is a heterobivalent DNA-binding protein able to catalyze site-specific recombination between a pair of target sequences, termed att sites, in the absence of high-energy cofactors (34). The target sequences (attP in the bacteriophage genome, attB in the bacterial genome) comprise a pair of 7-bp inverted core-binding sites separated by a 7-bp ‘overlap’ region (Figure 1). The core sequence is bound by a subdomain of the large C-terminal domain of Int [amino acid (aa)-residues 65–356]. In the much longer attP site, the core sequence is flanked by binding sites for accessory DNA-bending factors IHF, FIS and Xis. In addition to these accessory sites, several ‘arm’ binding sites for the N-terminal domain of Int (aa 1–64) also flank the attP core site. These ‘arm’ regions are essential for activating efficient DNA cleavage by the C-terminal catalytic domain of Int, and thus contribute to the regulation of recombination directionality (35,36).
Figure 1.
Sequence alignment of the core bacterial attB and human attH sequences. The 7 bp highlighted in grey represent the overlap sequence, which must be identical in the respective attP and attPH recombination partners shown below. The asterisks below the sequences indicate nucleotides that may interact with integrase (44).
Our results show that one particular Int variant that we have evolved by IVC is able to recombine a non-cognate target sequence bearing homology to the attB sequence (termed attH) more efficiently than the parental enzyme Int-h/218 recombines attB. Interestingly, the mutations conferring the altered specificity reside in a region of λ integrase hitherto not implicated in the control of substrate specificity. Our results thus demonstrate a first example for a strategy to generate Int variants with altered specificity, and thus may be applied to enlarge the tool box for controlled genome manipulations.
MATERIALS AND METHODS
Materials
Oligonucleotides were purchased from First Base (Singapore); restriction enzymes and T4 polynucleotide kinase were from NEB; Accuzyme DNA polymerase and T7 RNA polymerase were from Bioline (UK); DNA purification kits were from Qiagen and chemical reagents were from Sigma. The following primers were used:
IntNDE-F: 5′-CACACATATGGGAAGAAGGCGAAGTCATGAGCGC-3′
IntECO-R: 5′-CTCTGAATTCTCATTATTTGATTTCAATTTTGTCCCACTCCCTGCC-3′
attB-petF: 5′-CTGCTTTTTTATACTAACTTGGTGATGCCGGCCACGATGCGTC-3′
attH-petF: 5′-CTGCTTTCTTATACCAAGTGGATGCCGGCCACGATGCGTC-3′
aTT-petR: 5′-CGCCACAGGTGCGGTTGCTG-3′
Pet-F2: 5′-CATCGGTGATGTCGGCGAT-3′
Pet-RC: 5′-CGGATATAGTTCCTCCTTTCAGCA-3′
pCMVattPH-QC1: 5′-CATTTTACGTTTCTCGTTCAGCTTTCTTATACTAAGTTGGCATTATAAAAAAGCATTGC-3′
pCMVattPH-QC2: 5′-GCAATGCTTTTTTATAATGCCAACTTAGTATAAGAAAGCTGAACGAGAAACGTAAAATG-3′
pCMVattP-F3: 5′-CCAAAAACGAGGAGGATTTG-3′
pCMVattP-R1: 5′-ACTCAGACAATGCGATGCAA-3′
Int-FRev-R: 5′-CATGACTTCGCCTTCTTCCCAT-3′
IntY342A-QC1: 5′-CGGACACCATGGCATCACAGGCTCGTGATGACAGAGGCAGGGAG-3′
IntY342A-QC2: 5′-CTCCCTGCCTCTGTCATCACGAGCCTGTGATGCCATGGTGTCCG-3′
IntECOHIS-R: 5′-CTCTGAATTCTCATTAGTGGTGATGGTGATGATGTTTGATTTCAATTTTGTCCCACTCCCTGCC-3′
IntECO-F: 5′-CGGAATTCCGATGGGAAGAAGGCGAAGTCATGAGCGC-3′
IntXBA-R: 5′-GCTCTAGAGCTCAGTGATGGTGATGGTGATGTAATTTG-3′
attP-PHQC1: 5′-CGTTTCTCGTTCAGCTTTCTTATACCAAGTGGGCATATTAAAAAAGCATTGC-3′
attP-PHQC2: 5′-GCAATGCTTTTTTAATATGCCCACTTGGTATAAGAAAGCTGAACGAGAAACG-3′
attB-HQC1: 5′-AGCTAGCTGAAGCCTGCTTTCTTATACCAAGTGGAGCGAACGCAATTGAA-3′
attB-HQC2: 5′-TTCAATTGCGTTCGCTCCACTTGGTATAAGAAAGCAGGCTTCAGCTAGCT-3′
Rec-SYBR-F2: 5′-AGGTGTCCACTCCCAGGTC-3′
Rec-SYBR-R3: 5′-CGATCCTCTACGCCGGACGC-3′
Rec-SYBR-HAT 5′-CCATACGATGTTCCAGATTACGC-3′
SS-F 5′-GGCAGGCTTGAGATCTGG-3′
SYBRgDNA-R: 5′-AAGTCAGTCCCATCCCAGAA-3′.
Vector construction
Integrase cDNA was amplified from the vector pInt-h/218 (37) using primers IntNDE-F and IntECO-R and ligated into the NdeI/EcoRI sites of pET-22b to generate the vector pET-Int. The 21-bp attB or attH sites were then inserted upstream of the integrase gene using primer pairs attB-petF/att-petR and attH-petF/att-petR, respectively. Amplified products were phosphorylated using T4 polynucleotide kinase and self-ligated to generate the vectors pET-attB-Int and pET-attH-Int. A randomly mutagenized integrase cDNA library was made by error-prone polymerase chain reaction (PCR) (38,39) using primers IntNDE-F and IntECO-R and the parental pInt-h/218 as a template. Sequencing of 12 random library clones indicated that 25% of the clones had a single missense mutation, 33% had two missense mutations, 17% had three or more missense mutations and 25% were wild type (data not shown). None of the subsequently selected mutations was observed. Library DNA was restricted and ligated into pET-attH vector (pET-attH-Int with the parental Int gene removed) described above. The library was next amplified by PCR using primers Pet-F2 and Pet-RC to generate linear DNA templates for selection. The pCMVattPH vector was constructed by site-directed mutagenesis of the parental pCMVattPmut plasmid (32) using the Quikchange mutagenesis system (Stratagene) and primers pCMVattPH-QC1 and pCMVattPH-QC2. attPH substrate for selections was generated by PCR using primers pCMVattP-F3 and pCMVattP-R1 and the pCMVattPH vector template. Competitor attB and attP substrates were generated by PCR using primer pairs pet-F2/IntFRev-R and pCMVattP-F3/pCMVattP-R1 and plasmid templates pET-attB-Int and pCMVattPmut, respectively. The parental pCMVattPmut plasmid was additionally modified to enable independent detection of attB and attH recombination events via real-time PCR (see below). This was achieved by mutating the PCR priming site Rec-SYBR-F2 (5′-AGGTGTCCACTCCCAGGTC-3′) to Rec-SYBR-HAT (5′-CCATACGATGTTCCAGATTACGC-3′) using mutagenic PCR. The inactive mutant Y342A integrase (40) expression construct was made by Quickchange mutagenesis of pET-attH-Int using primers IntY432A-QC1 and IntY342A-QC2. For purification of recombinant proteins, integrase clones were subcloned into pET-22b with a C-terminal hexahistidine tag using primers IntNDE-F and IntECOHIS-R. For expression in HEK293 cells, integrase genes were amplified using primers IntECO-F and IntXBA-R and cloned in the EcoRI/XbaI sites of pcDNA3.1.
Green-fluorescent protein (GFP)-reporter vectors measuring attH × attPH and attH × attP recombination in cis were generated by Quickchange mutagenesis of the vector pλIR (32) using primer pairs attB-HQC1/attB-HQC2 and attP-PHQC-1/attP-PHQC-2.
In vitro selection of integrase mutants
In vitro coupled transcription–translation reactions were assembled on ice in 50 μl volumes and comprised 37% (v/v) T7 extract (Novagen), 30 ng (38.1 fmol, ∼1.5 × 1010 integrase variants) mutant library expression template (for round 1 of selection; 5, 1, 0.5 ng used in subsequent rounds), 20 ng (50.7 fmol) attPH substrate (for round 1 of selection; 5, 1, 0.5 ng used in subsequent rounds) and 0.5 µl (2.5 U) T7 RNA polymerase. Eight-hundred nanograms each of competing attB and attP substrates (respectively 4.9 and 2 pmol) (Supplementary Figure S1) was also added (for rounds 1 and 2 of selection; 1200 ng were used in rounds 3 and 4). Reactions were emulsified by dropwise addition of ice-cooled in vitro reaction mixtures (one drop per 5 s) to 450 µl of the oil phase [4.5% (v/v) Span 80, 0.5% (v/v) Tween-80 in mineral oil] in a 1.8-ml CryotubeTM vial (Nunc) under constant stirring (1150 r.p.m.) using a magnetic stir bar (8 × 3 mm, Jencons). Stirring was continued for 5 min after addition of the last drop and emulsions incubated at 30°C for 45 min. The emulsion was disrupted by ether extraction as previously described (25) and the aqueous phase purified using the DNA Clean & ConcentratorTM-5 Kit (Zymo Research). The purified selection products were amplified by up to three rounds of PCR with the sequentially nested primer pairs SS-F and PetRC, SS-F and IntECO-R, Rec-SYBR-F2 and IntECO-R and ligated into pET-attH as described above. Expression templates for subsequent rounds of selection were then amplified via PCR using primers pET-F2 and pET-RC. After round 4 of selection, the integrase mutant library was rediversified by staggered extension process (StEP) PCR shuffling (41) of the four most active clones with the starting library.
Screening of clones
Clones were screened by in vitro coupled transcription–translation followed by real-time PCR. The individual integrase clones coupled to attH were first amplified via PCR using primers pET-F2 and pET-RC and then purified using the DNA Clean & ConcentratorTM-5 Kit as per manufacturer’s instructions. Each 25 µl reaction comprised 20 ng of linear attH-integrase construct and 20 ng of linear attPH in the EcoProTM T7 System (Novagen). Reactions were incubated for 45 min at 37°C. Prior to real-time PCR analysis, all reactions were purified with the DNA Clean & ConcentratorTM-5 Kit and eluted in 12 µl water. Real-time PCR was performed with 200 nM each of primers Rec-SYBR-F2 and Rec-SYBR-R3 (to detect for attH × attPH recombination) and primers Rec-SYBR-HAT and Rec-SYBR-R3 (to detect for attB × attP recombination) using 3 µl of the eluate in a 20 µl final volume with the SYBR® GreenERTM qPCR Supermix for iCycler® (Invitrogen) on a Bio-Rad iCycler IQTM5. The cycling parameters were 95°C for 15 s followed by 60°C for 60 s (40 cycles). Reactions assaying for integration into endogenous attH in genomic DNA comprised 300 ng EcoRI-restricted human genomic DNA, 20 ng linear integrase construct and 10 ng linear attPH substrate in a 25 µl in vitro transcription–translation reaction. Reactions were incubated and processed as above. The primers used for real-time quantification were Rec-SYBR-F2 and SYBRgDNA-R.
Western blot analysis
Ten nanograms of linear substrate containing the parental or mutant integrase genes was combined with the Novagen EcoProTM T7 extract in a 20 µl reaction, and incubated for 30 min at 30°C. For western blot analysis, 3 µl of the above reactions was diluted 5× with water and subsequently size fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) on 10% bis-Tris NuPAge gels in MOPS-SDS running buffer (Invitrogen) at 150 V for 90 min, and transferred to Hybond-P® PVDF membrane (GE Healthcare) in NuPage transfer buffer (Invitrogen) with 10% methanol at 40 V for 60 min at 4°C. The membrane was probed with rabbit polyclonal to the 6 × his tag (ab1187, Abcam). Antibody–protein complexes were identified by ECL-Plus (Amersham), detected on the Bio-Rad VersaDocTM Imaging System and quantified using Quantity One 4.6.5 software.
Protein expression and purification
Two-milliliter cultures of Escherichia coli BL21 (DE3) cells carrying the various pet22 integrase vectors were grown for 16 h at 37°C in Luria–Bertani (LB) broth supplemented with Ampicillin (100 µg/ml; Sigma). One-hundred milliters of fresh LB broth was then inoculated with 1 ml of these cultures and grown for 2–3 h to an A600 of ∼0.5. Integrase expression was induced with 0.1 mM IPTG (Invitrogen) for 3 h; cultures were shaken at 30°C during induction. The cells were pelleted, resuspended in binding buffer [10 mM Tris–HCl (pH 8), 150 mM NaCl, 10 mM imidazole and protease inhibitor cocktail tablet; complete mini EDTA free (Roche)] and lysed by sonication. Cell debris was removed by centrifugation at 13 000 r.p.m. for 45 min at 4°C. Cleared lysates containing the 6 × His-tagged integrase proteins were mixed with 1.5 ml of nickel-charged resin (Ni–NTA Agarose; QIAGEN) which had been pre-equilibrated in binding buffer. The beads were subsequently rotated with the protein extracts for 3 h at 4°C. The Ni–NTA resins were then washed six times with chilled wash buffer (PBS + 80 mM imidazole). Two sequential elution steps were performed, each with 120 µl of elution buffer [250 mM imidazole, 50 mM Tris–HCl (pH 8), 1 mM DTT, 150 mM NaCl and 10 mM EDTA]. The amount of protein released was determined using the Quick Start™ Bradford Reagent (Bio-Rad Laboratories) as per manufacturer’s instructions. The purity of all proteins was determined by Coomassie Blue staining of polyacrylamide gels (NuPAGE® Novex 4–12% Bis–Tris Gel; Invitrogen).
In vitro recombination assay for purified proteins
One microgram of purified recombinant integrase protein was incubated with 200 ng of linear attH substrate and 100 ng of linear attPH substrate for 30 min at 37°C in recombination buffer (100 mM Tris pH 7.5, 500 mM NaCl, 25 mM DTT, 10 mM EDTA, 5 mg/ml bovine serum albumin). The reaction volume was 25 µl. One microliter of this reaction was subsequently used for real-time PCR quantification of recombination efficiency as described above.
Cell culture and fluorescence-activated cell sorting analysis
All cell culture reagents and culture plastics were obtained from Invitrogen/Gibco and Nunc, respectively, unless otherwise specified. Cell cultures were maintained at 37°C with 5% CO2. HEK-293 (ATCC: CRL-1573) cells were maintained in Dulbecco's; modified Eagle's; medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine and 1% (v/v) penicillin/streptomycin. Co-transfection of integrase constructs in pcDNA 3.1 and pλIR reporter plasmid into 293 cells was performed in 6-well plates. Twenty-four hours before transfection, 293 cells were seeded at a density of 800 000 cells per well. One microgram of parental or mutant integrase construct in pcDNA 3.1 was transfected per well, together with 2 µg of pλIR using Lipofectamine 2000 (Invitrogen) as per manufacturer’s instructions. All transfections were carried out in duplicate. Cells were incubated for 48 h prior to FACS analysis on the Facs Aria (Beckton Dickinson). Thirty-thousand cells were analyzed for GFP expression.
RESULTS
Selection of λ integrase variants with altered binding specificities by IVC
A search for the presence of the 21-bp sequence comprising bacterial attB in the human genome returned no perfect matches (our unpublished data). However, we identified a 21-bp sequence, termed attH, which differs from the bacterial attB site at one position in the 7-bp overlap sequence and three positions in the right arm (B′) core binding sequence (Figure 1). Two of the latter residues are known to be critical for binding of attB by wild-type integrase (42). This ‘attH’ site, which is present in exon 5 of the MCT5 gene (gene accession no. SLC16A4), was subsequently used as a new target recombination site. We used the previously described λ integrase mutant Int-h/218, subsequently referred to as the ‘parental’ enzyme, to construct a variant library. Int-h/218 carries the E174K and E218K mutations that enable λ integrase to function in the absence of accessory protein cofactors and negative supercoiling of attP (43).
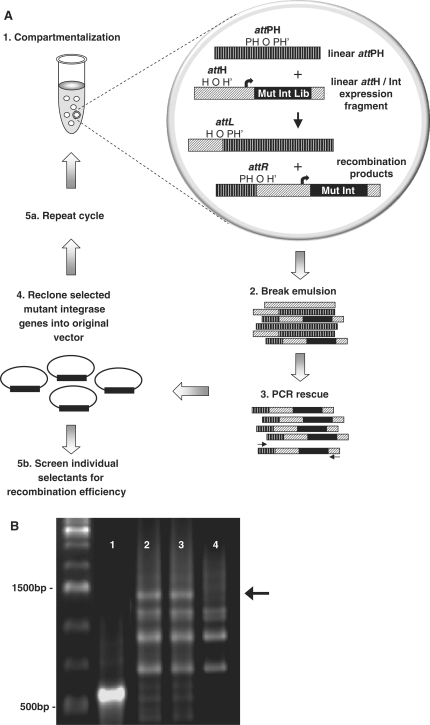
The selection strategy schematized in Figure 2A is based on the ability of in vitro expressed integrase variants to recombine attPH carrying an adjusted overlap sequence (provided in trans) with an attH site (Figure 1) tethered to the integrase expression construct. Emulsification ensures that an active integrase will only recombine attH tethered to its encoding gene, thus maintaining the essential genotype–phenotype linkage. Recombined products are resolved by PCR amplification using one primer specific for the integrase cassette and another for the recombined target DNA, i.e. attPH. Starting with a library of ∼1.5 × 1010 integrase variants, five rounds of selection were carried out. In order to direct preferential recombination of attH and attPH sites, competitor wild-type recombination substrates attB (100-fold excess) and attP (40-fold excess) were added. Figure 2B depicts an example of PCR-rescue of integrase variants capable of recombining attH and attPH after the first round of selection. A nested primer pair scoring for recombination, but not amplifying the complete integrase gene (Supplementary Figure S2), produced a discrete 500-bp band indicating correct recombination by library members (lane 1). Amplification of the complete integrase gene using a different primer pair also produced the expected band (∼1.4 kb), although several other non-specific products were co-amplified (lane 2). While inclusion of attB and attP competitor substrates appears to have a marginal effect (lane 3), the end-point 30-cycle PCR reaction most likely overrepresents the true recombination level in the presence of competitors. The levels of these competitors were, however, increased by up to 11 500- and 2400-fold excess over attH and attPH, respectively, in subsequent rounds of selection. Importantly, recombination was not observed in the absence of transcription/translation (lane 4).
Figure 2.
Selection of novel integrase mutants by IVC. (A) Schematic of the IVC protocol for selecting novel integrase mutants. (1) A library of integrase mutants is constructed using error-prone PCR. Linear mutant integrase expression constructs with appended attH sites are segregated into the aqueous compartments of a water-in-oil emulsion, together with a separate linear attPH construct. Compartmentalization ensures that after translation, an active integrase mutant only recombines into the attH sequence appended to it, and not that of other integrase mutants. (2,3,4) The emulsion is disrupted, and genes encoding the active integrase mutants are amplified by PCR for characterization and further rounds of selection. PCR primers (arrows) only amplify recombination products containing the genes of integrase mutants capable of performing the attH × attPH recombination event. (B) PCR-rescue of integrase selectants generated by IVC. DNA encoding active integrase mutants was amplified by PCR after round 1 of the selection. Lanes 1 and 2: integrase mutant library plus attPH substrate. The primer pair used in Lane 1 produces a short ∼500 bp amplicon that does not contain the full-length integrase gene, while the pair used in lane 2 produces a longer ∼1400 bp amplicon that contains the full-length integrase gene. Lane 3: integrase mutant library plus attPH substrate and competing attB and attP substrates. Lane 4: negative control selection (identical to lane 3 but with inactive IVC extract). PCR amplification in lanes 2–4 was performed with the same primer pair.
Selected λ integrase variants display altered substrate specificities
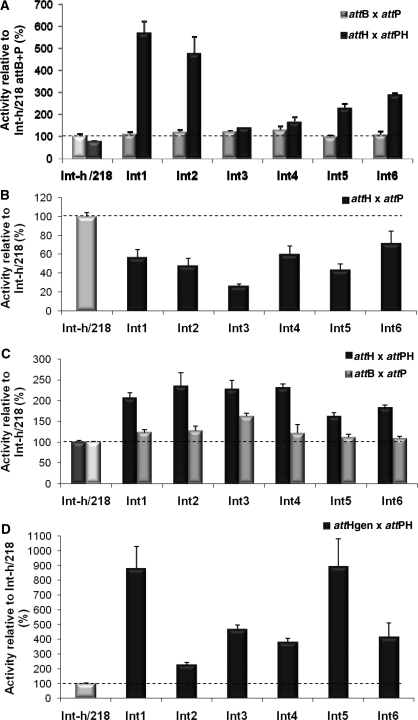
Integrase selectants (n = 74) were subjected to a secondary screen measuring attH x attPH recombination by in vitro expressed integrase using real-time PCR. Control experiments indicated that the primer pair used only amplified recombined DNA sequences (Supplementary Figure S3). In addition, further controls using the inactive Y342A Int mutant (40) and omitting the Int expression construct discounted recombination due to activity of a component in the expression extract (data not shown). Thirty variants showed improved attH × attPH recombination (Supplementary Figure S4) with the most active (Int1) giving 7-fold improved efficiency over the parental Int-h/218 (Figure 3A). Sequence analysis of the six most active variants revealed each to have between one and six mutations (Table 1). Several mutations are conserved between variants, notably I43F and H61R (resident in the N-terminal ‘arm’ DNA binding domain) and K122R, present in the core-binding (CB) domain. Interestingly, two of the mutants consistently showing a strong phenotype (Int1 and Int5) carry mutations only in the N-terminal domain. When tested for recombination with an attH x attP substrate pair, all six mutants showed moderately reduced recombination efficiency (between 25 and 70% of Int-h/218), suggesting they have evolved to preferentially recombine attH × attPH with perfectly matched overlap sequences (Figure 3B). Consistent with this, the mutants still recombined attH × attPH more efficiently than Int-h/218 in the presence of a 50-fold excess of wild-type attB and attP sites (Figure 3C).
Figure 3.
Improved attH × attPH recombination efficiency of integrase mutants translated in vitro. (A) The recombination efficiency of 74 integrase selectants on attP × attPH substrates was assayed using real-time PCR (Supplementary Figure S1) and results for the six best clones are shown. All activities are presented relative to activity of parental Int-h/218 with the attB × attP substrates (100%). Error bars indicate standard deviation of two independent experiments. (B) The efficiency of the six best selectants was also investigated for recombination between the non-cognate attH × attP substrate pair. (C) Recombination efficiency of the six selectants using attH × attPH substrates in the presence of 50-fold excess attB and attP competitors. Recombination of the competing attB. and attP substrates within the same reaction mix was also measured. Activities are presented relative to activity of parental Int-h/218 with the attP × attPH and attB × attP substrates (100%). (D) Recombination of endogenous attH site (attHgen) in genomic DNA by integrase selectants and Int-h/218. Activities are presented relative to activity of parental Int-h/218 (100%).
Table 1.
Location and number of mutations present in Int variants selected by IVC
 |
We next assayed recombination into the attH site present in EcoRI-restricted human genomic DNA extracted from HEK293 cells. All six mutants recombined into the endogenous attH site (attHgen) more proficiently than Int-h/218 with Int1 and Int5 showing a 9-fold increase (Figure 3D). Importantly, western analysis showed no difference in the in vitro expression levels of these two mutants compared to Int-h/218 (Supplementary Figure S5A).
Altered substrate specificity of recombinant λ integrase variants
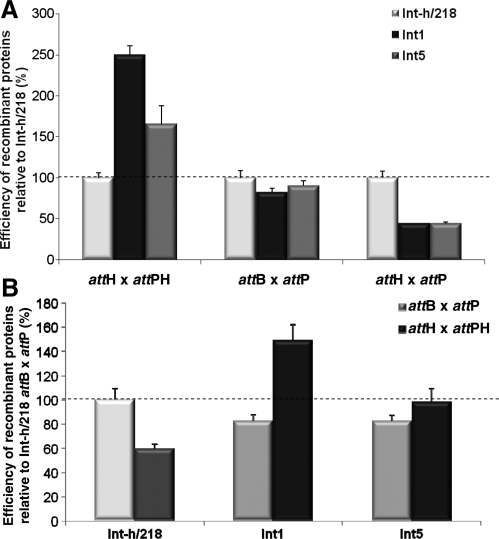
In order to discount possible confounding effects of factors present in the in vitro expression extract, we purified recombinant Int1 and Int5 proteins (Supplementary Figure S5B) and assayed their recombination proficiencies. As shown in Figure 4A, both variants preferentially recombine attH × attPH over attB × attP or the non-matching attH × attP pair, which is consistent with our previous findings. Of note, Int1 recombined attH × attPH 40% more efficiently than Int-h/218 recombined the attB × attP pair (Figure 4B). Int5 processed the attH × attPH pair with the same efficiency as Int-h/218 recombined its cognate attB × attP pair. Taken together, the data show an additive effect of the I43A and H61R mutations as the double mutant (Int1) consistently outperforms the single I43A (Int5) mutant.
Figure 4.
Improved recombination efficiency of recombinant integrase mutant proteins. (A) Real-time PCR was used to measure recombination by integrase enzymes. Recombinant mutant integrase proteins (Int1, Int5) are more efficient at performing the attH × attPH recombination reaction than recombinant parental integrase protein. They are also less efficient at recombining the non-cognate attH × attP substrate pair. Activities are presented relative to activity of Int-h/218 on each substrate pair (100%). (B) The Int1 clone is more efficient at the attH × attPH reaction than the parental integrase is at the attB × attP reaction. Activities are presented relative to activity of Int-h/218 on the attB × attP substrate pair (100%). Error bars indicate standard deviation of two independent experiments.
Recombination of episomal substrates in HEK293 cells
We next tested recombination by Int1 and Int5 in HEK293 cells using a reporter construct wherein recombination of att sites flanking an inversely orientated GFP cassette results in expression of GFP (Supplementary Figure S6) (32). Consistent with the in vitro phenotype (Figure 4B), Int1 and Int5 processed the attB × attP substrate less efficiently that In-h/218 (respectively 50% and 65% activity of Int-218 as measured by FACS analysis) (Figure 5). Int1 recombined attH × attPH ∼37% more efficiently than attB × attP, again in agreement with the in vitro phenotype. While Int-h/218 and Int1 showed similar efficiencies for attH × attPH as judged by numbers of GFP-positive cells, the average GFP intensity for Int5 was ∼1.6-fold higher, thus indicating improved intracellular substrate processing. Int5 preferentially recombined attB × attP over attH × attPH in this assay, which is not consistent with the behavior of the recombinant purified protein measured for intermolecular recombination in vitro (Figure 4B). In order to assay for non-specific recombination by Int1 and Int5 arising from relaxed specificity, we also tested an attH × attP GFP reporter. This substrate was poorly processed by all the integrases tested (<1% activity, data not shown).
Figure 5.
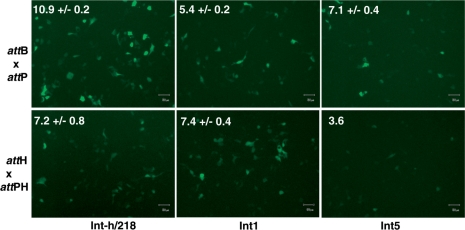
Preferential recombination of episomal attH × attPH substrates in HEK293 cells. Cells were transfected with GFP reporter constructs assaying attB × attP recombination (top panel) or attH × attPH recombination (lower panel) along with Int-h/218 and Int1/Int5 expression constructs. GFP expression was imaged 48 h post-expression. Cells (n = 30 000) were subsequently analyzed by FACS analysis. The percentage GFP-positive cells is indicated in upper left of each pane and represents average ± SD of two independent experiments.
DISCUSSION
In this article, we have applied IVC to select for integrase variants with improved recombination activities on a non-cognate substrate pair. In vitro selection methodologies are not restricted by transformation efficiencies, and thus allow the interrogation of exceptionally large variant libraries for desired phenotypes. IVC, therefore, enabled us to select from a considerably larger starting library (∼1.5 × 1010 variants) compared to bacteria-based recombinase selection systems (∼1 × 106 variants) (10). Mutants selected after five rounds showed up to 9-fold increased recombination of the non-cognate attH × attPH substrate pair, and one particular variant (Int1) catalyzed recombination more efficiently than the parental Int-h/218 enzyme recombined the standard attB × attP substrate.
Previous studies have shown relaxed substrate specificity to be the immediate consequence of selection pressure for altered specificity in lambda, Flp and Cre recombinases (10,12,17,18). The Int1 and Int5 mutants selected here are less proficient at recombining in vitro both attB × attP and the non-matching attH × attP substrate pair when compared to the parental Int-h/218 enzyme. Along with the latter, they are also inefficient at recombining attH × attP in the GFP reporter assay. These results suggest that the mutational drift is on a pathway toward a more restricted target site specificity. We anticipate, therefore, that more rounds of IVC with additional selection pressure (increased competitor substrates and reduced incubation times) will yield mutants displaying further restricted specificities.
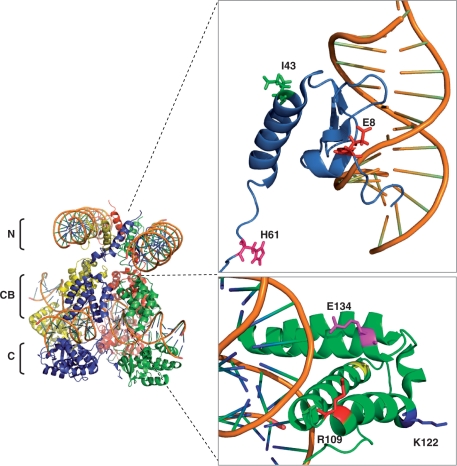
λ Integrase comprises three distinct domains that collaborate within a higher-order tetrameric structure to form a dynamic recombinogenic complex (Figure 6) (44). The N-terminal DNA-binding domain (residues 1–63) binds ‘arm-type’ DNA sequences flanking the attP core site. The Int DNA CB (residues 75–175) primarily recognizes the 7bp attP × attB core DNA sequence motifs and is joined to the C-terminal catalytic domain (residues 176–356). Binding of the N-domain to arm sites allosterically modulates the coupled CB and catalytic domain to increase the affinity to core sites, which ultimately enables DNA strand cleavage and productive recombination of attB × attP (44). Previous studies have identified mutations in the CB and catalytic domains of λ integrase which impact on recombination site specificity (17,18). Surprisingly, the mutations in the most active variants identified in our study, I43F and H61R, reside in the N-terminal domain. Within this domain, neither I43 nor H61 interact directly with arm-site DNA (Figure 6). I43 resides on an α-helix buttressing a three-stranded antiparallel B-sheet and flexible N-terminal tail that interact with the major and minor groove of arm site DNA, respectively. In the tetrameric structure, it is orientated away from the DNA toward the opposing helix of an adjacent Int protomer. Modeling indicates that mutation to the bulkier phenylalanine potentially induces a steric clash with I43 of the opposing protomer’s α-helix (not shown). This may influence the dynamics of the N-terminal allosteric rearrangement so as to license the recombination of non-cognate substrates. Interestingly, the R42L mutant in the closely related bacteriophage Hong Kong 022 integrase displays binding to non-cognate core-sites but is recombination deficient (19). Together, these results indicate a regional hotspot in the vicinity of I43 that plays a role in recombinase specificity. The R42L mutant additionally displays enhanced binding to arm-site DNA. In the absence of binding to arm-site DNA, the N-terminal domain intrinsically inhibits binding of λ integrase to core sites (45). The presence of arm-site sequences has also been shown to increase recombination of core-sites by Int-h/218 in vivo (32). Enhanced tethering to the ancillary arm sites might therefore maintain the CB and catalytic domains in association with non-cognate substrates for extended periods in order to promote the assembly and subsequent processing of viable recombinogenic complexes. It will therefore be worthwhile assaying the arm-binding affinities of the I43F and other N-terminal mutants described in this study.
Figure 6.
Location of selectant mutations in λ integrase. Left panel: quaternary structure of the λ integrase tetramer bound to core and arm site DNA. N: N-terminal domain; CB: core-binding domain; C: cayalytic domain. Upper right panel: Selectant mutations mapped onto the N-terminal domain. Bottom right panel: IVC-selected mutations in the CB domain that are in the vicinity of target DNA. Structures adapted from refs. 44 and 50. Image created using Pymol.
H61 resides in a loop region identified as a potential swivel about which the N-domains can rotate (46). While we did not analyze the H61R mutation in isolation, our data indicate that it potentiates the I43F phenotype. It may therefore act in concert with I43F to further impact on the N-domain conformations driving allosteric control of integrase activity. Furthermore, H61 is proximal to an α-helical coupler (residues 64–74) that connects to the CB domain. Mutations in the coupler (L64A in particular) affect the directionality of recombination to favor excision over integration (47). The parental enzyme Int-h/218 shows increased specificity for attB × attP derivatives over attL × attR, i.e. for integrative over excisive recombination (48). In our IVC assay, we screened for the accumulation of intermolecular recombination products, thus inadvertently further enriching for mutants with such a phenotype. Further studies combining coupler mutations along with the mutations identified here will be useful to delineate potential links between control of recombination direction and specificity.
Several mutations present in the CB domain of the less-active Int2 variant lie in the vicinity of the core site DNA. Notably, R109 is in contact with the phosphate backbone of the adenine residue base paired to thymidine-20 in the attB sequence (Figure 6). Loss of this contact through mutation to glycine might influence substrate specificity to tolerate the guanidine present at this position in attH (Figure 1). Similarly, E134 is in the vicinity of the cytosine base-paired to the guanidine-21 in the attB sequence, and mutation to glycine might confer acceptance of the non-canonical guanidine-20 in attH. Analysis of these novel mutations in isolation is warranted in order to further understand their contributions to substrate specificity.
A subtle difference in substrate type may account for the weaker attH × attPH recombination phenotype displayed by Int1 and Int5 in the episomal cell-based assay when compared with the results obtained in vitro. In the cell-based assay, recombination occurs in cis, i.e. both att sites are present on the same DNA molecule. Our selection strategy employed target sites present in trans and additionally involved substantial amounts of competitor DNA. An interesting possibility to be explored further is that the newly selected Int variants favor intermolecular over intramolecular recombination. Another possibility for the observed discrepancy is the reduced dynamic range inherent to the GFP reporter assay compared to the real-time PCR quantitation assay. While the mutant integrases were able to recombine in vitro the endogenous attH site of purified genomic DNA (Figure 3D), they could not do so in vivo (data not shown). There are several possible reasons for this observation. First, as suggested for the weaker phenotype observed in the episomal recombination assay, the linear attH substrates may not faithfully mimic the endogenous site, for example, with respect to higher-order chromatin structure(s). Second, the MCT5 gene locus may be a transcriptionally sub-active region, and hence inherently recalcitrant to recombination. Third, it is not known if MCT5 is a dispensable gene. Further selections utilizing different substrate types and alternative target sites will shed light on the suitability of IVC-generated integrases for in vivo applications.
Recombinase engineering thus far has mainly focused on the Cre and Flp recombinases. Although related to Int, these enzymes are devoid of an N-terminal domain and do not require ancillary arm sites for recombination. While this makes them good candidates for engineering, the intrinsic predisposition of λ integrase toward integrative over excessive recombination (48) makes it an attractive enzyme to pursue applications which require intermolecular recombination, in particular genomic transgene insertions. However, future selections using the exceptionally large Cre and Flp libraries afforded by IVC may also yield useful variants.
The selection platform detailed here should complement existing methodologies for generating recombinase enzymes with novel properties. Furthermore, by being completely in vitro, the IVC selection platform can potentially be used to engineer other desirable features into recombinases, such as thermal stability and altered salt/pH tolerance. These properties cannot be readily engineered using bacterial systems and would be desirable in reagent tools. In this regard, the improved in vitro recombination by Int1 using the attH × attPH substrate pair indicates that it may be a useful reagent tool for recombination-based cloning applications (49).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Agency for Science, Technology and Research (Singapore).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Drs Michael Entzeroth and William Sun for useful discussions. All experiments were carried out at the Experimental Therapeutics Centre (Singapore).
REFERENCES
- 1.Metzger D, Feil R. Engineering the mouse genome by site-specific recombination. Curr. Opin. Biotechnol. 1999;10:470–476. doi: 10.1016/s0958-1669(99)00012-9. [DOI] [PubMed] [Google Scholar]
- 2.Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- 3.Seibler J, Schubeler D, Fiering S, Groudine M, Bode J. DNA cassette exchange in ES cells mediated by Flp recombinase: an efficient strategy for repeated modification of tagged loci by marker-free constructs. Biochemistry. 1998;37:6229–6234. doi: 10.1021/bi980288t. [DOI] [PubMed] [Google Scholar]
- 4.Birling MC, Gofflot F, Warot X. Site-specific recombinases for manipulation of the mouse genome. Methods Mol. Biol. 2009;561:245–263. doi: 10.1007/978-1-60327-019-9_16. [DOI] [PubMed] [Google Scholar]
- 5.Irion S, Luche H, Gadue P, Fehling HJ, Kennedy M, Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat. Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- 6.Coates CJ, Kaminski JM, Summers JB, Segal DJ, Miller AD, Kolb AF. Site-directed genome modification: derivatives of DNA-modifying enzymes as targeting tools. Trends Biotechnol. 2005;23:407–419. doi: 10.1016/j.tibtech.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Collins CH, Yokobayashi Y, Umeno D, Arnold FH. Engineering proteins that bind, move, make and break DNA. Curr. Opin. Biotechnol. 2003;14:665. doi: 10.1016/j.copbio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Zhou R, Droge P. High-precision surgery in the genome of human stem cells. Curr. Genomics. 2006;7:427–433. [Google Scholar]
- 9.Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 proviral DNA excision using an evolved recombinase. Science. 2007;316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 10.Buchholz F, Stewart AF. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat. Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 11.Sclimenti CR, Thyagarajan B, Calos MP. Directed evolution of a recombinase for improved genomic integration at a native human sequence. Nucleic Acids Res. 2001;29:5044–5051. doi: 10.1093/nar/29.24.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolusani S, Ma CH, Paek A, Konieczka JH, Jayaram M, Voziyanov Y. Evolution of variants of yeast site-specific recombinase Flp that utilize native genomic sequences as recombination target sites. Nucleic Acids Res. 2006;34:5259–5269. doi: 10.1093/nar/gkl548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akopian A, He J, Boocock MR, Stark WM. Chimeric recombinases with designed DNA sequence recognition. Proc. Natl Acad. Sci. USA. 2003;100:8688–8691. doi: 10.1073/pnas.1533177100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordley RM, Gersbach CA, Barbas CF., III Synthesis of programmable integrases. Proc. Natl Acad. Sci. USA. 2009;106:5053–5058. doi: 10.1073/pnas.0812502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porteus MH. Mammalian gene targeting with designed zinc finger nucleases. Mol. Ther. 2006;13:438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 17.Dorgai L, Yagil E, Weisberg RA. Identifying determinants of recombination specificity: construction and characterization of mutant bacteriophage integrases. J. Mol. Biol. 1995;252:178–188. doi: 10.1006/jmbi.1995.0486. [DOI] [PubMed] [Google Scholar]
- 18.Yagil E, Dorgai L, Weisberg RA. Identifying determinants of recombination specificity: construction and characterization of chimeric bacteriophage integrases. J. Mol. Biol. 1995;252:163–177. doi: 10.1006/jmbi.1995.0485. [DOI] [PubMed] [Google Scholar]
- 19.Cheng Q, Swalla BM, Beck M, Alcaraz R., Jr, Gumport RI, Gardner JF. Specificity determinants for bacteriophage Hong Kong 022 integrase: analysis of mutants with relaxed core-binding specificities. Mol. Microbiol. 2000;36:424–436. doi: 10.1046/j.1365-2958.2000.01860.x. [DOI] [PubMed] [Google Scholar]
- 20.Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- 21.Calmels C, de Soultrait VR, Caumont A, Desjobert C, Faure A, Fournier M, Tarrago-Litvak L, Parissi V. Biochemical and random mutagenesis analysis of the region carrying the catalytic E152 amino acid of HIV-1 integrase. Nucleic Acids Res. 2004;32:1527–1538. doi: 10.1093/nar/gkh298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths AD, Tawfik DS. Miniaturising the laboratory in emulsion droplets. Trends Biotechnol. 2006;24:395–402. doi: 10.1016/j.tibtech.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Tawfik DS, Griffiths AD. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 1998;16:652–656. doi: 10.1038/nbt0798-652. [DOI] [PubMed] [Google Scholar]
- 24.Cohen HM, Tawfik DS, Griffiths AD. Altering the sequence specificity of HaeIII methyltransferase by directed evolution using in vitro compartmentalization. Protein Eng. Des. Sel. 2004;17:3–11. doi: 10.1093/protein/gzh001. [DOI] [PubMed] [Google Scholar]
- 25.Fen CX, Coomber DW, Lane DP, Ghadessy FJ. Directed evolution of p53 variants with altered DNA-binding specificities by in vitro compartmentalization. J. Mol. Biol. 2007;371:1238–1248. doi: 10.1016/j.jmb.2007.05.099. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Y, Roberts RJ. Selection of restriction endonucleases using artificial cells. Nucleic Acids Res. 2007;35:e83. doi: 10.1093/nar/gkm410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghadessy FJ, Ong JL, Holliger P. Directed evolution of polymerase function by compartmentalized self-replication. Proc. Natl Acad. Sci. USA. 2001;98:4552–4557. doi: 10.1073/pnas.071052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghadessy FJ, Ramsay N, Boudsocq F, Loakes D, Brown A, Iwai S, Vaisman A, Woodgate R, Holliger P. Generic expansion of the substrate spectrum of a DNA polymerase by directed evolution. Nat. Biotechnol. 2004;22:755–759. doi: 10.1038/nbt974. [DOI] [PubMed] [Google Scholar]
- 29.Campbell AM, editor. Episomes. New York: Academic Press; 1962. [Google Scholar]
- 30.Nash HA. Purification of bacteriophage λ Int protein. Nature. 1974;247:923–929. doi: 10.1038/247543a0. [DOI] [PubMed] [Google Scholar]
- 31.Suttie JL, Chilton M, Que Q. 2008 US patent no. 7351877. [Google Scholar]
- 32.Christ N, Corona T, Droge P. Site-specific recombination in eukaryotic cells mediated by mutant lambda integrases: implications for synaptic complex formation and the reactivity of episomal DNA segments. J. Mol. Biol. 2002;319:305–314. doi: 10.1016/S0022-2836(02)00327-3. [DOI] [PubMed] [Google Scholar]
- 33.Droge P, Enenkel B. 2005 EP patent no.1565562 (A1) [Google Scholar]
- 34.Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu. Rev. Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 35.Radman-Livaja M, Biswas T, Ellenberger T, Landy A, Aihara H. DNA arms do the legwork to ensure the directionality of lambda site-specific recombination. Curr. Opin. Struct. Biol. 2006;16:42–50. doi: 10.1016/j.sbi.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radman-Livaja M, Shaw C, Azaro M, Biswas T, Ellenberger T, Landy A. Arm sequences contribute to the architecture and catalytic function of a lambda integrase-Holliday junction complex. Mol. Cell. 2003;11:783–794. doi: 10.1016/s1097-2765(03)00111-4. [DOI] [PubMed] [Google Scholar]
- 37.Christ N, Droge P. Genetic manipulation of mouse embryonic stem cells by mutant lambda integrase. Genesis. 2002;32:203–208. doi: 10.1002/gene.10031. [DOI] [PubMed] [Google Scholar]
- 38.Vartanian JP, Henry M, Wain-Hobson S. Hypermutagenic PCR involving all four transitions and a sizeable proportion of transversions. Nucleic Acids Res. 1996;24:2627–2631. doi: 10.1093/nar/24.14.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaccolo M, Gherardi E. The effect of high-frequency random mutagenesis on in vitro protein evolution: a study on TEM-1 beta-lactamase. J. Mol. Biol. 1999;285:775–783. doi: 10.1006/jmbi.1998.2262. [DOI] [PubMed] [Google Scholar]
- 40.Kazmierczak RA, Swalla BM, Burgin AB, Gumport RI, Gardner JF. Regulation of site-specific recombination by the C-terminus of λ integrase. Nucleic Acids Res. 2002;30:5193–5204. doi: 10.1093/nar/gkf652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao H, Giver L, Shao Z, Affholter JA, Arnold FH. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat. Biotechnol. 1998;16:258–261. doi: 10.1038/nbt0398-258. [DOI] [PubMed] [Google Scholar]
- 42.Ross W, Landy A. Patterns of lambda Int recognition in the regions of strand exchange. Cell. 1983;33:261–272. doi: 10.1016/0092-8674(83)90355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorbach E, Christ N, Schwikardi M, Droge P. Site-specific recombination in human cells catalyzed by phage lambda integrase mutants. J. Mol. Biol. 2000;296:1175–1181. doi: 10.1006/jmbi.2000.3532. [DOI] [PubMed] [Google Scholar]
- 44.Biswas T, Aihara H, Radman-Livaja M, Filman D, Landy A, Ellenberger T. A structural basis for allosteric control of DNA recombination by lambda integrase. Nature. 2005;435:1059–1066. doi: 10.1038/nature03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkar D, Radman-Livaja M, Landy A. The small DNA binding domain of lambda integrase is a context-sensitive modulator of recombinase functions. EMBO J. 2001;20:1203–1212. doi: 10.1093/emboj/20.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hazelbaker D, Azaro MA, Landy A. A biotin interference assay highlights two different asymmetric interaction profiles for lambda integrase arm-type binding sites in integrative versus excisive recombination. J. Biol. Chem. 2008;283:12402–12414. doi: 10.1074/jbc.M800544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warren D, Lee SY, Landy A. Mutations in the amino-terminal domain of lambda-integrase have differential effects on integrative and excisive recombination. Mol. Microbiol. 2005;55:1104–1112. doi: 10.1111/j.1365-2958.2004.04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan SM, Droge P. Comparative analysis of sequence-specific DNA recombination systems in human embryonic stem cells. Stem Cells. 2005;23:868–873. doi: 10.1634/stemcells.2005-0044. [DOI] [PubMed] [Google Scholar]
- 49.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fadeev EA, Sam MD, Clubb RT. NMR structure of the amino-terminal domain of the lambda integrase protein in complex with DNA: immobilization of a flexible tail facilitates beta-sheet recognition of the major groove. J. Mol. Biol. 2009;388:682–690. doi: 10.1016/j.jmb.2009.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]