Abstract
High-resolution mapping of chromatin features has emerged as an important strategy for understanding gene regulation and epigenetic inheritance. We describe an in vivo tagging system coupled to chromatin purification for genome-wide epigenetic profiling in Caenorhabditis elegans. In this system, we coexpressed the Escherichia coli biotin ligase enzyme (BirA), together with the C. elegans H3.3 gene fused to BioTag, a 23-amino-acid peptide serving as a biotinylation substrate for BirA, in vivo in worms. We found that the fusion BioTag::H3.3 was efficiently biotinylated in vivo. We developed methods to isolate chromatin under different salt extraction conditions, followed by affinity purification of biotinylated chromatin with streptavidin and genome-wide profiling with microarrays. We found that embryonic chromatin is differentially extracted with increasing salt concentrations. Interestingly, chromatin that remains insoluble after washing in 600 mM salt is enriched at 5′ and 3′ ends, suggesting the presence of large protein complexes that render chromatin insoluble at transcriptional initiation and termination sites. We also found that H3.3 landscapes from these salt fractions display consistent features that correlate with gene activity: the most highly expressed genes contain the most H3.3. This versatile two-component approach has the potential of facilitating genome-wide chromatin dynamics and regulatory site identification in C. elegans.
INTRODUCTION
The DNA of eukaryotes is compacted by histones to form chromatin. The basic unit of chromatin is the nucleosome, containing 146 bp of DNA wrapped around the histone core, an octamer containing two copies each of histone H2A, H2B, H3 and H4 proteins. Nucleosomes are important not only for compacting the genome, but also as candidates to transmit epigenetic information (1–4). Most histones are expressed during S phase and are assembled into nucleosomes behind the DNA replication fork. In contrast, variant histones are expressed and incorporated into chromatin throughout the cell cycle by a distinct set of nucleosome assembly proteins. Particular histone variants have been found to be associated with diverse cellular functions such as DNA repair and mitosis.
Histone H3.3 is a universal histone variant that is expressed throughout the cell cycle. It is incorporated at sites of active transcription (5), and is enriched in histone modifications associated with transcriptional activity (6,7). Previous studies have documented the presence of H3.3 in the germline of diverse organisms, including Caenorhabditis elegans, Drosophila melanogaster and mice (8–11). H3.3 is incorporated into the sex (XY) body during meiotic sex chromosome inactivation in mice (11), and is depleted from the X chromosome, but not the autosomes, during meiosis in C. elegans (9). Importantly, H3.3 is retained in mature C. elegans sperm (9). Taken together, these results make H3.3 an excellent candidate to be involved in transmission of epigenetic information via the germline. This possibility is further strengthened by the observation in Xenopus that incorporation of H3.3 in the absence of transcription is required for the epigenetic memory of gene transcription during embryonic development after nuclear transplantation (12).
In recent years, many advances have been made in describing how chromatin differences might contribute to epigenetic processes in cell-line systems (13,14). However, the establishment and transmission of chromatin features through normal development and especially through the germline, remain elusive. For a better understanding of these processes, better tools are needed to dissect epigenetic events at a genome-wide level within the context of an entire organism.
Here, we describe a system to profile C. elegans chromatin genome wide. Caenorhabditis elegans is a promising model organism to dissect histone-based epigenetic processes, with an invariant developmental pattern and excellent genetics. Furthermore, adult worms provide an abundant source of germ cells, as almost half of the adult is occupied by the germline, making it an especially favorable organism to probe epigenetic processes in the totipotent germ cells. Caenorhabditis elegans has a small (∼100 Mb) genome that lacks DNA methylation, simplifying the study of chromatin-based processes by eliminating the need to parse out the interplay between histone- and DNA methylation-based mechanisms. Epigenetic events have been documented in C. elegans, including imprinting of the paternal X chromosome in early embryos (15), and specific loss of the paternal X chromosome during development when worms are exposed to stress (16).
We have developed a chromatin purification system in C. elegans for epigenomic profiling that involves in vivo biotinylation of a tagged histone. The biotin–streptavidin interaction, with a Kd of 10−15 M, is the strongest non-covalent interaction known, and it allows the recovery of essentially all biotinylated chromatin from samples. The use of biotinylated chromatin purification has been successful for Drosophila S2 cell lines and Arabidopsis plants (17,18). This system is especially suitable for H3.3, which differs from S-phase H3 by only 4-amino-acid residues and for which highly specific antibodies are not available. We have developed methods to purify chromatin under different salt conditions from worm embryos, followed by affinity pull-down with streptavidin and recovery of DNA from the pulled-down nucleosomes for genome-wide epigenetic profiling. We show that zygotic H3.3 is enriched in gene bodies, and H3.3 abundance correlates with gene activity: the more highly expressed genes have more H3.3, indicating that H3.3 incorporation is a good indicator of chromatin disruption associated with gene activity.
MATERIALS AND METHODS
Nematode strains and maintenance
Nematodes were cultured and manipulated genetically as described (19). All strains were grown at 20–23°C. The wild-type strain was the N2 (Bristol) strain, and LGIII: unc-119(ed3) was used for transformation. The following transgenic strains were created for this study: JJ2059, containing zuIs235 [(his-721-kb 5′ UTR::BIOTAG::3XHA::HIS-72::his-721-kb 3′ UTR); unc-119(ed3)]; JJ2060, containing zuIs236 [(his-721-kb 5′ UTR::BIRA::GFP:: his-721-kb 3′ UTR); unc-119(ed3)] and JJ2061, homozygous for both zuIs235 and zuIs236. unc-119 was used as marker to identify transformants based on its Unc+ phenotype. To obtain biochemical quantities of worms, we used peptone-rich plates seeded with NA22 bacteria (Supplementary Data 1).
Plasmids
Plasmids pSO220 (his-721-kb::BioTag::3XHA::HIS-72::his-721-kb) and PSO221(his-721-kb::BIRA::GFP::his-721-kb) were constructed using a two-step polymerase chain reaction (PCR) fusion method. In pSO220, the BioTag peptide sequence is MASSLRQILDSQKMEWRSNAGGS (corresponding to the DNA sequence of ATGGCTTCTTCTCTTCGTCAGATCCTCGACTCTCAGAAGATGGAGTGG CGTTCTAACGCTGGAGGATCT), and the 3XHA peptide sequence is YPYDVPDYAGYPYDVPDYAGSYPYDVPDYA (corresponding to the DNA sequence of TACCCATACGACGTTCCAGACTATGCCGGCTACC CCTATGATGTCCCGGACTATGCAGGATCTTATCCATATGACGT CCCAGATTACGCT). Both the BIOTAG and 3XHA sequences were optimized for worm codon usage. The BIOTAG::3XHA::HIS-72 fusion ORF was flanked by 1 kb of his-72 5′ and 3′ untranslated regions (UTRs) that have previously been shown to be capable of expression in the germline. In pSO221, the Escherichia coli BirA gene (M10123, GI145430) was fused at its C-terminus to green fluorescent protein (GFP) (containing introns to facilitate gene expression) using the linker peptide sequence SRPVAT (TCGAGACCGGTAGCTACT). Like pSO220, the BIRA::GFP ORF was also flanked by 1 kb of his-72 5′ and 3′ UTRs for expression.
Biolistic bombardment and transgenes
Plasmids pSO220 and pSO221 were introduced into unc-119(ed3) worms to generate lines expressing zuIs235 (BIOTAG::3XHA::HIS-72) and zuIs236 (BIRA::GFP) independently via microparticle bombardment (20). For zuIs235 (BIOTAG::3XHA::HIS-72), we obtained transformants with and without germline expression. For zuIs236 (BIRA::GFP), we only obtained transformants without germline expression. Thus, BIRA::GFP expression in the embryos represents mainly zygotic expression. As expected, different lines expressing the same transgene displayed similar somatic expression patterns. Based on its Unc+ phenotype, both BIOTAG::HIS-72 and BIRA::GFP are most likely integrated transgenes, and show a transmission rate of >90%. To generate JJ2060, we crossed zuIs235 (BIOTAG::3XHA::HIS-72) and zuIs236 (BIRA::GFP) to each other and identified progeny that is homozygous for both transgenes for subsequent experiments.
Microscopy, immunofluorescence and image analysis
Worm fixation procedures, immunofluorescence and GFP fluorescence image analysis were performed as previously described (9). For immunofluorescence, the primary anti-HA antibody used was mouse anti-HA (1: 1000 dilution, HA.11, MMS-101R from Covance, USA) and the secondary antibody used was Cy3 goat anti-mouse antibody (1: 500 dilution, 115-165-146 from Jackson ImmunoResearch Laboratories, USA).
Western blot analysis
Antibodies used for western blot analysis include the anti-HA antibody described above (at 1: 1000 dilution) and anti-mouse IgG-HRP (1: 10000 dilution, NA9310V from GE Healthcare UK Limited), as well as streptavidin-HRP (1: 5000 dilution, RPN1231V from GE Healthcare UK Limited). Note that streptavidin-HRP was resuspended in PBST (PBS with 0.1% Tween-20) containing 2% bovine serum albumin, instead of the conventionally used 5% milk. This step is necessary because milk powder contains molecules that interact with streptavidin, resulting in background HRP activity. In addition, the streptavidin-HRP incubation step was restricted to 30 min at room temperature. For streptavidin–Sepharose-bound nucleosomes, we first denatured the protein sample in 8 M urea, followed by the addition of 3XSDS containing β-mercaptoethanol, boiling for 10 min at 100°C and storing at −20°C until use.
Biotinylated chromatin affinity pull-down with streptavidin
A detailed version of our biotinylated chromatin affinity pull-down with streptavidin protocol is available as Supplementary Data 1. As biotinylated proteins are known to exist in vivo (21), we performed control experiments to ensure that (i) our streptavidin pull-down system specifically recovers DNA bound by biotinylated histone H3.3 variant containing nucleosomes; (ii) streptavidin–Sepharose alone will not pull down any DNA; and (iii) there are no biotinylated proteins in worms that bind DNA under the conditions used. We did not recover any DNA while performing a mock pull-down (Supplementary Figure S1). In addition, western blot analysis of histone octamers obtained during salt extraction from the 2.5 M eluate showed that none of the core histones was detectably biotinylated in worms (Supplementary Figure S2). These data further show that our system is specific for the introduced transgenes. In addition, there are several known peptides that serve as BirA targets, and we have also generated a separate transgenic line expressing a different BioTag (Biotag2 with the amino acid sequence of MAGGLNDIFEAQKIEWHEDTGGS) peptide fused to H3.3. Western blot analysis showed that this peptide sequence is biotinylated just as efficiently by BirA in vivo in worms (Supplementary Figure S3).
Worm growth
To obtain biochemical quantities of worm embryos for chromatin isolation followed by streptavidin–Sepharose pull-down, we amplified worm cultures in three steps. Worms were grown on peptone-rich media plates seeded with NA22 (PR-NA22) bacteria, which allowed us to reproducibly observe biotinylation of BioTag::H3.3. Note that it remains to be determined as to whether the medium, the bacteria or a combination of both is responsible for the observed reproducible biotinylation of BioTag::H3.3. On Day 1, we seeded three plates (100 ×15 mM) of unsynchronized BioTag::H3.3/BIRA::GFP worms onto five ‘big’ (150 × 15 mM) PR-NA22 plates, and worms were grown until the gravid adult stage at 22°C. On Day 4, we bleached the worms and expanded the culture to seven big PR-NA22 plates, and incubated the worms at 22°C until the gravid adult stage. On Day 7, worms were again bleached, but now the culture was expanded to 30 PR-NA22 plates at a concentration of 1 million embryos per plate, and incubated at 22°C until they reached the gravid adult stage. This translates to a total of ∼30 million embryos in 30 plates. Assuming that each embryo becomes an adult worm containing about 10 embryos, our growth procedure yields ∼10 million embryos per plate or 300 million embryos in total.
Worm embryo and blastomere preparation
Next, we collected gravid adults from all 30 plates for embryo isolation, followed by chitinase treatment to remove the eggshell of the embryos to obtain blastomeres. Worms were bleached in 800 ml of a solution containing 10% bleach/0.5 N KOH/M9 for 8 min. This high volume ensures efficient bleaching. Worms were then pelleted, resuspended in M9 buffer (19) and subjected to Miracloth filtration to separate embryos from incompletely digested worms. The embryos were then pelleted, and subjected to chitinase treatment (∼15 ml in 0.2 unit/ml of chitinase from Streptomyces griseus, Sigma C6137) to remove the eggshell surrounding worm embryos. Next, chitinase-treated embryos were washed three times in buffer A (15 mM Tris–HCl pH 7.5/2 mM MgCl2/0.34 M sucrose/0.15 mM spermine/0.5 mM spermidine/1 mM DTT/0.5 mM PMSF).
Worm embryonic nuclei preparation
Blastomeres were resuspended in 7.5 ml of Buffer A containing 0.25% NP-40 substitute (Fluka 74385) and 0.1% Triton X-100 and subjected to 15 strokes each of pestle A and pestle B homogenization. The homogenate was then subjected to a low speed spin (100g) to separate nuclei (supernatant) from incompletely homogenized blastomeres (pellet). The pellet was then re-extracted using 15 strokes each of pestle A and pestle B homogenization again, followed by a low-speed spin to recover nuclei in the supernatant. Next, nuclei from both extraction steps were pelleted using a high-speed spin (1000g). Nuclei of high quality are creamy or white in appearance and easy to resuspend.
Embryonic nuclei micrococcal nuclease treatment
Next, we used micrococcal nuclease (MNase) to digest the nuclei, stopped the MNase treatment with EGTA, followed by solubilization of chromatin at different salt concentrations. We used 0.8 U of MNase to digest about 150 µl of nuclei resuspended in 1 ml of buffer A for 10 min. We stopped the reaction by adding 2 mM EGTA. We performed a centrifugation step to separate the digested sample from MNase containing buffer. We then resuspended the pellet in 2 ml of 80-mM salt buffer (10 mM Tris-HCl pH 7.5/2 mM MgCl2/70 mM NaCl/2 mM EGTA/0.5 mM PMSF/0.1% Triton X-100) and extracted chromatin at 4°C for 1–4 h. We then performed a centrifugation step to separate the 80-mM salt-soluble chromatin from the pellet, which was then resuspended in 600 mM salt buffer (10 mM Tris-HCl pH 7.5/2 mM MgCl2/585 mM NaCl/2 mM EGTA/0.5 mM PMSF/0.1% Triton X-100) and extracted overnight at 4°C. A centrifugation step was then performed to separate the 600-mM salt-soluble chromatin from the 600-mM salt-insoluble pelleted material.
Biotinylated nucleosome affinity pull-down with streptavidin-Sepharose
We used the 80-mM and 600-mM salt-soluble chromatin, and also the 600-mM salt-washed pellet for affinity pull-down with streptavidin–Sepharose. We first equilibrated the streptavidin–Sepharose in 80 mM and 600-mM buffer. Next, we performed a centrifugation step at 4000g for 10 min to separate the 80-mM and 600-mM salt-soluble chromatin from the residual pellet. The supernatant from this centrifugation step was then used as input for affinity purification. Twenty percent of the salt-soluble material was saved as input and the rest used for the streptavidin–Sepharose pull-down. Streptavidin–Sepharose (100 µl) was added to 1 ml of each salt-soluble sample, and binding to streptavidin–Sepharose was performed at 4°C for 1 h. Next, we washed the beads in buffer containing the same salt concentrations (80 mM or 600 mM) three times. The unbound and 600-mM pellet fractions were also saved for DNA recovery. We then prepared DNA from the different chromatin isolation and affinity pull-down steps and quantified recovery of DNA by Nanodrop (Thermo Fisher, Inc.) measurements.
Microarray-based profiling
DNA samples were labeled with Cy3 or Cy5 by random priming following the NimbleGen manufacturer’s protocol. For samples containing mononucleosomes, we performed the labeling step with the following modifications as previously described (22): 50-μl strand-displacement reactions were performed at 37°C overnight and stopped by addition of 5-μl 0.5 M EDTA, transferred to a 1.5-ml tube, mixed with 5.7-μl 5-M NaCl and precipitated by addition of 60-μl isopropanol. After dissolving in water and measuring A260-levels, equal amounts of Cy3- and Cy5-labeled samples were mixed, and volumes were dried or reduced to ∼12.3 μl and delivered to the Fred Hutchinson Center Genomics Shared Resource for hybridization to NimbleGen microarrays. Profiling was performed on single custom-designed high-density 2.1 million feature isothermal microarrays (NimbleGen C_elegans_WS170_Tiling_Iso_HX1 tiling design, GEO GPL7098) purchased from NimbleGen, Inc., and was hybridized and scanned by the Fred Hutchinson Center Genomics Facility using NimbleGen protocols. The design was based on WormBase release WS170. This is an isothermal (target Tm = 76) design microarray, with a mean probe length of 54 (range 50–75) with all probes on the forward strand. The mean distance between probes is −6, with a range of −18 to 1111.
Data analysis
For all data figures, log-ratios of the Cy3 and Cy5 channels were computed using the method of Peng et al. (23). Ends plots were calculated as standard deviates of the Peng log-ratios to facilitate comparisons between arrays. All data sets were aligned with C. elegans genome release WS190 obtained from Wormbase (http://www.wormbase.org) for mapping. Ends analysis was performed essentially as previously described (17) except that we used 25-bp intervals for averaging to take advantage of the denser tiling (averaging ∼65 bp). We used the 8244 genes and 1151 operons that were well annotated in WS190. We aligned the genes and the operons at their 5′ and 3′ ends and rank-ordered them based on embryonic gene expression, measured as log-ratios of cDNA/genomic DNA using Nimblegen, Inc. tiling microarrays, as described (22). Tracks were displayed with SignalMap (NimbleGen, Inc.) as the Peng et al. log-ratios after smoothing using a window of three probes with weights of (1,3,1). Tracks displayed in Figure 6 from other studies were obtained as log-ratios and aligned with WS190, then divided into 25-bp intervals. For heat-map analysis of transposon families, we used consensus sequences obtained from RepBase version 14.09 (http://www.grinst.org) and searched against the WS190 genomic sequence. We aligned the 10 longest highest scoring hits at their 5′ ends and created heat maps using Java TreeView v. 1.1.0 using a contrast level of 2.0 (24). Data are available from GEO (Acc# 18898).
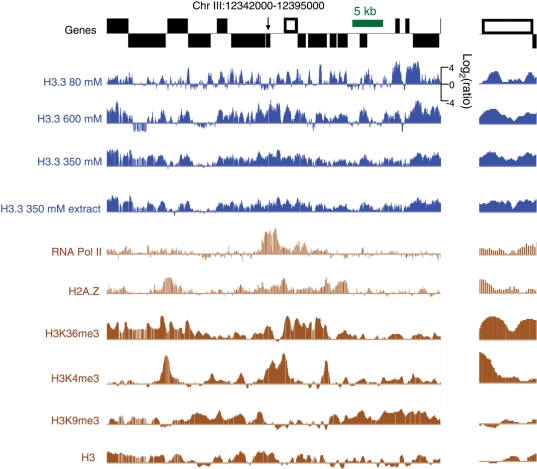
Figure 6.
H3.3 landscapes compared to other chromatin marks. The same gene-rich region displayed in Figure 4 is shown at lower and higher magnification. Blue tracks display H3.3 landscapes and brown tracks display landscapes based on data from previous studies: RNA polymerase II and H2A.Z (35) and H3K36me3, H3K4me3, H3K9me3 and H3 (34).
RESULTS AND DISCUSSION
In vivo biotinylation for chromatin affinity purification in C. elegans
The in vivo biotinylation system consists of two components. The first component is a transgenic worm line (BioTag::H3.3) expressing C. elegans H3.3 (HIS-72) fused to a BioTag, a ϵ-amino-acid peptide containing a lysine residue whose ϵ-amino group serves as substrate for biotinylation by BirA (25,26) (Figure 1A). The BioTag used in this study had been isolated from a synthetic peptide library screened for BirA-mediated biotinylation, first described by de Boer and co-workers (25). We also introduced three tandem hemagglutinin epitope (3XHA) tags between the BioTag and H3.3 to serve as a linker sequence and for immunofluorescence purposes.
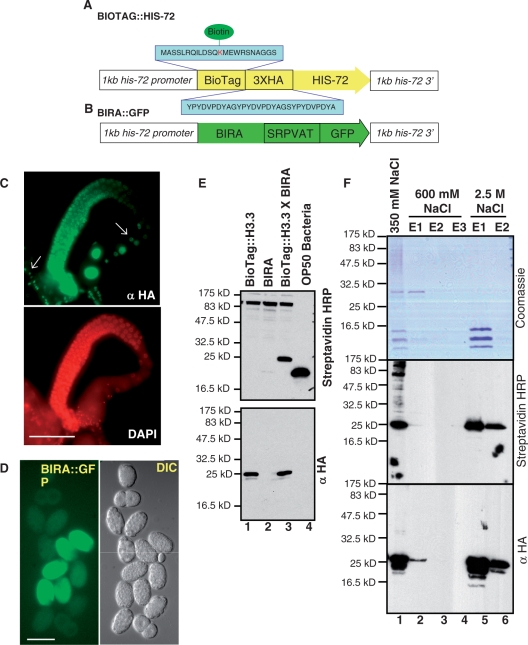
Figure 1.
In vivo chromatin biotinylation in C. elegans. Diagrams of (A) BioTag::H3.3 and (B) BirA::gfp transgenes used in this study. The H3.3 transgene was fused to the BioTag and the three hemagglutinin (3XHA) epitope tag at its N-terminus. This BioTag is a ϵ-amino-acid peptide containing a lysine whose ϵ-amino group serves as substrate for biotinylation by the E. coli Biotin Ligase (BirA). A linker amino acid sequence (SRPVAT) was used to fuse BIRA to GFP. Each transgene included 1 kb upstream and 1 kb downstream of the his-72 coding sequence. (C) An adult hermaphrodite fixed and stained for HA and DNA. BioTag::H3.3 is present in the germline and somatic cells (white arrows) of adult hermaphrodites and co-localizes with DNA, except in the mature oocytes, where it is stored in the nucleus as a maternal factor. Scale bar, 50 µm. (D) GFP fluorescence and DIC images of dissected embryos expressing BIRA::GFP. GFP fluorescence could be detected starting from the 50–80-cell stage embryos, and is distributed in both the cytoplasm and nuclei of the embryo. Scale bar, 50 µm. (E) In vivo biotinyation of BioTag::H3.3 is efficient in C. elegans. Western blot analysis using Streptavidin-HRP (top panel) and an HA antibody (bottom panel) for mixed stage worm strains expressing BioTag::H3.3 alone (lane 1), GFP::BIRA alone (lane 2), BioTag::H3.3 in combination with BIRA::GFP (BioTag::H3.3/BIRA::GFP, lane 3) and OP50 Bacteria (lane 4). Biotinylation of BioTag::H3.3 (∼21.4 kDa) could be detected only in worm strains expressing both BioTag::H3.3 and BIRA::GFP. The band residing between 16 and 25 kDa in the OP50 bacteria (food for the worm used to prepare the samples) lane is the endogenous biotinylated protein in bacteria. The anti-HA western blot experiment confirmed that the band detected by streptavidin::HRP is the transgene, since they have the same migration rate in the gel. (F) BioTag::H3.3 is incorporated into chromatin. Histones were extracted with increasing salt concentrations from embryos expressing BioTag::H3.3/BIRA::GFP. Core histones elute with 2.5 M NaCl but not with 600 mM NaCl. High salt (2.5 M) extraction was used to separate core histones from chromatin-associated proteins. Coomassie blue staining (top panel) and western blot analysis using Streptavidin-HRP (middle panel) and an HA antibody (bottom panel) are shown for samples retrieved during the core histone extraction procedure. Like core histones, BioTag::H3.3 elutes primarily at 2.5 M NaCl. Lane 1, nuclei resuspended in 350 mM NaCl; lanes 2–4: respectively the first, second and third eluates of the nuclear pellet resuspended in 600 mM NaCl; and lanes 5 and 6: respectively the first and second eluates of nuclei resuspended in 2.5 M NaCl.
The second component consists of a transgenic worm line (BIRA::GFP) expressing the E. coli BirA enzyme (Figure 1B), which catalyzes the transfer of biotin to the ϵ-amino group of lysine residue in its substrate. BirA was fused to GFP at its C-terminus to serve as a visualization marker to facilitate analysis of BIRA localization within the animals. Both BioTag::H3.3 and BIRA::GFP transgenes were driven by the his-72 promoter, a nearly ubiquitous promoter with germline expression capability derived from the endogenous his-72 H3.3 gene (9). These two transgenic lines were then crossed to each other, and worms homozygous for both transgenes were identified and used for all experiments.
The expression pattern of N-terminally tagged BioTag::H3.3 essentially recapitulates that of the previously published H3.3 transgene fused at its C-terminus to H3.3 also expressed under the his-72 promoter (9). BioTag::H3.3 displays punctate nuclear staining indicating incorporation into chromatin, and its expression could be detected in the germline and in the nuclei of all somatic cell types except in those of the intestines (Figure 1C). In contrast, the second component, BIRA::GFP fluorescence displayed a cytoplasmic and nuclear expression pattern (Figure 1D). That BIRA::GFP has a cytoplasmic distribution makes it likely that BIRA::GFP can biotinylate BioTag::H3.3 in the cytoplasm, prior to nucleosome assembly, minimizing potential bias caused by chromatin accessibility differences between loci.
BioTag::H3.3 is efficiently biotinylated in vivo
Next, we assayed for in vivo biotinylation of BioTag::H3.3 using proteins extracted from mixed stage worms on western blots probed with streptavidin-horseradish peroxidase (streptavidin-HRP). We were able to detect an abundant endogenous biotinylated protein, which is most likely acetyl-CoA carboxylase, a known biotin-dependent protein (21). We also detected a second biotinylated band in a worm strain expressing both BioTag::H3.3 and BIRA::GFP, showing that BioTag::H3.3 is biotinylated in vivo (Figure 1E). Furthermore, the intensity of the biotinylated BioTag::H3.3 band is comparable to that of the endogenous biotinylated protein. The identity of the biotinylated BioTag::H3.3 is further confirmed by western blot analysis, probing with an anti-HA antibody, which also detected a band at the same molecular weight (∼21.5 kDa).
Biotinylation of BioTag::H3.3 was detected only in worm strains expressing both BioTag::H3.3 and BIRA::GFP, but not in those expressing either transgene (Figure 1E), which indicates that the biotinylation of BioTag::H3.3 is specific and requires the presence of both components. This result also indicated that the endogenous C. elegans biotin ligase is not capable of biotinylating the BioTag, and that the E. coli BirA cannot detectably biotinylate any endogenous worm proteins. Thus, the BioTag is the sole target of E. coli BirA in C. elegans.
To confirm that BioTag::H3.3 is functional, we performed a salt fractionation experiment to separate nucleosomes (containing histone octamers) from non-nucleosomal proteins (27). Extracts from lysed embryonic nuclei in 350 mM salt were applied to hydroxyapatite, which binds DNA, and eluted at increasing salt concentrations. Non-nucleosomal proteins are predicted to elute at low salt concentrations, while core histones elute in 2.5 M NaCl. Coomassie blue staining of protein extracts during salt extraction showed that C. elegans histone octamers are enriched in the 2.5 M eluate fractions. The majority of BioTag::H3.3, predicted to be about 21.5 kDa, stained positively with streptavidin-HRP and anti-HA antibody in the 2.5 M NaCl eluate fractions, indicating that BioTag::H3.3 is incorporated into nucleosomes (Figure 1F). BioTag::H3.3 is evidently present at levels that are too low to detect by Coomassie blue staining. In summary, our results show that BioTag::H3.3 is biotinylated in vivo by BirA in C. elegans, and that it is incorporated into nucleosomes (Figure 1 and Supplementary Figures S1–3).
Biotinylated H3.3 chromatin purification from C. elegans embryos
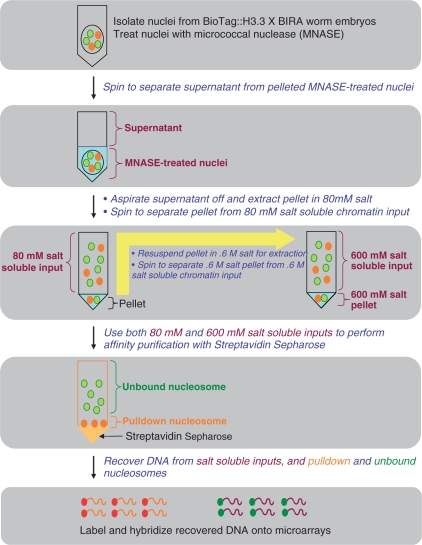
The general scheme of our biotinylated BioTag::H3.3 chromatin purification method is outlined in Figure 2. We first isolate mixed stage worm embryos, which are then treated with chitinase to remove the eggshells from the embryos. Next, we prepare nuclei using a non-ionic detergent in combination with glass pestle homogenization, followed by micrococcal nuclease (MNase) treatment of the nuclei to fragment the chromatin into nucleosomes. The MNase-digested chromatin is then extracted using different salt concentrations and these salt-solubilized nucleosome fractions are then used for affinity pull-down with streptavidin–Sepharose. DNA is then recovered from the samples, labeled by strand-displacement using Klenow fragment exo− DNA polymerase with Cy3- and Cy-5 containing 9-mers and hybridized to NimbleGen tiling microarrays.
Figure 2.
The scheme for purification of biotinylated chromatin in C. elegans embryos. We first prepared a biochemical quantity of gravid adult worms expressing biotinylated BioTag::H3.3. After obtaining nuclei from worm embryos, we subjected the nuclei to micrococcal nuclease digestion in buffer A (containing ∼17 mM salt). Next, we performed centrifugation to separate the supernatant from the MNase-treated nuclei. The nuclear pellet was then resuspended and extracted in low salt buffer (80 mM), followed by centrifugation to separate 80-mM salt-soluble chromatin from the pellet. The pellet was then resuspended in high salt buffer (600 mM), followed by centrifugation to separate the 600-mM salt-soluble chromatin from the 600-mM salt-insoluble pellet. In the next step, we used the 80-mM and 600-mM salt-soluble chromatin as inputs to perform affinity pull-down with streptavidin–Sepharose. We then recovered DNA from different steps during the streptavidin pull-down procedure, including the inputs, pulled-down and unbound nucleosomes, labeled the DNA, and hybridized the labeled DNA onto micorarrays.
In the first experiment, we obtained nuclei from mixed stage worm embryos expressing biotinylated BioTag::H3.3 and performed chromatin extraction as previously described (17). We treated the nuclei with MNase in 100 mM salt for 10 min and stopped the reaction with EDTA, followed by centrifugation. The resulting pellet was then resuspended in 350 mM NaCl for overnight extraction, and the soluble extract was affinity-purified with streptavidin–Sepharose.
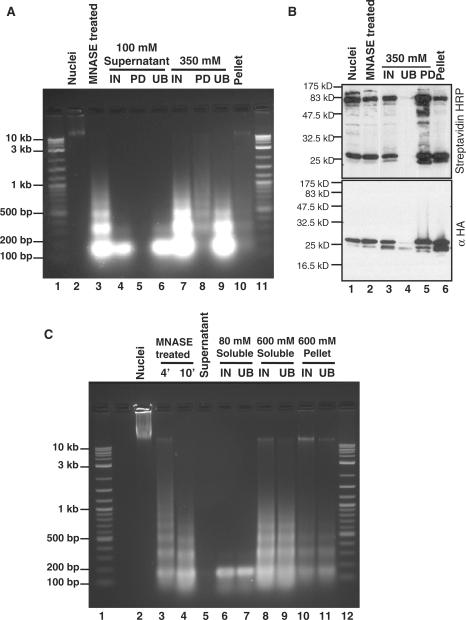
DNA recovered from MNase-digested chromatin produced a typical nucleosome ladder, where the majority of DNA is of mononucleosome size (∼150 bp), with progressively decreasing amounts of DNA of di-, tri- and oligo-nucleosome size (Figure 3A, lane 3). In the 350-mM chromatin extract, we observed a typical nucleosome ladder. Nucleosomal DNA was present in the streptavidin pull-down from the 350-mM chromatin extract, and the pulled-down chromatin was enriched for oligonucleosomes (Figure 3A, lane 8), which are more likely than mononucleosomes to contain at least one BioTag. In addition, we found that during the purification process, a small amount of mononucleosomes remained in the 100-mM supernatant after centrifugation (Figure 3A, lane 4), probably due to leakage of mononucleosomes from the EDTA-treated nuclei.
Figure 3.
DNA and protein characterizations of C. elegans embryo chromatin fractions. (A) DNA recovered from different chromatin fractions was resolved on a 1.5% agarose gel and stained with ethidium bromide. MNase-digested chromatin produced a typical nucleosomal ladder (lane 3). A sufficient amount of DNA was recovered from the 350-mM salt pull-down sample, and the streptavidin–Sepharose pull-down DNA is oligonucleosomal in nature (lane 8). Under this chromatin isolation condition, we found that a portion of the mononucleosomes leaks out into the 0.1 M salt supernatant fraction (lane 4). Lanes 1 and 11: DNA molecular weight standard; lane 2: nuclei; lane 3: an MNase-treated sample; lanes 4–6: the input, streptavidin pull-down and unbound fractions of the 0.1 M supernatant, respectively; lanes 7–9: the input, streptavidin pull-down and unbound fractions of 350-mM salt-soluble samples, respectively; lane 10: 350-mM salt-insoluble pellet. (B) Protein samples from different steps of the biotinylated chromatin purification were subjected to electrophoresis on an 18% SDS–PAGE gel and probed with streptavidin-HRP and an anti-HA antibody. Biotinylated BioTag::H3.3 could be detected in the 350-mM salt-soluble streptavidin pull-down fraction, but not in the unbound lane. Lane 1: nuclei; lane 2: an MNase-treated sample; lanes 3–5: respectively the input, unbound and streptavidin pull-down fractions of 350-mM salt-soluble samples; lane 6: the 350-mM salt-insoluble pellet. (C) Biotinylated chromatin isolation and affinity purification were performed. Nuclei were resuspended in buffer A containing ∼17 mM salt for MNase digestion, and the digestion was stopped using EGTA. Next, centrifugation was performed to separate the supernatant containing MNase from the pellet containing nuclei. The nuclei were then resuspended in 80-mM salt, extracted, then the pellet was resuspended in 600-mM salt and extracted again. The 80-mM and 600-mM salt-soluble chromatin fractions, together with the 600-mM salt-washed pellet, were used for affinity purification with streptavidin–Sepharose. DNA recovered from different chromatin fractions was resolved on a 1.5% agarose gel and stained with ethidium bromide. MNase digested chromatin produced a typical nucleosomal ladder (lane 4). Under this chromatin isolation condition, no mononucleosomes leak into the supernatant during the centrifugation step performed after MNase digestion (lane 5). The 80-mM salt-soluble fraction consists of only mononucleosomes (lane 6), while the 600-mM salt-soluble fraction shows a typical nucleosomal ladder (lane 8). A low amount of DNA remains in the pellet (lane 10). Lanes 1 and 12: DNA molecular weight standards; lane 2: nuclei; lanes 3 and 4: respectively samples treated with MNase for 4 or 10 min; lane 5: supernatant; lanes 6 and 7: respectively the input and streptavidin-unbound component of the 80-mM fraction; lanes 8 and 9: respectively the input and streptavidin-unbound component of the 600-mM fraction; lanes 10 and 11: respectively the input and streptavidin-unbound fraction of 600-mM salt-washed pellet.
We performed western blot analysis on proteins extracted from pulled-down chromatin. Protein analysis with streptavidin-HRP indicated that essentially all biotinylated nucleosomes in the input were being captured by streptavidin–Sepharose, because there are no biotinylated proteins remaining in the unbound fraction (Figure 3B, lane 4). Therefore streptavidin pull-down of BioTag::H3.3 is extremely efficient. In addition, western blot analysis with an anti-HA antibody shows that only an extremely low percentage of the BioTag::H3.3 resides in the unbound fraction and so is not biotinylated.
One of the common challenges during chromatin extraction is to minimize preferential losses and obtain an unbiased population of nucleosomes. To this end, we applied the classical salt fractionation protocol originally introduced over 30 years ago (28), which we had previously adapted for Drosophila cells (22). This procedure yields one or more low-salt ‘active’ chromatin fractions (29–32), a 600-mM fraction and an insoluble pellet. Using this protocol, we prepared nuclei and performed MNase digestion in a buffer that contained only ∼12–17-mM salt. After MNase digestion, we used EGTA instead of EDTA to chelate Ca++ and terminate digestion while still leaving sufficient Mg++ unchelated to maintain nuclear integrity (28). We then extracted chromatin in an 80 mM NaCl buffer, followed by extraction in a 600-mM NaCl buffer, saving the salt-washed pellet. We performed streptavidin affinity purification using the 80-mM and 600-mM fractions.
We found that DNA recovered from MNase-digested chromatin using this protocol provides a typical nucleosome ladder (Figure 3C, lane 4). We consistently observed no loss of mononucleosomes into the supernatant fraction (Figure 3C, lane 5). DNA recovered from the 80-mM fraction was almost entirely mononucleosomal (Figure 3C, lane 6), consistent with it being derived from the classical ‘active’ chromatin fraction obtained in numerous previous studies, which is enriched for mononucleosomes (22,28–32). In contrast, DNA recovered from the 600-mM fraction showed an MNase ladder of mono- and oligo-nucleosomes (Figure 3C, lane 8). We also observed a small amount of chromatin in the insoluble pellet. The overall yields (relative to the MNase-digested chromatin) of the 80-mM fraction, the 600-mM fraction and the remaining pellet were 16–24%, 39–48% and 8–14% respectively. Therefore, our chromatin fractionation method recovers >80% of bulk nucleosomes for affinity purification.
Characterization of nucleosomes solubilized at different salt concentrations
Our group previously showed that native Drosophila S2 cell chromatin solubilized from intact nuclei with increasing levels of NaCl resulted in fractions that differ radically in their genome-wide landscapes (22). To determine whether the protocol that we have developed for C. elegans also yields salt-dependent nucleosome profiles, we performed genome-wide tiling microarray analysis by hybridizing with Cy3- and Cy5-labeled samples to obtain log-ratio measurements. We used C. elegans high-density tiling arrays containing 2.1 million isothermal 50–75-mer probes with an average overlap of successive probes of 6 bp.
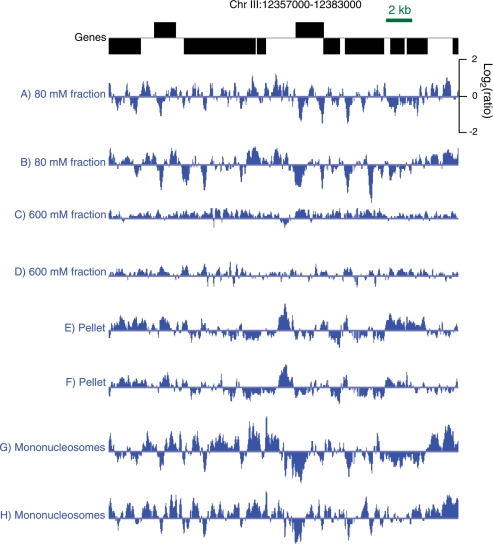
Nucleosomes were obtained using successive extractions of intact MNase-digested nuclei with 80 mM and 600 mM NaCl. In addition, we profiled DNA extracted from the remaining insoluble pellet. For a typical gene-rich region of the genome, a reproducible pattern of sharply defined peaks was seen for the 80-mM salt fraction (Figure 4A and B), whereas the 600-mM salt fraction displayed a flatter landscape (Figure 4C and D). Interestingly, the insoluble pellet fraction showed a different pattern from those of the other fractions (Figure 4E and F).
Figure 4.
Profiles of chromatin obtained from salt fractions, nuclear extracts and gel-purified mononucleosomes. A typical gene-rich region of the C. elegans genome is shown with annotated (WS190) 5′-to-3′ ends shown for genes oriented left to right above the line and from right to left below. Tracks in blue represent log-ratios after triangular smoothing by averaging each probe with its neighboring probe on either side. Pairs of biological replicates are displayed in successive tracks: (A, B) 80-mM salt fractions; (C, D) 600-mM salt fractions; (E, F) 600-mM insoluble pellet; (G, H) gel-excised mononucleosomes.
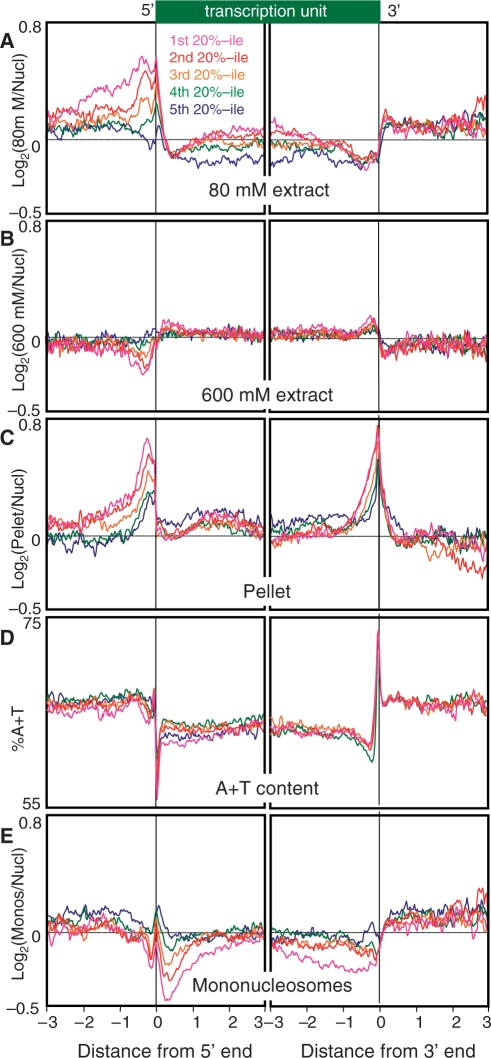
To obtain a genome-wide view of chromatin landscapes for these salt fractions, we aligned all well-annotated genes at their 5′ and 3′ ends and divided them into five subsets based on expression levels. About 70% of the transcripts in C. elegans are subjected to trans-splicing to SL1 and SL2 spliced leader RNAs; thus, the 5′ end of a transcript usually corresponds to SL1 or SL2 acceptor sites, rather than the true transcriptional start site. Nevertheless, this ends analysis reveals that the 80-mM salt fraction is enriched in intergenic regions and depleted in genic regions essentially, regardless of gene expression level (Figure 5A). A broad peak of enrichment is centered over ∼−250 from the 5′ end, which decreases with decreasing transcription and disappears for genes in the lowest expression quintile. Superimposed over this broad peak is a sharp peak immediately over the 5′ end. Within gene bodies, the 80-mM salt fraction shows a gradual increase for the more highly expressed genes that gradually decreases approaching the 3′ end, whereupon a sharp increase is seen. In contrast, the 600-mM salt fraction shows a flatter profile overall, with features opposite to those for the 80-mM profile as expected for depletion of the 80-mM active chromatin fraction (Figure 5B). Interestingly, the pellet fraction shows a strong peak of enrichment centered at ∼−250 and a sharp 3′-end peak that begins with a gradual rise at ∼−1 kb upstream (Figure 5C).
Figure 5.
Ends analysis profiles of salt fractions and extracts. Genes with both ends annotated were aligned at their 5′ and 3′ ends and rank-ordered by gene expression, then divided into quintiles for averaging. Means of two biological replicates are shown as log-ratios. (A) Eighty-millimolar salt fractions; (B) 600-mM salt fractions; (C) 600-mM insoluble pellet- (D) A + T content; (E) gel-excised mononucleosomes. The X-axis interval unit is 100 bp.
Similar salt-fractionation patterns were seen for the subset of transcription units annotated as operons (Supplementary Figure S4A–C).
We wondered whether some of the features that we observed for salt fractions represent inherent properties of chromatin or are instead introduced as a result of our chromatin extraction procedure. One possibility is that some of the features that we observed for the salt fractions reflect DNA base-pair compositional differences, such as the G+C/A+T ratio. For example, the overall genic depletion observed for the 80-mM salt fraction might have resulted from the well-known preference of MNase for AT-rich DNA, and thus better solubilization of AT-rich sequences. Indeed, when the same quintile alignment is done for the A+T content over the genome, intergenic regions are seen to be more AT-rich than genic regions, with sharp transitions over 5′ and 3′ ends of genes (Figure 5D). It is possible that the extreme transition from AT-richness to GC-richness over 5′ ends accounts for the sharp promoter peak in the 80-mM fraction. However, it is unlikely that the extreme AT-richness of the 3′ ends can entirely account for the 3′ peak in the pellet fraction, because pellet enrichment is nearly maximal at 150 bp upstream of the 3′ end, which is where the A+T content drops to a minimum (compare Figure 5C and D, right panels). Alternatively, features such as genic depletion and 5′- and 3′-end features might reflect an inherent preference of nucleosomes for packaging sequences of non-random base composition. To distinguish these possibilities, we resolved the 80-mM salt fraction on an electrophoretic gel, excised DNA of mononucleosomal size (∼150 bp) and profiled it versus the total MNased-treated material from which it was derived. If any of these patterns had resulted from MNase bias, then we would expect that the nucleosomes with AT-rich linkers on either side would be enriched by purification of DNA cleaved on both sides of a mononucleosome relative to the bulk MNase digest. This is because the bulk digest also includes oligonucleosomes, which by definition are cleaved only on one side or not at all. Consistent with this interpretation, the genic depletion seen for the 80-mM salt fraction is reproduced in part in the gel-extracted mononucleosomes (Figures 4G and H, and 5E). Furthermore, the peak over the 5′ end that is seen for the 80-mM salt fraction is seen as well for gel-extracted mononucleosomes. Therefore, these features probably result from the A+T bias of MNase, rather than our salt-fractionation procedure. In contrast, the transcription-coupled enrichments upstream of the 5′ end and within gene bodies seen in the 80-mM profile show opposite trends in the mononucleosome profile, where more highly expressed genes show depletion, not enrichment (compare Figure 5A with 5E). These trends are also seen in profiles of mononucleosomes isolated from mixed stage adults (Supplementary Figures S5 and S6), which indicates that they represent constitutive properties of the C. elegans epigenome.
The striking average enrichment of insoluble chromatin just upstream of the 5′ ends of C. elegans genes is reminiscent of promoter enrichment seen for insoluble chromatin obtained from Drosophila S2 cells [Figure 5C and Henikoff et al. (22)]. In the Drosophila case, evidence was presented that the pellet was enriched in RNA polymerase II and depleted of nucleosomes relative to the other chromatin fractions. It seems likely that the same interpretation applies to the C. elegans insoluble chromatin, in which case, the 5′ upstream peak might represent poised RNA polymerase II and regulatory complexes. Our finding of a prominent peak of insoluble chromatin at genic 3′ ends for C. elegans (but not from Drosophila) is intriguing, and might represent a 3′-end processing complex that would render this chromatin insoluble. This could be due to trans-splicing in worms, which necessitates 3′ end processing of transcripts residing within an operon. If so, then the mapping of insoluble salt-washed chromatin represents a simple method for mapping regulatory features of the C. elegans epigenome.
Zygotic H3.3 is enriched in gene bodies and its abundance correlates with gene expression
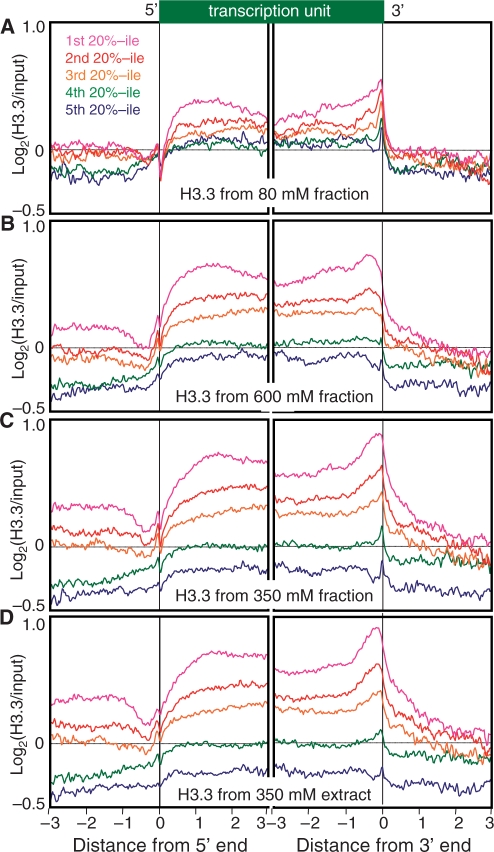
We next performed genome-wide profiling of biotinylated H3.3-containing chromatin. Based on immunofluorescence of the BioTag::H3.3/GFP::BIRA worm strain, biotinylated H3.3 could be detected in embryos and in somatic cells, but not in the germline of adults (Supplementary Figure S7). Thus, the H3.3 incorporation that we observed in embryos represents zygotic incorporation of H3.3 that occurred during embryogenesis, as opposed to maternally loaded H3.3. To characterize the zygotic H3.3 landscape genome wide, we performed biotinylated H3.3 profiling for the 80-mM and 600-mM salt fractions from intact nuclei in the presence of excess Mg++. We also similarly profiled chromatin extracted from a nuclear lysate using 350 mM NaCl either after EDTA treatment, which causes partial lysis, or on intact nuclei using EGTA (28). Very similar results were obtained in all four cases (Figure 6, blue tracks). As can be seen for a typical gene-rich region, sharp peaks of H3.3 from the 80-mM fraction in general correspond to broader peaks seen for the 600-mM fraction and for the 350-mM extract. The higher resolution seen for the 80-mM fraction is attributable to the mononucleosome size of this chromatin fraction. High resolution is an inherent advantage of using native mononucleosomes as opposed to sonicated and cross-linked chromatin (33), and is especially valuable for genome-wide profiling.
Ends analysis profiles reveal that H3.3 becomes gradually enriched over gene bodies relative to intergenic regions on either side, with a prominent enrichment near the 3′ end (Figure 7). This enrichment correlates with expression level: the most highly expressed genes have the most H3.3, and genes with the lowest expression have the least H3.3. H3.3 profiles are quite similar for 80-mM and 600-mM salt fractions, for 350-mM salt fractions from intact nuclei and for 350-mM extracts. Therefore, chromatin with distinct solubility properties that result from differential extractions nevertheless display very similar H3.3 landscapes in C. elegans. Similar H3.3 profiles were also seen for the subset of transcription units annotated as operons (Supplementary Figure S4D–G). Consistent with previous results for Drosophila cultured cells (17), our results show that H3.3 incorporation is a good indicator of transcriptional elongation activity within the context of a developing animal.
Figure 7.
H3.3 ends analysis profiles. (A) From chromatin isolated from intact nuclei using 80-mM salt; (B) from chromatin isolated from intact nuclei using 600-mM salt after depletion of the 80-mM salt fraction; and (C) from EDTA-treated nuclei using 350-mM salt. See the legend to Figure 5 for details.
In order to determine whether H3.3 corresponds to known chromatin marks in the C. elegans genome, we compared our H3.3 landscapes to chromatin landscapes published in two previous studies of C. elegans chromatin (34,35). We detected little, if any, similarity between H3.3 and any other chromatin marks (Figure 6, compare blue tracks with brown tracks), including RNA polymerase II, H2A.Z, H3K36me3, H3K4me3, H3K9me3 and total H3. Consequently, H3.3 patterns provide a unique signature of the epigenome.
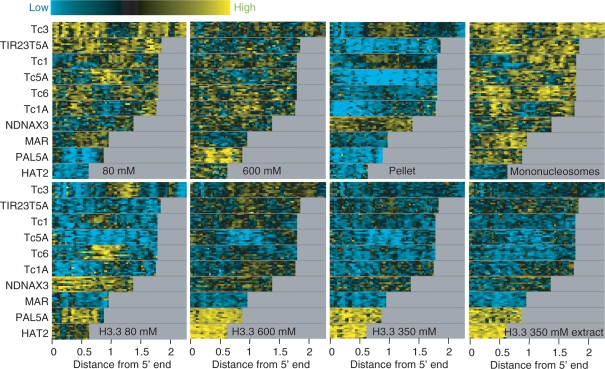
To determine whether H3.3 also marks dynamic regions outside of genic regions, we examined the profiles of transposon families genome-wide. As we had previously found for Drosophila transposons (22), worm transposons show salt-fractionation chromatin patterns that vary from one family to the next and differ dramatically between fractions (Figure 8, top row). In contrast, H3.3 enrichment patterns were relatively consistent between salt fractions, although the 80-mM salt fraction showed generally higher levels of enrichment and lower levels of depletion than were seen for the other salt extracts (Figure 8, bottom row). Therefore, H3.3 provides a consistent epigenomic signature of histone replacement that can be used to identify dynamic regions throughout the genome.
Figure 8.
Enrichment of chromatin salt fractions and H3.3 locally within transposons. Heat-map stacks of 10 transposon families for salt fractions, mononucleosomes and H3.3. (Top row) Successive 80-mM and 600-mM salt-extracted fractions and the insoluble pellet profiled versus MNased nuclear starting material, and gel-purified mononucleosomes/input bulk DNA. (Second row) H3.3/input levels for successive 80-mM and 600-mM or 350-mM pull-down fractions and for a 350-mM extract. Vertical gray streaks result from alignment gaps.
CONCLUSION
We have described a system for epigenomic profiling of native chromatin for C. elegans and have applied it to the determination of H3.3 landscapes. Chromatin profiling methods have been previously described for C. elegans (34,35). These methods used non-native formaldehyde-fixed and sonicated chromatin, followed by immunoprecipitation with antibodies. Our strategy differs from these methods in several ways. Our adoption of a native chromatin procedure that solubilizes ∼90% of chromatin and yields salt fractions provides additional chromatin profiles based on physical properties of nucleosomes. By using 80-mM salt extraction, we obtain mononucleosomes that provide the highest resolution possible both for profiling classical active chromatin and for profiling H3.3 and potentially other histone variants and modifications. By profiling the salt-washed insoluble pellet that remains after the large majority of chromatin has been solubilized, we have found enrichment for gene regulatory sites, including a novel genic 3′-end enrichment. Our use of biotin-tagging for chromatin profiling allows for essentially complete recovery of labeled nucleosomes, avoiding epitope-masking and other antibody-related issues that might otherwise bias a chromatin landscape. Importantly, the two-component nature of our system makes it adaptable for isolation of chromatin from any cell type, including the germ cells, at any stage during development and throughout the C. elegans life cycle. This can be done by expressing BioTag::H3.3 or BIRA::GFP in a cell-type-specific manner. Other uses of this system include combining the powerful genetic tools available in C. elegans such as RNA interference assays to deplete chromatin-remodeling factors and observe their effects on the epigenetic profiles. Therefore, this versatile system has the potential of generally facilitating chromatin and epigenetic research in C. elegans.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health-NHGRI modENCODE project (U01 HG004274). Funding for open access charges: National Institutes of Health U01 HG004274.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Deal, T. Furuyama, P. Maddox, J. Priess and the Henikoff and Priess lab members for advice and discussions, R. Deal, T. Furuyama, J. Priess, F. Steiner and P. Talbert for critical reading of the manuscript, and the FHCRC Genomics facility for NimbleGen array processing. S.O. performed all the worm culture experiments in the Priess laboratory.
REFERENCES
- 1.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Henikoff S, McKittrick E, Ahmad K. Epigenetics, histone H3 variants, and the inheritance of chromatin states. Cold Spring Harb. Symp. Quant. Biol. 2004;69:235–243. doi: 10.1101/sqb.2004.69.235. [DOI] [PubMed] [Google Scholar]
- 3.Ooi SL, Henikoff S. Germline histone dynamics and epigenetics. Curr. Opin. Cell. Biol. 2007;19:257–265. doi: 10.1016/j.ceb.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 4.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell. Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 6.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl Acad. Sci. USA. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waterborg JH. Sequence analysis of acetylation and methylation in two histone H3 variants of alfalfa. J. Biol. Chem. 1990;265:17157–17161. [PubMed] [Google Scholar]
- 8.Akhmanova A, Miedema K, Wang Y, van Bruggen M, Berden JH, Moudrianakis EN, Hennig W. The localization of histone H3.3 in germ line chromatin of Drosophila males as established with a histone H3.3-specific antiserum. Chromosoma. 1997;106:335–347. doi: 10.1007/s004120050255. [DOI] [PubMed] [Google Scholar]
- 9.Ooi SL, Priess JR, Henikoff S. Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS Genet. 2006;2:e97. doi: 10.1371/journal.pgen.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 2006;50:455–461. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- 11.van der Heijden GW, Derijck AA, Posfai E, Giele M, Pelczar P, Ramos L, Wansink DG, van der Vlag J, Peters AH, de Boer P. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat. Genet. 2007;39:251–258. doi: 10.1038/ng1949. [DOI] [PubMed] [Google Scholar]
- 12.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat. Cell. Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 13.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 14.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bean CJ, Schaner CE, Kelly WG. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat. Genet. 2004;36:100–105. doi: 10.1038/ng1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prahlad V, Pilgrim D, Goodwin EB. Roles for mating and environment in C. elegans sex determination. Science. 2003;302:1046–1049. doi: 10.1126/science.1087946. [DOI] [PubMed] [Google Scholar]
- 17.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 18.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jitrapakdee S, Wallace JC. The biotin enzyme family: conserved structural motifs and domain rearrangements. Curr. Protein Pept. Sci. 2003;4:217–229. doi: 10.2174/1389203033487199. [DOI] [PubMed] [Google Scholar]
- 22.Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng S, Alekseyenko AA, Larschan E, Kuroda MI, Park PJ. Normalization and experimental design for ChIP-chip data. BMC Bioinform. 2007;8:219. doi: 10.1186/1471-2105-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl Acad. Sci. USA. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology. 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- 27.Stein A, Mitchell M. Generation of different nucleosome spacing periodicities in vitro. Possible origin of cell type specificity. J. Mol. Biol. 1988;203:1029–1043. doi: 10.1016/0022-2836(88)90127-1. [DOI] [PubMed] [Google Scholar]
- 28.Sanders MM. Fractionation of nucleosomes by salt elution from micrococcal nuclease-digested nuclei. J. Cell. Biol. 1978;79:97–109. doi: 10.1083/jcb.79.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Annunziato AT, Schindler RK, Thomas CA, Jr, Seale RL. Dual nature of newly replicated chromatin. Evidence for nucleosomal and non-nucleosomal DNA at the site of native replication forks. J. Biol. Chem. 1981;256:11880–11886. [PubMed] [Google Scholar]
- 30.Davie JR, Saunders CA. Chemical composition of nucleosomes among domains of calf thymus chromatin differing in micrococcal nuclease accessibility and solubility properties. J. Biol. Chem. 1981;256:12574–12580. [PubMed] [Google Scholar]
- 31.Rocha E, Davie JR, van Holde KE, Weintraub H. Differential salt fractionation of active and inactive genomic domains in chicken erythrocyte. J. Biol. Chem. 1984;259:8558–8563. [PubMed] [Google Scholar]
- 32.Rose SM, Garrard WT. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J. Biol. Chem. 1984;259:8534–8544. [PubMed] [Google Scholar]
- 33.O’Neill LP, Turner BM. Immunoprecipitation of native chromatin: NChIP. Methods. 2003;31:76–82. doi: 10.1016/s1046-2023(03)00090-2. [DOI] [PubMed] [Google Scholar]
- 34.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whittle CM, McClinic KN, Ercan S, Zhang X, Green RD, Kelly WG, Lieb JD. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet. 2008;4:e1000187. doi: 10.1371/journal.pgen.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.