Abstract
The heterotrimeric CCAAT-binding complex is evolutionary conserved in eukaryotic organisms. The corresponding Aspergillus nidulans CCAAT- binding factor (AnCF) consists of the subunits HapB, HapC and HapE. All of the three subunits are necessary for DNA binding. Here, we demonstrate that AnCF senses the redox status of the cell via oxidative modification of thiol groups within the histone fold motif of HapC. Mutational and in vitro interaction analyses revealed that two of these cysteine residues are indispensable for stable HapC/HapE subcomplex formation and high-affinity DNA binding of AnCF. Oxidized HapC is unable to participate in AnCF assembly and localizes in the cytoplasm, but can be recycled by the thioredoxin system in vitro and in vivo. Furthermore, deletion of the hapC gene led to an impaired oxidative stress response. Therefore, the central transcription factor AnCF is regulated at the post-transcriptional level by the redox status of the cell serving for a coordinated activation and deactivation of antioxidative defense mechanisms including the specific transcriptional activator NapA, production of enzymes such as catalase, thioredoxin or peroxiredoxin, and maintenance of a distinct glutathione homeostasis. The underlying fine-tuned mechanism very likely represents a general feature of the CCAAT-binding complexes in eukaryotes.
INTRODUCTION
CCAAT sequences are present in the promoter regions of a large number of eukaryotic genes, in general located 50–200 bp upstream from the transcriptional start point (1). They are bound by different distinct transcription factors in order to modulate the transcription rate (2–8). A heterotrimeric complex binding to CCAAT sequences was first discovered in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe (6,9–12). Since this discovery, the particular regulatory complex has been found in all eukaryotes, which have been analyzed so far. It has been designated Hap complex in S. cerevisiae (6,11), Kluyveromyces lactis (13), and Arabidopsis thaliana (14), Php in S. pombe (10), AnCF in Aspergillus species (15), CBF in Xenopus laevis (16) and NF-Y in mammals (17,18), respectively. The Hap complex of S. cerevisiae comprises four subunits, Hap2p, Hap3p, Hap4p and Hap5p. Hap2/3/5 p form the core CCAAT-binding complex, which is responsible for DNA binding, while Hap4p is involved in transcriptional activation (11). Moreover, S. cerevisiae Hap2p, A. nidulans HapB and human NF-YA are functionally interchangeable, which was demonstrated by the complementation of an S. cerevisiae hap2 and an A. nidulans hapB deletion mutant by a human NF-YA cDNA (19,20). Recombinant rat CBF-C is able to form a stable protein-DNA complex together with the yeast Hap2p and Hap3p (21) supporting that the CCAAT-binding complex is evolutionary conserved. The respective A. nidulans CCAAT-binding factor (AnCF), consisting of the Hap2/3/5 p orthologs HapB/C/E, activates the expression of numerous genes including the anabolic penicillin biosynthesis genes ipnA and aatA (22,23) and the catabolic acetamidase encoded by amdS (24). On the other side, AnCF-mediated repression of gene expression was also found: the homoaconitase-encoding lysF and the hapB gene were repressed by AnCF (25,26). However, a comprehensive picture of the CCAAT-binding complex regulon in fungi is missing so far. For this reason, one of the key questions related to the CCAAT-binding complex is the regulation of the activity of the core complex. The stoichiometrical distribution of the different subunits seems to be essential for an efficient complex formation, supported by the AnCF-mediated auto-regulation of the hapB gene (25) and by the quantity control mechanism of HapE levels by HapC (27) in A. nidulans. In addition, however, there is growing evidence that the expression of certain CCAAT-regulated genes needs the interaction of the heterotrimeric core complex with additional proteins. In S. cerevisiae, the Hap2/3/5 p complex together with Hap4p acts as an activator of genes involved in oxidative phosphorylation in response to growth on non-fermentable carbon sources (6). In A. nidulans, AnCF represses the transcription of genes that encode iron-containing proteins during iron-replete growth and via physical interaction with the iron-sensing subunit HapX during iron-depleted conditions (28). HapX displays no similarity to S. cerevisiae Hap4p apart from an N-terminal 17 amino acid-motif, which is essential for interaction with the Hap/AnCF core complex (29). Nakshatri et al. (30) showed in vitro that formation of intra- and intermolecular disulfide bridges within and between NF-YB subunits prevents NF-YB from associating with NF-YC, the first step of the CCAAT- binding complex assembly. Consistently, different redox active proteins such as the human ADF, or Ref-1 increased the DNA-binding activity of recombinant NF-Y to its CCAAT box. These data suggested that the activity of CCAAT-binding complexes is regulated by the cellular redox status, as demonstrated for several other transcription factors (31).
The CCAAT-binding complex is a global regulator in all eukaryotes, but the regulation of its activity is still poorly understood. Here, we demonstrate that the CCAAT-binding complex AnCF is regulated at the post-transcriptional level by the redox status of the cell, thereby serving as a redox sensor coordinating the cellular oxidative stress response in the genetic model organism A. nidulans. The strict conservation of the redox status-sensing cysteine residues within a domain of HapC that was described as histone fold motif (HFM) for human NF-YB, and which was shown to be indispensable for the first step of NF-Y assembly, i.e. interaction with the second histone-like protein, NF-YC (32,33), suggests evolutionary conservation of this function.
MATERIALS AND METHODS
Strains, plasmids, oligonucleotides, media, growth conditions and transformation
Aspergillus nidulans strains used in this study are listed in Table 1. Plasmids and oligonucleotides used in this study are listed in Supplementary Tables S1 and S2, respectively. A. nidulans strains were grown at 37°C in Aspergillus minimal medium (AMM), according to Pontevorvo et al. (34), containing 1% (w/v) glucose as carbon source, 20 mM glutamine as nitrogen source, 30 µM FeSO4 as iron source and the appropriate supplements. To overcome the auxotrophy for organic sulfur of the ΔtrxA strains, 1 mM cysteine was added to the AMM. Oxidative stress conditions were induced with H2O2. Mycelia were harvested before and after 30 min treatment with H2O2. Transformation of A. nidulans was performed by the method of Balance et al. (35).
Table 1.
Fungal strains used in this study
| Strain designation in text | Strain | Genotype | Reference |
|---|---|---|---|
| Northern blot analysis and glutathione measurements | |||
| Wild type | TN02A7 | pyrG89; pyroA4; riboB2; argB2; ΔnkuA::argB; Arg+ | (77) |
| ΔhapC | Nat24 | pyrG89; pabaA1; riboB; ΔhapC::riboB; RiboB+ | (78) |
| ΔtrxA | AnTrxAKO | pyrG89; pyroA4; riboB2; argB2; ΔnkuA::argB; Arg+; ΔtrxA::pyr-4; PyrG+ | (36) |
| ΔnapA | AnYap1KO | pyrG89; pyroA4; nkuA::argB; riboB2; anyap1::pyr-4; PyrG+ | unpublished |
| Fluorescence analysis | |||
| AXB4A2 | pyrG89; pabaA1; argB2; fwA1; bga0; argB2::pAXB4A (acvAp-uidA, ipnAp-lacZ), ArgB+ | (79) | |
| ΔC-HapCegfp | pyrG89; pabaA1; riboB; ΔhapC::riboB; RiboB+; pHapC-EGFP; HapC+; pyr-4::pKTB1; PyrG+ | (62) | |
| ΔC-HapCegfp-SSS | pyrG89; pabaA1; riboB; ΔhapC::riboB; RiboB+; pyr-4::pKTB1-HapC3CS-EGFP; PyrG+; HapC3CS+ | This study | |
| ΔtrxA-HapCegfp | pyrG89; pyroA4; riboB2; argB2; ΔnkuA::argB; Arg+; ΔtrxA::pyr-4; PyrG+; pyroA::pHapC-EGFP-pyro; Pyro+ | This study | |
| yN | pyrG89; pabaA1; argB2; fwA1; bga0; argB2::pAXB4A (acvAp-uidA, ipnAp-lacZ), ArgB+; pyr-4::pYN-pyr4; PyrG+ | This study | |
| yC | pyrG89; pabaA1; argB2; fwA1; bga0; argB2::pAXB4A (acvAp-uidA, ipnAp-lacZ), ArgB+; pyr-4::pYC-pyr4; PyrG+ | This study | |
| yCN | pyrG89; pabaA1; argB2; fwA1; bga0; argB2::pAXB4A (acvAp-uidA, ipnAp-lacZ), ArgB+; pyr-4::pYC-pyr4; PyrG+; pEYFPN | This study | |
| yHapE-C | pyrG89; pabaA1; argB2; fwA1; bga0; argB2::pAXB4A (acvAp-uidA, ipnAp-lacZ), ArgB+; pyr-4::pYC-pyr4; PyrG+; pHapE-YN | This study | |
| yHapC-N | pyrG89; pabaA1; argB2; fwA1; bga0; argB2::pAXB4A (acvAp-uidA, ipnAp-lacZ), ArgB+; pyr-4::pHapC-YC-pyr4; PyrG+; pEYFPN | This study | |
| yHapC-HapE | pyrG89; pabaA1; argB2; fwA1; bga0; argB2::pAXB4A (acvAp-uidA, ipnAp-lacZ), ArgB+; pyr-4::pHapC-YC-pyr4; PyrG+; pHapE-YN | This study | |
| yTrxA-C | pyrG89; pabaA1; argB2; fwA1; bga0; argB2::pAXB4A (acvAp-uidA, ipnAp-lacZ), ArgB+; pyr-4::pYC-pyr4; PyrG+; pTrxA-YN | This study | |
| yHapC-TrxA | pyrG89; pabaA1; argB2; fwA1; bga0; argB2::pAXB4A (acvAp-uidA, ipnAp-lacZ), ArgB+; pyr-4::pHapC-YC-pyr4; PyrG+; pTrxA-YN | This study | |
| ΔhapE | ΔE-89 | pyrG89; pabaA1; argB; ΔhapE::argB; ArgB+ | (62) |
| yHapC-TrxA-ΔE | pyrG89; pabaA1; argB; ΔhapE::argB; ArgB+; pabaA1::pHapC-YC-paba; Paba+; pyr-4::pTrxA-YN-pyr4; PyrG+ | This study | |
Purification of recombinant proteins
The three AnCF-forming subunits, i.e. HapB, wild-type HapC and HapE were overproduced in E. coli Rosetta 2 (DE3) and purified as described elsewhere (28). The cysteine to serine exchanges in HapC were created by applying the QuikChange® II Site-Directed Mutagenesis Kit (Stratagene) using the HapC-encoding plasmid pMalC2TEVHapC for mutagenesis. The His6-tagged proteins TrxA, TrxA(C39S) and TrxR were overproduced in E. coli BL21 (DE3) and purified as previously described (36).
Thioredoxin system-dependent reduction of oxidized HapC
Two milligrams of the lyophilized HapC proteins HapC or HapC(CCS) were dissolved in 1 ml 0.1 M potassium phosphate buffer, pH 7.5 and incubated with either 15 mM diamide or H2O2 for 3 h. To remove the excess of the respective oxidizing agent, the reaction mixtures were then applied to a NAP-10 column (GE Healthcare). The oxidized proteins were eluted with 0.1% (v/v) trifluoracetic acid (TFA) in 40% (v/v) acetonitrile and freeze dried.
For the thioredoxin-dependent reduction assay, aliquots of the freeze dried oxidized HapC proteins were dissolved in 0.1 M potassium phosphate buffer, pH 7.5. The reduction assay was carried out in a final volume of 1 ml containing 40 µM of oxidized HapC, i.e. HapC or HapC(CCS), 1 µM TrxR and 3 µM TrxA [TrxA or TrxA(C39S)]. After starting the reaction by the addition of NADPH, the NADPH consumption was followed by recording the decrease in absorbance at 340 nm. Furthermore, samples of the reduction assay were taken at different time points, directly transferred into denaturating SDS sample buffer and finally analyzed by non-reducing SDS–PAGE. If no change in absorbance was detectable any longer, 500 µl of the reduction assay were transferred into 0.1% (v/v) trifluoracetic acid (TFA) in 40% (v/v) acetonitrile for a further lyophilization.
Native PAGE analysis
Lyophilized fractions of reduced, oxidized and thioredoxin system-treated HapC were dissolved in water to give a final concentration of 0.1 mM. Two microliters (0.2 nmol) of these different HapC solutions were incubated with equimolar amounts of HapE or HapE and HapB for 5 min. After transferring the incubation mixtures into 2 × native Tris–Glycine sample buffer, the probes were analyzed by native PAGE on a 14% Tris–Glycine gel (Invitrogen).
Surface plasmon resonance (Biacore) measurements
Real-time analyses were performed on Biacore 2000 (AnCF complex assembly) and T100 (AnCF–DNA interaction) systems at 25°C. Data were processed with the BIA evaluation software versions 4.1 or 1.1.1 (Biacore). Refractive index errors due to bulk solvent effects were corrected by subtracting responses from non-coated flow cell 1. Sample injection and dissociation times were set to 2.5 or 5 min at a flow rate of 30 µl/min. For measuring AnCF complex formation, N-biotinylated HapB was captured on an SA sensor chip on flow cell 2 (80 RU). Samples containing equimolar amounts of HapC (wild-type and mutants) and HapE were prepared by mixing 0.1 M solutions of each subunit in water. Samples for SPR analysis were generated by 500-fold dilution of this stock solution in running buffer [10 mM HEPES pH 7.4, 0.15 M NaCl, 1 mM DTT and 0.005% (v/v) surfactant P20] followed by serial 2-fold dilution. Injection was performed at concentrations from 6.25 to 200 nM HapC/HapE. Regeneration of the biosensor surface was carried out with 10 mM glycine/HCl, pH 2.0 for 1 min. Dissociation constants (KD) were calculated from the concentration-dependent steady-state binding of HapC/HapE. DNA duplexes containing CCAAT boxes from the promoter regions of sreA, trxA, catB, prxA and napA were produced by annealing, using a 5-fold molar excess of the non-biotinylated oligonucleotide. The dsDNA was injected on flow cells of a streptavidin (SA)-coated CM3 sensor chip at a flow rate of 10 µl/min until 40–80 RU had been bound. AnCF complexes were preformed from the single HapC, HapE and HapB proteins as described above and were injected in running buffer [10 mM phosphate buffer pH 7.4, containing 2.7 mM KCl, 137 mM NaCl, 1 mM DTT and 0.005% (v/v) surfactant P20] at concentrations from 1.56 to 100 nM. Regeneration was achieved with running buffer containing 0.5 M NaCl and 0.005% (w/v) SDS for 1 min. Dissociation constants were calculated from both the kinetic rate constants for AnCF–DNA complex formation and dissociation derived from a 1:1 interaction model including a mass transfer term or the concentration-dependent steady-state binding of AnCF complexes using the 1:1 steady-state affinity model.
Fluorescence microscopy
Generation of the encoding plasmids and strains, which were used for eGFP or BiFC analysis, is described in detail in the Supplementary Data. For nuclear staining 20 µl of 4′,6-diamidino-2-phenylindol (DAPI) solution (1 µg/ml) were added for 1 min. A. nidulans hyphae were analyzed by light and fluorescence microscopy using Leica DM4500 B digital fluorescence microscope provided with Leica DFC480 digital camera and suitable Leica filter cubes (Leica Microsystems).
Isolation of RNA and northern blot analysis
For isolation of total RNA, fungal strains were grown for 26–28 h until half of the glucose was consumed. Glucose concentrations in AMM were measured with the BIOSEN C-line, GP+ system (EKF diagnostics). Mycelia were harvested and washed with 200 ml of sterile distilled water and were broken using liquid nitrogen as described previously (37). Total RNA was prepared using the RNeasy Plant minikit (QIAGEN). The amount of RNA was determined both spectrophotometrically and by agarose gel electrophoresis. Northern blot analysis was carried out as described previously (38). Detection of the hybridized fragments was performed using CDP-Star detection reagent (Roche). Fluorescence detection, mediated by the digoxigenin labled probes and image analysis were carried out with the Molecular® Imager VersaDOCTM MP imaging system (BioRad).
Glutathione measurements
Determination of GSH and GSSG was carried out according to a modified GSH-recycling method described by Tietze (39). In detail, for measuring GSH/GSSG ratios 400–500 mg of wet mycelia in 400 µl ice-cold 5% (w/v) 5′-sulfo-salicylic-acid (SSA) were homogenized in innuPrep Lysis Tubes (Analytik Jena) using the FastPrep® Cell Disruptor FP120-system (Qbiogene). After centrifugation at 14 000 rpm for 10 min at 4°C, the resulting supernatant was further diluted in six volumes of ice-cold 5% (w/v) SSA and stored at −80°C for further analysis. For measuring total glutathione concentrations, homogenization of wet mycelia was carried out without previous SSA treatment. After centrifugation, aliquots were taken for protein concentration determination and the remaining supernatant was also diluted in six volumes of ice-cold 5% (w/v) SSA. After a further centrifugation step at 14 000 rpm for 10 min at 4°C, the resulting protein-free supernatant was stored at −80°C for further analysis.
The GSH samples in 5% (w/v) SSA were neutralized with 0.5 N NaOH and diluted 5-fold in assay buffer (0.125 M sodium phosphate, 6.3 mM EDTA, pH 7.5). To measure GSSG by the enzymatic method in samples containing both GSH and GSSG, free GSH was masked with a 3–4-fold molar excess of the thiol-scavenger 1-methyl-2-vinyl-pyridinium trifluoromethane sulfonate (MV2P) (Natutec). Measurements were carried out in assay buffer containing 0.6 mM 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) and 0.3 mM NADPH using the UV–Vis/NIR Spectrophotometer V-630 (Jasco). The reaction was started with one unit of recombinant yeast glutathione reductase (Sigma).
Proteome analysis
Proteome analysis of A. nidulans, including sample preparation, 2D-SDS–PAGE, protein visualization, quantification and MS identification of proteins, was essentially carried out as previously described for A. fumigatus (40). Gel images (n = 3) were analyzed using the software Delta 2D 3.6 (Decodon). Afterwards, background subtraction spot volumes were normalized against total spot volume and total spot area. Spot values were regarded as differentially regulated with a t-test value P < 0.05. The MALDI-TOF data were used to search the NCBI database and Central Aspergillus Data Repository (CADRE), using the Mascot algorithm (Matrix Science), with the following parameters: Cys as S-carbamidomethyl derivative and Met in oxidized form (variable), one missed cleavage site, peptide mass tolerance of 200 ppm.
RESULTS
The formation of HapC multimers is dependent on redox sensitive cysteine residues
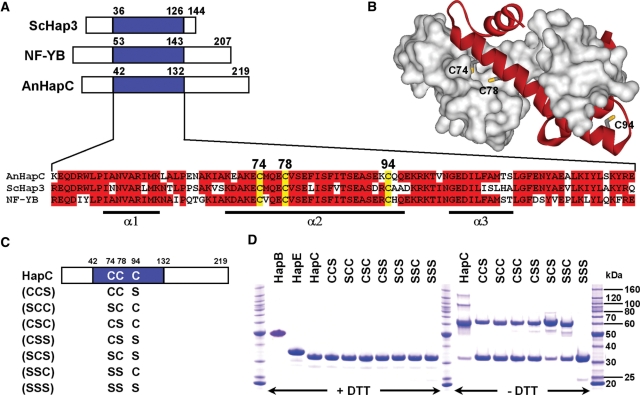
All identified HapC orthologs exhibit three conserved cysteine residues in close proximity at conserved positions (Figure 1A and data not shown). Remarkably, these cysteine residues are located within a HFM core region that is indispensable for NF-YB/NF-YC and CBF-A/CBF-C heterodimer formation in men and rat, respectively (41,42). This HFM displays moderate amino acid sequence similarity to histone H2B (32). Most HFMs consist of a central long α-helix that is flanked by two shorter α-helices and a HFM is also present in all NF-YC (HapE) orthologs, which have sequence similarity to histone H2A (33). The solved crystal structure of the NF-YB/NF-YC subcomplex resembles the head-to-tail association of H2A and H2B (43). Comparative structure modeling of the HapC/HapE dimer by using SWISS-MODEL workspace (44) and the NF-YB/NF-YC structure as template revealed that cysteine residues 74 and 78 of HapC are buried in the HapC/HapE subcomplex interface (Figure 1B).
Figure 1.
Alignment of HapC orthologs and redox behavior of HapC and its serine mutant proteins. (A) Amino acid sequence alignment of the evolutionarily conserved HapE subunit interaction domains (blue boxes) of HapC orthologs. Conserved cysteine residues are numbered according to the A. nidulans HapC sequence. Invariant amino acids present in at least two of the three sequences are boxed in red. The numbers indicate the first and last residues of the domains depicted. Abbreviations: ScHap3, S. cerevisiae Hap3; NF-YB, human NF-YB; AnHapC, A. nidulans HapC. α-helices observed in the NF-YB structure are shown below the alignment. (B) Homology model of the HapC/HapE subcomplex based on the NF-YB/NF-YC structure (PDB accession code 1N1J). A surface representation of the HapE molecule (residues L78–P163) is colored white and a ribbon representation of HapC (residues L48–R131) is colored red. Cysteine residues of HapC are shown as sticks. (C) Schematic structure of the recombinant HapC protein and all possible Cys to Ser mutants. The positions of the cysteine and serine residues are indicated by the letters ‘C’ and ‘S’, respectively. (D) SDS–PAGE analysis of purified HapB, HapE, HapC and its serine mutants after 96 h in the absence and presence of 25 mM DTT using samples containing 0.2 nmol of each Hap subunit.
Purified HapC formed dimers and multimers consistent with possible formation of intramolecular and even intermolecular disulfide bridges as response to oxidation (Figure 1D). To analyze whether and which of the cysteine residues of HapC are redox sensitive, HapC mutant proteins were generated and purified, which contain cysteine to serine exchanges in all possible combinations (Figure 1C). All mutant proteins, except of the one lacking all three cysteine residues (SSS), had lost their ability to form multimers but were still able to form dimers (Figure 1C). The oxidized forms of HapC (HapCox) could be reduced by the addition of DTT (Figure 1D) showing that formation of dimers and multimers is reversible.
The thioredoxin system can reduce HapCox in vitro
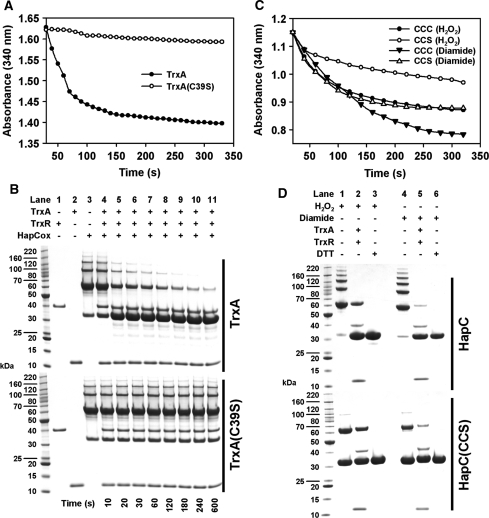
To study whether the thioredoxin system accepts HapCox as substrate, HapCox (oxidized by diamide) was incubated with recombinant A. nidulans thioredoxin (TrxA), A. nidulans thioredoxin reductase (TrxR) and NADPH. Reduction was analyzed photometrically as described previously (36) or by SDS–PAGE (Figure 2A and B). TrxA, but not the inactive TrxA (C39S) version, was able to reduce oxidized HapC in vitro, underlining the specificity.
Figure 2.
Reduction of oxidized HapC by the A. nidulans thioredoxin system. (A) The NADPH-dependent reduction of H2O2-oxidized wild-type HapC by TrxA or AnTrxA(C39S) was monitored by the decrease in absorbance at 340 nm. (B) After NADPH addition, aliquots were taken from the reaction mixtures containing TrxA (upper panel) or TrxA(C39S) (lower panel) at the indicated time points, denaturated in sample buffer and further analyzed by SDS–PAGE under non-reducing conditions. (C) Wild-type HapC or the mutant HapC(CCS) were treated with either H2O2 or diamide. Reduction by TrxA was followed as described in (A). The degree of HapC reduction was verified by SDS–PAGE (D) including a control under reducing conditions (lanes 3 and 6).
The formation of an intramolecular disulfide bridge between Cys85 and Cys89 in the human HapC ortholog, NF-YB, prevents assembly of the CCAAT-binding complex (30). Therefore, the redox behavior of the HapC mutant protein HapC(CCS) in comparison with the wild-type HapC was analyzed. Both proteins were treated with either H2O2 or diamide (Figure 2D). Irrespective of the used oxidant, both protein versions could be reduced by the thioredoxin system (Figure 2C and D). Assuming that the two HapC versions were fully oxidized and the subsequent reduction by the thioredoxin system was complete, the NADPH-dependent reduction of 40 µM HapCox and 40 µM HapC(CCS)ox should lead to changes in absorbance of ∼0.37 and 0.25, repectively, which matched the values observed for the diamide-oxidized proteins (Figure 2C). The complete reduction was further confirmed by SDS–PAGE analysis (Figure 2D). In contrast, the thioredoxin system reduced only ∼70% of H2O2-oxidized HapCox and HapC (CCS)ox (Figure 2C), which was supported by high amounts of non-reduced HapC dimers in SDS–PAGE analysis (Figure 2D). A possible explanation of these results might be the fact that diamide treatment results in the oxidation of thiol groups to form disulfide bonds, which are susceptible to reduction by thioredoxin, whereas H2O2 exposure can also lead to additional oxidation products such as sulfinic-, sulfenic- and even sulfonic-acids, which are not targets for thioredoxin. Moreover, H2O2 is also able to produce methionine sulfoxides from methionine residues leading to tertiary structures that are not accessible by TrxA.
Although the treatment of HapC(CCS) with diamide or H2O2 led to the formation of HapC(CCS) dimers, there was still a considerable amount of monomer present (Figure 2D). By contrast, the NADPH consumption indicated that HapC(CCS) was always fully oxidized, indicating oxidation by intramolecular disulfide bridge formation. Moreover, thioredoxin affinity chromatography (45,46) demonstrated that the thioredoxin mutant version TrxA(C39S) recognizes all artifical protein-TNB adducts that resulted from the reaction of HapC(CSS), HapC(SCS) and HapC(SSC) with DTNB (Supplementary Figure S1), which underlines that TrxA can recognize and reduce all disulfide bridges of HapC formed involving either Cys74, Cys78 or Cys94.
Only reduced HapC participates in AnCF assembly
The HapC and HapC(CCS) proteins were further analyzed for their ability to form a stable heterodimer with HapE or to form the heterotrimeric complex together with HapE and HapB by native PAGE. The non-oxidized HapC proteins were able to form a stable HapC/HapE heterodimer (Supplementary Figure S2A) and a functional AnCF (Supplementary Figure S2B), whereas oxidized HapC and HapC(CCS) did not. Consistently, subsequent reduction of the oxidized HapC proteins by the A. nidulans thioredoxin system led to restoration of active HapC, which again was able to interact with HapE (Supplementary Figure S2A) or HapE and HapB (Supplementary Figure S2B). When diamide- or H2O2-treated HapC(CCS) protein was analyzed for its ability to form a HapC/HapE heterodimer (Supplementary Figure S2A) or HapC/HapE/HapB heterotrimer (Supplementary Figure S2B) by native PAGE analysis, HapC(CCS) failed to interact with both of the other Hap-proteins. However, the SDS–PAGE results from Figure 2D, as well as the native PAGE analysis in Supplementary Figure S2 show that there was still a high amount of HapC(CCS) monomer present (40–50%) after oxidation with diamide or H2O2, which failed to interact with HapE and showed minimal activity in formation of AnCF. These data suggest that already the formation of an intramolecular disulfide bridge within the HFM of HapC leads to an inactive tertiary HapC structure unable to interact with HapE and to form AnCF, as HapB requires the heterodimeric HapC/HapE-structure to complete the assembly of AnCF.
HapC cysteine residues 74 and 78 are essential for both stable heterodimer formation with HapE and AnCF stability
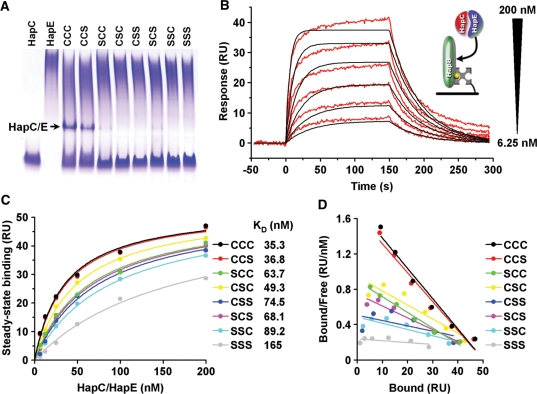
To characterize the impact of HapC cysteine to serine mutations on HapC/HapE heterodimer formation as well as AnCF assembly and stability quantitatively, surface plasmon resonance (SPR) equilibrium binding measurements with highly purified HapC and all seven HapC cysteine to serine mutant proteins were performed. All attempts to measure the interaction between HapC and HapE directly on the surface of a SA-coated sensor chip failed, irrespective of which subunit was immobilized or of the mode of immobilization (data not shown). However, HapC/HapE heterodimer formation in free solution was detectable by native PAGE analysis when HapC or HapC(CCS) proteins were mixed with HapE resulting in a final subunit concentration of 10 µM in the sample (Figure 3A). Therefore, an experimental setup was chosen consisting of the biotinylated HapB subunit fixed to the SA chip and preformed HapC/HapE complexes, generated by mixing equimolar amounts of both subunits, which were applied by injection. Concentration dependent SPR responses of HapC/HapE heterodimer binding to HapB fitted a simple 1:1 interaction model (Figure 3B). Equilibrium analysis led to the calculation of a dissociation constant (KD) of 35.3 nM. The heterodimer composed of the HapC(CCS) mutant and HapE bound with an almost identical affinity to HapB (KD = 36.8 nM), whereas any single substitution of the cysteine residues at positions 74 or 78 of HapC to serine decreased the HapB binding affinity of the respective heterodimers to ∼50% of the HapC/HapE heterodimer (Figures 3C and D). Furthermore, mutation of all three conserved cysteine residues caused a substantial loss of HapC(SSS)/HapE-binding affinity to HapB. Taken together, we concluded that particularly HapC cysteine residues 74 and 78 are required for both the formation of a stable HapC/HapE heterodimer, and the stable interaction of the HapC/HapE heterodimer with HapB.
Figure 3.
HapC/HapE affinity to HapB and HapC/HapE heterodimer formation are impaired by subsitution of HapC cysteine residues 74 and 78 to serine. (A) Native PAGE analysis of heterodimer formation between HapC or all possible HapC cysteine to serine mutants and HapE using samples containing 10 µM of each Hap subunit. (B) SPR analysis of A. nidulans AnCF complex formation from wild-type HapC, HapE and sensor-immobilized HapB, as shown schematically in the inset. Responses of 200, 100, 50, 25, 12.5 and 6.25 nM HapC/HapE heterodimer binding (red lines) to 80 RU of HapB are shown overlaid with the fit derived of a 1 : 1 interaction model (black lines). (C) Fit of the equilibrium data for HapB binding of HapC/HapE containing wild-type HapC or all possible HapC cysteine to serine mutants. Dissociation constants (KDs) were calculated from the concentration-dependent equilibrium binding using a 1 : 1 interaction model. (D) Scatchard plot from the responses shown in (C).
HapC cysteine residues contribute to high affinity DNA binding of AnCF
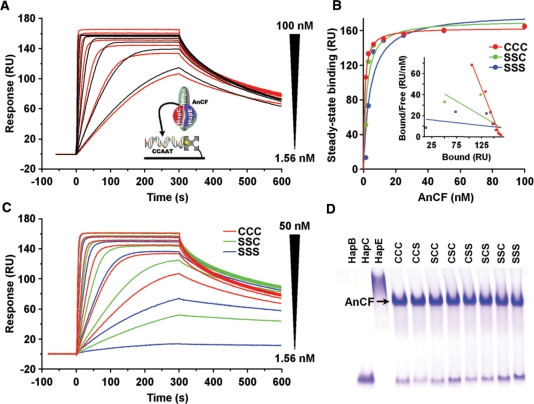
Previous SPR real time protein–DNA interaction studies have demonstrated that AnCF bound with high affinity to a 50-bps DNA duplex representing the CCAAT box at position −1235 of the sreA promoter (28). Despite their susceptibility to oxidation, cysteine residues of Hap3 and NF-YB were previously shown not to be essential for DNA binding (30). To determine the effect of HapC cysteine to serine exchanges on the AnCF DNA-binding affinity, SPR analyses of AnCF binding to this DNA duplex were performed using either HapC or the HapC(SSC) and HapC(SSS) mutants for AnCF reconstitution. Kinetic analysis of the responses of HapC/HapE/HapB heterotrimer binding to the CCAAT box-containing duplex led to the calculation of a KD of 0.42 nM (Figure 4A). Equilibrium analysis of the association-phase responses reaching a steady-state plateau revealed an apparent KD of 1.36 nM, indicating that the measured KD values are valid (Figure 4B). Neither the HapC/HapE heterodimer, nor any other combination of two of the three AnCF subunits, or any subunit alone, bound to DNA in our SPR experiments (data not shown). By contrast, exchange of HapC against HapC(SSS) or HapC(SSC) for AnCF reconstitution impaired DNA association rates below 6.25 nM (Figure 4C) leading to a set of sensor responses, which cannot be fitted by a global 1:1 interaction model. Furthermore, DNA binding of HapC(SSS)/HapE/HapB is nearly abolished at a concentration of 1.56 nM, which reveals an increasing instability of this complex with decreasing concentrations. At higher non-physiological AnCF concentrations (10 µM), however, all seven HapC cysteine to serine mutant proteins are able to form a stable AnCF heterotrimer with HapE and HapB detectable by native PAGE analysis (Figure 4D).
Figure 4.
SPR analysis of AnCF binding to DNA containing a CCAAT box from the promoter region of A. nidulans sreA. (A) Responses of 100, 50, 25, 12.5, 6.25, 3.13 and 1.56 nM AnCF binding (red lines) to 40 RU of DNA duplex is shown overlaid with the fit derived from a 1: 1 interaction model (black lines). (B) Fit of the equilibrium data for DNA binding of AnCF containing wild-type HapC or the mutants HapC(SSC) and HapC(SSS). The inset shows the Scatchard plot from both responses. (C) Responses of AnCF-DNA binding using complexes containing HapC, HapC(SSC) or HapC(SSS), respectively. (D) Native PAGE analysis of AnCF complex formation from HapC or HapC mutants using samples containing 10 µM of each Hap subunit.
Nuclear import of AnCF requires cysteine residues of HapC and is dependent on their redox state
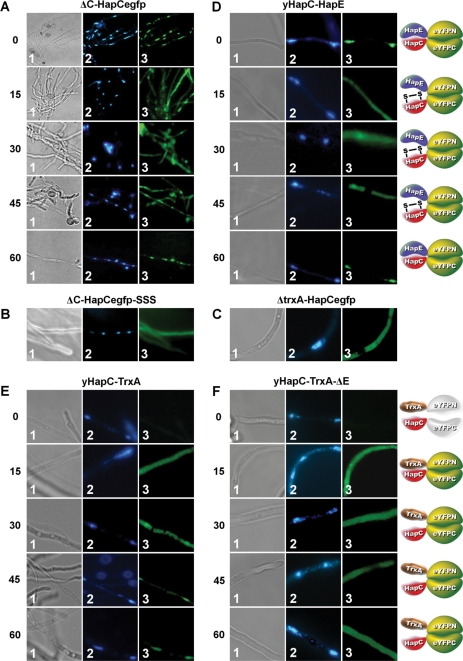
To analyze whether the redox status influenced the cellular localization of AnCF, eGFP and bimolecular fluorescence complementation (BiFC) studies were carried out. The hapC deletion (Δ) strain ΔC-HapCegfp carries a deletion of the endogenous hapC gene and, in addition, encodes a hapC-egfp gene fusion complementing the ΔhapC phenotype to wild type. Under normal conditions, the HapC-eGFP-derived fluorescence was located in the nucleus, as expected for a transcription factor (Figure 5A). Addition of H2O2, however, led to cytoplasmic localization of the HapC-eGFP-derived fluorescence. The cytoplasmic localization lasted up to 45 min after H2O2 addition, whereas fluorescence reappeared in the nucleus after 60 min exposition (Figure 5A). By contrast, a HapC-eGFP fusion, in which all HapC cysteines were exchanged to serines (strain ΔC-HapCegfp-SSS), was located in the cytoplasm in the absence of H2O2, which is in agreement with the impaired stability of the HapC(SSS)/HapE heterodimer (Figure 5B).
Figure 5.
Cellular localization of HapC, HapC/E or HapC/TrxA-dependent fluorescence under oxidative stress conditions. Samples were analyzed by light microscopy (panels 1) and fluorescence microscopy (panels 2 for DAPI staining of nuclei and panels 3 for eGFP or eYFP localization). (A) Localization of HapC-eGFP in a ΔhapC strain at 0, 15, 30, 45 and 60 min after addition of 2.5 mM H2O2. (B) Localization of HapC(SSS)-eGFP in a ΔhapC strain and (C) of HapC-eGFP in a ΔtrxA strain. (D) Cellular co-localization and visualization of HapC and HapE under oxidative stress conditions using BiFC in A. nidulans strains producing HapC and HapE fused with the C- and N-terminal split fragments of eYFP, respectively. (E) Cellular co-localization and visualization of transient HapC-TrxA interaction under oxidative stress conditions by BiFC in A. nidulans wild-type and (F) ΔhapE strains producing HapC and TrxA fused with the C- and N-terminal split fragments of eYFP, respectively.
BiFC assays were previously shown to be valuable tools to define in vivo interaction of proteins in the homologous host (28,47–50). Therefore, either the N-terminal region (aa 1–154) or the C-terminal region (aa 155–238) of the eYFP were fused as described previously (48) to HapC and HapE. Fluorescence was only visible when HapC-YC and HapE-YN were present in the cell at the same time (Figure 5D). In the absence of H2O2, fluorescence was clearly visible in the nucleus indicative of the AnCF complex present in the nucleus. This result indicates that HapC and HapE interact with each other and thereby nucleate the assembly of the eYFP chromophore. In the presence of H2O2, the eYFP fluorescence was visible mainly in the cytoplasm, and after 60 min in the nuclei again (Figure 5D). Although this localization pattern was identical to that observed for a HapC-eGFP fusion, it is important to consider that formation of the bimolecular fluorescence complex is essentially irreversible and protein–protein interaction is not required to maintain a reassembled eYFP (51). Based on these limitations of the BiFC assay, it is likely that upon oxidation HapC-YC and HapE-YN do not interact any longer and the observed fluorescence is due to the reconstitution of functional eYFP before H2O2 treatment. As a consequence of the abolished HapC/HapE heterodimer formation, HapC-YC and HapE-YN, and therefore also the fluorescence is confined to the cytoplasm. Hence, the non-natural split fragment-derived HapC-YC/HapE-YN heterodimer does not allow the recruitment of HapB, which is necessary for nuclear localization.
On the basis of in vitro results and these in vivo fluorescence data, we conclude that oxidative stress-mediated oxidation of HapC impairs its interaction with HapE and consequently HapB-mediated piggy back transport into the nucleus. However, because nuclear relocalization 60 min after H2O2 treatment occurred, this finding indicated recycling of the oxidized HapC subunit. To analyze the involvement of TrxA in this process, the strain yHapC-TrxA encoding the eYFP split fragment fusion proteins HapC-YC and TrxA-YN was generated. In the absence of ROS, no significant fluorescence was visible, whereas H2O2 treatment for 15 min induced strong fluorescence (Figure 5E) indicative of an inter-protein thiol-disulfide exchange between oxidized HapC-YC and the TrxA part of TrxA-YN, followed by formation of a functional eYFP. Noteworthy, this fluorescence does not reflect transcriptional regulation because the gene fusions are under control of the constitutive gpdA promoter. Due to the fact that the interaction between TrxA and HapC is only transient, reduced HapC-YC is now able to form a heterotrimer with endogenous HapE and HapB. Furthermore, as the HapC and TrxA fusion fragments are still linked by the eYFP split parts, the eYFP chromophore enters with the AnCF complex the nucleus leading again to a nuclear fluorescence 60 min post treatment. This assumption was confirmed by the same BiFC experiment carried out in a hapE deletion strain (Figure 5F). Here, HapC was also only bound by TrxA upon oxidative stress, but after reduction the HapC-YC/TrxA-YN complex was not transported to the nucleus anymore as HapE is lacking.
On the basis of HapC-YC/TrxA-YN interaction and the relocalization of HapC-eGFP in the nucleus 45–60 min after initiating oxidative stress, we suggest that the activation of stress response genes upon exposure to oxidative stress leads to recycling of the oxidized HapC subunit by the thioredoxin system. Consistently, in an A. nidulans ΔtrxA mutant strain (36), the HapC-eGFP fusion was constitutively located in the cytoplasm and possibly also in the nucleus, which cannot be distinguished because of the bright fluorescence of the hyphae (Figure 5C). Taken together, these data demonstrate that TrxA is capable to reduce oxidized HapC in vivo and the importance of the thioredoxin system for AnCF functionality.
The redox-regulated AnCF is involved in redox regulation
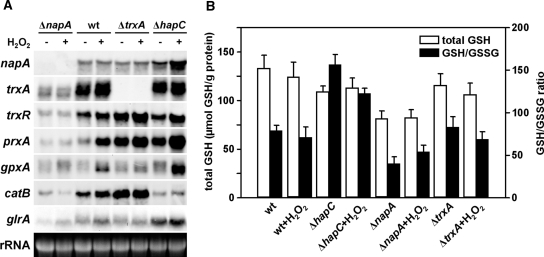
Several redox-regulated transcription factors are known to be involved in redox regulation as well. The most prominent members of such oxidative stress regulators in fungi are the proteins of the so called Yap1-family (yeast activator protein 1) with S. cerevisiae Yap1 being the best-studied member (52) and its A. nidulans ortholog NapA (53). Upon oxidative stress, Yap1 accumulates in the nucleus, where it functions as a transcriptional activator for many stress response genes (52). To address a possible impact of the redox-regulated AnCF on the oxidative stress response in A. nidulans, we compared the expression profiles of certain stress genes of the wild type with ΔnapA, ΔtrxA and ΔhapC mutants (Figure 6A). Moreover, we measured the levels of total cellular glutathione and the GSH(reduced)/GSSG(oxidized) ratios in these strains (Figure 6B), because glutathione is one of the most important cellular antioxidants. Changes in this system indicate occurrence or deregulation of response to oxidative stress.
Figure 6.
Oxidative stress response in A. nidulans wild-type (wt), ΔnapA, ΔtrxA and ΔhapC strains. (A) Gene expression in A. nidulans wild-type, ΔnapA, ΔtrxA and ΔhapC strains. For northern analysis, total RNA was isolated from non-stressed mycelia and mycelia treated for 30 min with 10 mM H2O2. 28S rRNA bands are shown as loading control. (B) Total GSH content and GSH/GSSG ratios of A. nidulans wild-type, ΔhapC, ΔnapA and ΔtrxA mycelia. Total GSH levels and GSH/GSSG ratios of non-stressed mycelia and mycelia treated for 30 min with 10 mM H2O2 are compared.
Deletion of napA led to decreased basal expression of catB, which encodes the mycelial catalase B (54) and genes encoding components of the thioredoxin system, i.e. trxA, trxR and prxA (encoding thioredoxin-dependent peroxidase). The ΔnapA mutant also failed to induce these genes when treated with H2O2, which indicates direct control by NapA. Similarly, the basal transcript level of glutathione reductase-encoding glrA was decreased in ΔnapA but, in contrast to the other genes, H2O2 treatment caused transcriptional up-regulation of glrA. Consistently, napA deletion caused a decrease in the GSH/GSSG ratio under non-stress conditions, and the ratio slightly increased upon H2O2 treatment. These data suggest that the expression of glrA is not exclusively NapA-dependent. Moreover, the total glutathione content of ΔnapA was 1.6-fold decreased compared to the wild type.
A similar expression pattern of catB, as found for ΔnapA, was observed in the ΔhapC mutant. By contrast, mutation of trxA led to a completely different gene expression profile. Basal expression of the (remaining) thioredoxin system-encoding genes, e.g. trxR and prxA, and of catB was up-regulated and H2O2 treatment only had a minor additional increasing effect.
The expression of glrA was not affected by deletion of trxA and, consistently, the GSH/GSSG ratio during unstressed and stressed conditions was wild-type-like. However, in the ΔhapC strain, the expression profile of the thioredoxin system-encoding genes largely resembled that of ΔtrxA, i.e. compared to the wild-type up-regulation of the basal expression and reinforced up-regulation during oxidative stress of both trxR and prxA. In addition, deletion of hapC caused significantly higher expression of glrA, gpxA, napA and increased GSH/GSSG ratios during stressed and unstressed conditions compared to the wild type.
Taken together, deletion of either trxA, napA or hapC caused different changes in the expression profiles of genes involved in the oxidative stress response and of GSH/GSSG ratios. The GSH/GSSG ratios measured in the different strains during stressed and unstressed conditions well agreed with the respective glrA expression patterns. Notably, only free cellular GSH or GSSG can be measured with this assay and therefore, we can only speculate about the ‘real total glutathione content’ that also includes protein-bound glutathione (PrSH/PrSSG) or other GSH S-conjugates. Moreover, oxidative stress not only results in glutathione oxidation, but also in an extracellular export of GSSG or GSH S-conjugates (55,56), which might explain the decreased glutathione concentrations in the different mutant strains, especially in the ΔnapA strain.
The napA promoter and promoter regions of NapA target genes are bound by AnCF
Our northern blot analysis revealed a HapC-dependent expression of napA and NapA target genes. Therefore, the 5′-upstream regions of these genes were analyzed for AnCF consensus binding motifs (15). All of the promoter regions contained at least one core CCAAT box as putative binding motif for AnCF (Table 2). To evaluate the functionality of the found AnCF binding sites, AnCF–DNA interaction analysis was performed with DNA duplexes covering selected CCAAT boxes of the trxA, catB, prxA and napA promoters (Supplementary Figure S3A). Kinetic analysis of the responses of AnCF binding to the CCAAT box-containing duplexes led to the calculation of KD values ranging from 1.7 to 10.2 nM, indicating specific and high-affinity binding (Supplementary Figure S3B–G). Steady-state affinity analysis of the association-phase responses compared with the calculated maximum binding capacities of the respective duplexes revealed a 1:1 stoichiometry, as expected (Supplementary Figure S3H and I). Interestingly, a DNA duplex covering both the CCAAT boxes 1 and 2 of the napA promoter was bound by two AnCF complexes simultaneously. This was not the case for duplex prxApC34, probably due to preferred binding to CCAAT box 4 (which fits better to the consensus sequence RRCCAATMRCR) and/or steric hindrance.
Table 2.
Putative AnCF and NapA binding sites within selected A. nidulans genes and their response to inactivation of AnCF (ΔhapC ) and napA, respectively
| Gene IDa | Promoterb | AnCF CCAAT | NapA TKASTAAc | ΔhapC d | ΔnapAd | Proven in vitro AnCF promoter interactione |
|---|---|---|---|---|---|---|
| AN7513.4 | napA | 4 | 1 | + | na | + |
| AN0170.4 | trxA | 2 | 1 | nd | − | + |
| AN3581.4 | trxR | 2 | 2 | + | − | ni |
| AN8692.3 | prxA | 8 | 3 | + | − | + |
| AN2846.4 | gpxA | 1 | − | + | − | ni |
| AN9339.4 | catB | 3 | − | − | − | + |
| AN0932.4 | glrA | 2 | 3 | + | − | ni |
aCentral Aspergillus Data REpository (CADRE) (80).
bPromoters were defined as the intergenic region up to the next upstream gene.
d+, up-regulation; −, down-regulation; na, not applicable; nd, no difference (from Figure 6A).
e+, yes; ni, not investigated (from Supplementary Figure S3).
Proteomic response to inactivation of AnCF or TrxA
Up to now, ∼20 genes were verified to be regulated in an AnCF-dependent manner (28,57). To further characterize the response to AnCF inactivation and identify possible new targets of AnCF-dependent regulation, we examined the global effect of a hapC deletion at the proteomic level. Due to the fact that a hapC deletion resulted in an up-regulation of oxidative stress-associated genes even under non-stressed conditions, which was similar to the gene expression profile in the ΔtrxA strain, we also included this strain in our proteome analysis. For this purpose, protein extracts of the wild-type strain, a ΔhapC strain and ΔtrxA strain grown in AMM were compared by 2D-PAGE. Reproducibly, 36 protein spots displayed an increase and 35 protein spots displayed a decrease in intensity of more than 1.3-fold (2-fold standard deviation) in ΔhapC extracts (Supplementary Table S3). For ΔtrxA extracts, eight spots with higher and 28 spots with decreased intensities were identified (Supplementary Table S4). The PrxA protein appeared in the gels as more than one spot with the same apparent molecular mass, but with different pI values and abundance (Supplementary Figure S4). MALDI MS analysis revealed that this protein was post-translationally modified by hyperoxidation of cysteine residue 61 to its sulfinic (-SO2H) and sulfonic form (-SO3H). The sulfinic acid that forms in a peroxide-reducing peroxiredoxin can be reduced enzymatically by sulfiredoxins (58,59), but the sulfonic acid represents an irreversible form of protein oxidation. Both hyperoxidized forms of PrxA were found with higher abundance in ΔhapC extracts. Additionally, the production of choline oxidase (CodA) was up-regulated more than 4-fold. This enzyme is involved in glycine, serine and threonine metabolism and catalyzes the O2-dependent conversion of choline to betaine aldehyde and H2O2 (60). Thus H2O2 production might trigger oxidative stress. Furthermore, increased amounts of several proteins involved in protein ubiquitination (putative Thif domain protein) and degradation (the proteasome components Pre6 and Pre8 and the proteasome core alpha 1 component) were found in ΔhapC protein extracts. The ΔhapC strain also produces higher amounts of a novel putative glutathione S-transferase (GST); GSTs catalyze the conjugation of reduced glutathione to electrophilic xenobiotics and peroxidized lipids in order to detoxify these compounds (61). Taken together, 5 of the 36 proteins with higher levels in ΔhapC extracts have antioxidative functions and four of the 35 down-regulated proteins are most likely involved in the oxidative stress response, too.
As already mentioned, deletion of trxA and hapC resulted in a similar expression pattern of stress response genes both under normal and oxidative stress conditions. Consistently, the proteome analysis revealed that four of the eight proteins with higher abundance in protein extracts derived from the ΔtrxA strain were also found with higher abundance in ΔhapC extracts. Similarly, nine of the 28 proteins showing lower levels in ΔtrxA protein extracts were also less abundant in protein extracts of the ΔhapC strain.
DISCUSSION
The CCAAT-binding complex consists of three different subunits that are necessary for formation of a DNA-binding protein complex, for regulation of the CCAAT- binding complex and its import into the nucleus. The mode of protein interaction between the subunits, i.e. association of the nuclear localization signal (NLS)-carrying subunit (for A. nidulans HapB) with a pre-formed heterodimer composed of two histone-like subunits, appears to be similar in higher and lower eukaryotes (32,33). However, a comprehensive picture of the CCAAT-binding complex regulon is missing until now and one of the key questions concerns the regulation of its activity.
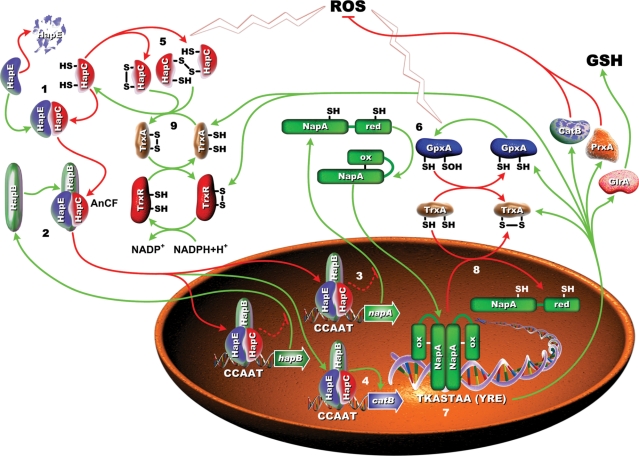
Here, we provide evidence that the CCAAT-binding complex is regulated by the redox status of the cell and regulates genes required for an adequate response to oxidative stress. Based on data presented here, we propose the following model for redox regulation of AnCF (Figure 7): the H2B-like subunit HapC with reduced thiol groups in its HFM is able to form a stable heterodimer with the H2A-like subunit HapE. This heterodimer is bound by the HapB subunit that carries the NLS. Thereby, HapB mediates the nuclear import of the whole AnCF via a piggy-back mechanism (62). Upon oxidation, HapC forms intra- and/or intermolecular disulfide bridges, leading to structural changes within the HFM of HapC that prevent the interaction with HapE. Consequently, nuclear localization is abolished, because HapB needs the pre-formed heterodimer for completion of complex formation.
Figure 7.
Proposed model for redox regulation of the CCAAT-binding complex AnCF and the oxidative stress response in A. nidulans coordinated by AnCF. The bold numbers in the figure refer to the following steps. (1) Cytoplasmic quantity control mechanism of the HapC/HapE heterodimer level by the HapC subunit. Reduced HapC stabilizes the HapE subunit by interaction through the histone fold motifs localized in each subunit. (2) HapB-mediated piggy back transport of AnCF into the nucleus and AnCF-mediated auto-regulation (repression) of the hapB gene. (3) During unstressed conditions, AnCF represses full expression of napA and some NapA target genes such as glrA and prxA, but is required (4) for basal expression of catB. (5) Reactive oxygen species (ROS) inactivate AnCF via oxidation of HapC and (6) activate NapA by the redox transducer GpxA (glutathione peroxidase-like protein A), leading to (7) increased expression of napA and NapA target genes. The ROS response triggered activation of the thioredoxin system leads to (8) inactivation of NapA activity and (9) reactivates AnCF in a feedback loop, which (4) enables production of CatB.
Due to the fact that H2B histones lack the conserved cysteine residues located within the HFM of HapC orthologs, it is conceivable that oxidation of these redox sensitive cysteines represents a key regulatory feature of CCAAT-binding complexes. The most likely rationale for redox regulation of a transcription factor is its involvement in oxidative stress response. The best-studied fungal redox regulator is the S. cerevisiae basic region leucine zipper (bZIP) transcription factor Yap1 that is activated upon peroxide stress (63–67). S. cerevisiae Yap1 and its orthologs were shown to increase the expression of several genes, whose protein products counteract oxidative stress (52). Thioredoxin (Trx) and thioredoxin reductase (TrxR)-encoding genes are the most prominent target genes of this type of regulators in various fungi (38,53,68,69). The activity of S. cerevisiae Yap1 is controlled by a thioredoxin-dependent negative feedback loop and therefore, deficiency of both cytosolic thioredoxins caused a constitutive activation and nuclear accumulation of Yap1 (64,70). As a result, Yap1 target genes such as trr1 and gsh1 (γ-glutamylcysteine synthetase) were up-regulated even under non-stressed conditions and glutathione reductase activity was increased (70). As we showed here, the same is true for A. nidulans. Deletion of trxA (thioredoxin A) up-regulated trxR, prxA and catB under non-stressed conditions. In contrast to S. cerevisiae and S. pombe (70–74), however, deletion of napA and trxA had only a minor or no effect, respectively, on the expression of glrA (glutathione reductase) and the glutathione homeostasis in A. nidulans. By contrast, the increased glrA expression in the hapC-deficient strain well agrees with an ∼2.0-fold increased GSH/GSSG ratio compared with the respective wild-type strain. This finding indicates A. nidulans glutathione homeostasis is predominantly regulated by AnCF which is consistent with the notion that several proteins showed the same altered regulation in both the A. nidulans ΔhapC and ΔglrA strain (75) (Supplementary see Table S3).
Remarkably, neither oxidative stress nor inactivation of the thioredoxin system affected expression of S. cerevisiae yap1 (67,70,73) or, as shown here, napA in A. nidulans. However, the increased napA expression in the ΔhapC strain and the in vitro interaction of AnCF with two CCAAT boxes in the napA promoter suggest direct repression of napA by AnCF. Even more, AnCF also interacts in vitro with the promoters of classical NapA target genes such as trxA, catB and prxA (see Supplementary Figure S3). Dual control, i.e. activation by NapA and repression by AnCF, of these genes is further indicated by the presence of both putative Yap1/NapA and AnCF responsive elements in the respective promoters (except catB) (Table 2). Nevertheless, since AnCF is most likely involved in the regulation of hundreds of genes, it cannot be excluded that some of the regulatory effects are indirectly caused by deletion of hapC, e.g. due to an altered metabolism in the mutant strain. For instance, the identified hyperoxidation of PrxA and the increased production of proteins involved in ubiquitination and degradation processes in non-stressed ΔhapC cultures could be caused by enhanced endogenous oxidative stress due to, e.g. the reduced catalase B activity and/or increased production of H2O2-generating enzymes such as choline oxidase. The S. cerevisiae homolog of PrxA, designated Tsa1p, can also undergo hyperoxidation that impairs its peroxidase activity but increases its chaperone activity (76). Similar to this observation could be our finding of an enhanced production of several proteins with chaperone-like activity in the ΔhapC strain such as the eta subunit of the T-complex protein 1 or the DnaJ domain protein Psi (Supplementary Table S3). However, it is unlikely that all alterations of the oxidative stress response due to the hapC deletion are caused indirectly by increased endogenous oxidative stress because, according to this assumption, hapC deletion would be expected to up-regulate expression of catB in concert with the other NapA target genes but not to down-regulate catB, as observed here. In addition to the decreased catB expression, several other proteins with antioxidative function were found by proteome analysis to be decreased in ΔhapC, e.g. methionine-R-sulfoxide reductase and a putative 1-Cys-peroxiredoxin. Moreover, the increased napA expression and the unusual high GSH/GSSG ratio in the ΔhapC strain were observed neither in the wild type upon oxidative stress nor in ΔnapA and ΔtrxA mutants under both standard and oxidative stress conditions. These findings clearly confirm a direct impact of AnCF on the oxidative stress response.
Taken together all data obtained in vitro and in vivo, we propose the following interplay of AnCF and NapA in redox regulation (see Figure 7 and Supplementary Figure S5): under standard conditions, AnCF prevents full expression of napA and some NapA target genes such as glrA and prxA. Oxidative stress leads to inactivation of AnCF via oxidation of HapC, which increases expression of napA and NapA target genes directly via the release of AnCF repression, and indirectly via NapA activation. This response includes the activation of the thioredoxin system, which represses NapA activity and reactivates AnCF in interconnected feedback loops (Supplementary Figure S5). Thereby, oxidative stress most likely also blocks AnCF-mediated repression of the hapB gene because no heterodimer is formed (25) that is indispensable for HapB-mediated nuclear import of AnCF (62). Because hapC and hapE are constitutively expressed, an increased cellular HapB level causes a rapid nuclear accumulation of AnCF as soon as HapC is reduced by thioredoxin A. Formation of the functional AnCF heterotrimer allows increased production of CatB leading to an accelerated response to oxidative stress. The strict conservation of the redox status-sensing cysteine residues within the HFM of all HapC orthologs indicates that redox regulation very likely represents a general feature of all eukaryotic CCAAT-binding complexes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (Priority Program 1152 to A.A.B); International Leibniz Research School for Microbial and Biomolecular Interactions (ILRS to A.A.B.); Austrian Science Foundation (FWF P-18606-B11 to H.H.). Funding for open access charge: Deutsche Forschungsgemeinschaft (DFG) and HKI.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Sylke Fricke and Silke Steinbach for excellent technical assistance. Robert Winkler is gratefully acknowledged for identification of proteins using mass spectrometry. They thank Birgit Hoff and Ulrich Kück for the gift of BiFC plasmids pEYFPN and pEYFPC.
REFERENCES
- 1.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 2.Barberis A, Superti-Furga G, Busslinger M. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell. 1987;50:347–359. doi: 10.1016/0092-8674(87)90489-2. [DOI] [PubMed] [Google Scholar]
- 3.Johnson PF, McKnight SL. Eukaryotic transcriptional regulatory proteins. Annu. Rev. Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 5.Morgan WD, Williams GT, Morimoto RI, Greene J, Kingston RE, Tjian R. Two transcriptional activators, CCAAT-box-binding transcription factor and heat shock transcription factor, interact with a human hsp70 gene promoter. Mol. Cell. Biol. 1987;7:1129–1138. doi: 10.1128/mcb.7.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinkham JL, Guarente L. Cloning and molecular analysis of the HAP2 locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1985;5:3410–3416. doi: 10.1128/mcb.5.12.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raymondjean M, Cereghini S, Yaniv M. Several distinct "CCAAT" box binding proteins coexist in eukaryotic cells. Proc. Natl Acad. Sci. USA. 1988;85:757–761. doi: 10.1073/pnas.85.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santoro C, Mermod N, Andrews PC, Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988;334:218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- 9.Forsburg SL, Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- 10.McNabb DS, Tseng KA, Guarente L. The Saccharomyces cerevisiae Hap5p homolog from fission yeast reveals two conserved domains that are essential for assembly of heterotetrameric CCAAT-binding factor. Mol. Cell. Biol. 1997;17:7008–7018. doi: 10.1128/mcb.17.12.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNabb DS, Xing Y, Guarente L. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 1995;9:47–58. doi: 10.1101/gad.9.1.47. [DOI] [PubMed] [Google Scholar]
- 12.Olesen JT, Fikes JD, Guarente L. The Schizosaccharomyces pombe homolog of Saccharomyces cerevisiae HAP2 reveals selective and stringent conservation of the small essential core protein domain. Mol. Cell. Biol. 1991;11:611–619. doi: 10.1128/mcb.11.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulder W, Scholten IH, de Boer RW, Grivell LA. Sequence of the HAP3 transcription factor of Kluyveromyces lactis predicts the presence of a novel 4-cysteine zinc-finger motif. Mol. Gen. Genet. 1994;245:96–106. doi: 10.1007/BF00279755. [DOI] [PubMed] [Google Scholar]
- 14.Edwards D, Murray JA, Smith AG. Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol. 1998;117:1015–1022. doi: 10.1104/pp.117.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brakhage AA, Andrianopoulos A, Kato M, Steidl S, Davis MA, Tsukagoshi N, Hynes MJ. HAP-Like CCAAT-binding complexes in filamentous fungi: implications for biotechnology. Fungal Genet. Biol. 1999;27:243–252. doi: 10.1006/fgbi.1999.1136. [DOI] [PubMed] [Google Scholar]
- 16.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko VV, Nakatani Y, Wolffe AP. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hooft van Huijsduijnen R, Li XY, Black D, Matthes H, Benoist C, Mathis D. Co-evolution from yeast to mouse: cDNA cloning of the two NF-Y (CP-1/CBF) subunits. EMBO J. 1990;9:3119–3127. doi: 10.1002/j.1460-2075.1990.tb07509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maity SN, Vuorio T, de Crombrugghe B. The B subunit of a rat heteromeric CCAAT-binding transcription factor shows a striking sequence identity with the yeast Hap2 transcription factor. Proc. Natl Acad. Sci. USA. 1990;87:5378–5382. doi: 10.1073/pnas.87.14.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker DM, Fikes JD, Guarente L. A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc. Natl Acad. Sci. USA. 1991;88:1968–1972. doi: 10.1073/pnas.88.5.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tüncher A, Spröte P, Gehrke A, Brakhage AA. The CCAAT-binding complex of eukaryotes: evolution of a second NLS in the HapB subunit of the filamentous fungus Aspergillus nidulans despite functional conservation at the molecular level between yeast, A.nidulans and human. J. Mol. Biol. 2005;352:517–533. doi: 10.1016/j.jmb.2005.06.068. [DOI] [PubMed] [Google Scholar]
- 21.Sinha S, Maity SN, Lu J, de Crombrugghe B. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc. Natl Acad. Sci. USA. 1995;92:1624–1628. doi: 10.1073/pnas.92.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Litzka O, Then Bergh K, Brakhage AA. The Aspergillus nidulans penicillin-biosynthesis gene aat (penDE) is controlled by a CCAAT-containing DNA element. Eur. J. Biochem. 1996;238:675–682. doi: 10.1111/j.1432-1033.1996.0675w.x. [DOI] [PubMed] [Google Scholar]
- 23.Then Bergh K, Litzka O, Brakhage AA. Identification of a major cis-acting DNA element controlling the bidirectionally transcribed penicillin biosynthesis genes acvA (pcbAB) and ipnA (pcbC) of Aspergillus nidulans. J. Bacteriol. 1996;178:3908–3916. doi: 10.1128/jb.178.13.3908-3916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Littlejohn TG, Hynes MJ. Analysis of the site of action of the amdR product for regulation of the amdS gene of Aspergillus nidulans. Mol. Genet. Genom. 1992;235:81–88. doi: 10.1007/BF00286184. [DOI] [PubMed] [Google Scholar]
- 25.Steidl S, Hynes MJ, Brakhage AA. The Aspergillus nidulans multimeric CCAAT binding complex AnCF is negatively autoregulated via its hapB subunit gene. J. Mol. Biol. 2001;306:643–653. doi: 10.1006/jmbi.2001.4412. [DOI] [PubMed] [Google Scholar]
- 26.Weidner G, Steidl S, Brakhage AA. The Aspergillus nidulans homoaconitase gene lysF is negatively regulated by the multimeric CCAAT-binding complex AnCF and positively regulated by GATA sites. Arch. Microbiol. 2001;175:122–132. doi: 10.1007/s002030000246. [DOI] [PubMed] [Google Scholar]
- 27.Kato M, Tateyama Y, Hayashi K, Naruse F, Oonishi R, Tanoue S, Tanaka A, Kobayashi T, Tsukagoshi N. A quantity control mechanism regulating levels of the HapE subunit of the Hap complex in Aspergillus nidulans: no accumulation of HapE in hapC deletion mutants. FEBS Lett. 2002;512:227–229. doi: 10.1016/s0014-5793(02)02266-4. [DOI] [PubMed] [Google Scholar]
- 28.Hortschansky P, Eisendle M, Al-Abdallah Q, Schmidt AD, Bergmann S, Thön M, Kniemeyer O, Abt B, Seeber B, Werner ER, et al. Interaction of HapX with the CCAAT-binding complex-a novel mechanism of gene regulation by iron. EMBO J. 2007;26:3157–3168. doi: 10.1038/sj.emboj.7601752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNabb DS, Pinto I. Assembly of the Hap2p/Hap3p/Hap4p/Hap5p-DNA complex in Saccharomyces cerevisiae. Eukaryot. Cell. 2005;4:1829–1839. doi: 10.1128/EC.4.11.1829-1839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakshatri H, Bhat-Nakshatri P, Currie RA. Subunit association and DNA binding activity of the heterotrimeric transcription factor NF-Y is regulated by cellular redox. J. Biol. Chem. 1996;271:28784–28791. doi: 10.1074/jbc.271.46.28784. [DOI] [PubMed] [Google Scholar]
- 31.Yoshioka J, Schreiter ER, Lee RT. Role of thioredoxin in cell growth through interactions with signaling molecules. Antioxid. Redox Signal. 2006;8:2143–2151. doi: 10.1089/ars.2006.8.2143. [DOI] [PubMed] [Google Scholar]
- 32.Maity SN, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 33.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 34.Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. The genetics of Aspergillus nidulans. Adv. Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 35.Ballance DJ, Buxton FP, Turner G. Transformation of Aspergillus nidulans by the orotidine-5′-phosphate decarboxylase gene of Neurospora crassa. Biochem. Biophys. Res. Commun. 1983;112:284–289. doi: 10.1016/0006-291x(83)91828-4. [DOI] [PubMed] [Google Scholar]
- 36.Thön M, Al-Abdallah Q, Hortschansky P, Brakhage AA. The thioredoxin system of the filamentous fungus Aspergillus nidulans: impact on development and oxidative stress response. J. Biol. Chem. 2007;282:27259–27269. doi: 10.1074/jbc.M704298200. [DOI] [PubMed] [Google Scholar]
- 37.Litzka O, Then Bergh K, Brakhage AA. Analysis of the regulation of the Aspergillus nidulans penicillin biosynthesis gene aat (penDE), which encodes acyl coenzyme A:6-aminopenicillanic acid acyltransferase. Mol. Gen. Genet. 1995;249:557–569. doi: 10.1007/BF00290581. [DOI] [PubMed] [Google Scholar]
- 38.Lessing F, Kniemeyer O, Wozniok I, Loeffler J, Kurzai O, Haertl A, Brakhage AA. The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot. Cell. 2007;6:2290–2302. doi: 10.1128/EC.00267-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tietze F. Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues. Anal. Biochem. 1969;27:502–522. doi: 10.1016/0003-2697(69)90064-5. [DOI] [PubMed] [Google Scholar]
- 40.Kniemeyer O, Lessing F, Scheibner O, Hertweck C, Brakhage AA. Optimisation of a 2-D gel electrophoresis protocol for the human-pathogenic fungus Aspergillus fumigatus. Curr. Genet. 2006;49:178–189. doi: 10.1007/s00294-005-0047-9. [DOI] [PubMed] [Google Scholar]
- 41.Kim IS, Sinha S, de Crombrugghe B, Maity SN. Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol. Cell. Biol. 1996;16:4003–4013. doi: 10.1128/mcb.16.8.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinha S, Kim IS, Sohn KY, de Crombrugghe B, Maity SN. Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol. Cell. Biol. 1996;16:328–337. doi: 10.1128/mcb.16.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romier C, Cocchiarella F, Mantovani R, Moras D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003;278:1336–1345. doi: 10.1074/jbc.M209635200. [DOI] [PubMed] [Google Scholar]
- 44.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nature protocols. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 45.Hisabori T, Hara S, Fujii T, Yamazaki D, Hosoya-Matsuda N, Motohashi K. Thioredoxin affinity chromatography: a useful method for further understanding the thioredoxin network. J. Exp. Bot. 2005;56:1463–1468. doi: 10.1093/jxb/eri170. [DOI] [PubMed] [Google Scholar]
- 46.Messens J, Van Molle I, Vanhaesebrouck P, Limbourg M, Van Belle K, Wahni K, Martins JC, Loris R, Wyns L. How thioredoxin can reduce a buried disulphide bond. J. Biol. Chem. 2004;339:527–537. doi: 10.1016/j.jmb.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 47.Hink MA, Bisselin T, Visser AJ. Imaging protein-protein interactions in living cells. Plant Mol. Biol. 2002;50:871–883. doi: 10.1023/a:1021282619035. [DOI] [PubMed] [Google Scholar]
- 48.Hoff B, Kück U. Use of bimolecular fluorescence complementation to demonstrate transcription factor interaction in nuclei of living cells from the filamentous fungus Acremonium chrysogenum. Curr. Genet. 2005;47:132–138. doi: 10.1007/s00294-004-0546-0. [DOI] [PubMed] [Google Scholar]
- 49.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 50.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Magliery TJ, Wilson CG, Pan W, Mishler D, Ghosh I, Hamilton AD, Regan L. Detecting protein–protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J. Am. Chem. Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 52.Toone WM, Morgan BA, Jones N. Redox control of AP-1-like factors in yeast and beyond. Oncogene. 2001;20:2336–2346. doi: 10.1038/sj.onc.1204384. [DOI] [PubMed] [Google Scholar]
- 53.Asano Y, Hagiwara D, Yamashino T, Mizuno T. Characterization of the bZip-type transcription factor NapA with reference to oxidative stress response in Aspergillus nidulans. Biosci., Biotechnol., Biochem. 2007;71:1800–1803. doi: 10.1271/bbb.70133. [DOI] [PubMed] [Google Scholar]
- 54.Kawasaki L, Wysong D, Diamond R, Aguirre J. Two divergent catalase genes are differentially regulated during Aspergillus nidulans development and oxidative stress. J. Bacteriol. 1997;179:3284–3292. doi: 10.1128/jb.179.10.3284-3292.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Penninckx MJ. An overview on glutathione in Saccharomyces versus non-conventional yeasts. FEMS Yeast Res. 2002;2:295–305. doi: 10.1016/S1567-1356(02)00081-8. [DOI] [PubMed] [Google Scholar]
- 56.Pocsi I, Prade RA, Penninckx MJ. Glutathione, altruistic metabolite in fungi. Adv. Microb. Physiol. 2004;49:1–76. doi: 10.1016/S0065-2911(04)49001-8. [DOI] [PubMed] [Google Scholar]
- 57.Kato M. An overview of the CCAAT-box binding factor in filamentous fungi: assembly, nuclear translocation, and transcriptional enhancement. Biosci., Biotechnol., Biochem. 2005;69:663–672. doi: 10.1271/bbb.69.663. [DOI] [PubMed] [Google Scholar]
- 58.Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 59.Woo HA, Chae HZ, Hwang SC, Yang KS, Kang SW, Kim K, Rhee SG. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science. 2003;300:653–656. doi: 10.1126/science.1080273. [DOI] [PubMed] [Google Scholar]
- 60.Ikuta S, Imamura S, Misaki H, Horiuti Y. Purification and characterization of choline oxidase from Arthrobacter globiformis. J. Biochem., Tokyo. 1977;82:1741–1749. doi: 10.1093/oxfordjournals.jbchem.a131872. [DOI] [PubMed] [Google Scholar]
- 61.Douglas KT. Mechanisms of action of glutathione-dependent enzymes. In: Meister A, editor. Advances in Enzymology and Related Areas of Molecular Biology. Vol. 59. New York: John Wiley and Sons, Inc.; 1987. pp. 103–167. [DOI] [PubMed] [Google Scholar]
- 62.Steidl S, Tüncher A, Goda H, Guder C, Papadopoulou N, Kobayashi T, Tsukagoshi N, Kato M, Brakhage AA. A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: piggy back transport of the pre-assembled complex to the nucleus. J. Mol. Biol. 2004;342:515–524. doi: 10.1016/j.jmb.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 63.Delaunay A, Isnard AD, Toledano MB. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 2000;19:5157–5166. doi: 10.1093/emboj/19.19.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 65.Kuge S, Jones N, Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997;16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tachibana T, Okazaki S, Murayama A, Naganuma A, Nomoto A, Kuge S. A major peroxiredoxin-induced activation of Yap1 transcription factor is mediated by reduction-sensitive disulfide bonds and reveals a low level of transcriptional activation. J. Biol. Chem. 2009;284:4464–4472. doi: 10.1074/jbc.M807583200. [DOI] [PubMed] [Google Scholar]
- 67.Wemmie JA, Steggerda SM, Moye-Rowley WS. The Saccharomyces cerevisiae AP-1 protein discriminates between oxidative stress elicited by the oxidants H2O2 and diamide. J. Biol. Chem. 1997;272:7908–7914. doi: 10.1074/jbc.272.12.7908. [DOI] [PubMed] [Google Scholar]
- 68.Kuge S, Jones N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee J, Godon C, Lagniel G, Spector D, Garin J, Labarre J, Toledano MB. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 1999;274:16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- 70.Izawa S, Maeda K, Sugiyama K, Mano J, Inoue Y, Kimura A. Thioredoxin deficiency causes the constitutive activation of Yap1, an AP-1-like transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:28459–28465. doi: 10.1074/jbc.274.40.28459. [DOI] [PubMed] [Google Scholar]
- 71.Grant CM, Maciver FH, Dawes IW. Stationary-phase induction of GLR1 expression is mediated by the yAP-1 transcriptional regulatory protein in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 1996;22:739–746. doi: 10.1046/j.1365-2958.1996.d01-1727.x. [DOI] [PubMed] [Google Scholar]
- 72.Lee J, Dawes IW, Roe JH. Isolation, expression, and regulation of the pgr1+ gene encoding glutathione reductase absolutely required for the growth of Schizosaccharomyces pombe. J. Biol. Chem. 1997;272:23042–23049. doi: 10.1074/jbc.272.37.23042. [DOI] [PubMed] [Google Scholar]
- 73.Takeuchi T, Miyahara K, Hirata D, Miyakawa T. Mutational analysis of Yap1 protein, an AP-1-like transcriptional activator of Saccharomyces cerevisiae. FEBS Lett. 1997;416:339–343. doi: 10.1016/s0014-5793(97)01233-7. [DOI] [PubMed] [Google Scholar]
- 74.Wu AL, Moye-Rowley WS. GSH1, which encodes gamma-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol. Cell. Biol. 1994;14:5832–5839. doi: 10.1128/mcb.14.9.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sato I, Shimizu M, Hoshino T, Takaya N. The glutathione system of Aspergillus nidulans involves a fungus-specific glutathione S-transferase. J. Biol. Chem. 2009;284:8042–8053. doi: 10.1074/jbc.M807771200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lim JC, Choi HI, Park YS, Nam HW, Woo HA, Kwon KS, Kim YS, Rhee SG, Kim K, Chae HZ. Irreversible oxidation of the active-site cysteine of peroxiredoxin to cysteine sulfonic acid for enhanced molecular chaperone activity. J. Biol. Chem. 2008;283:28873–28880. doi: 10.1074/jbc.M804087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papagiannopoulos P, Andrianopoulos A, Sharp JA, Davis MA, Hynes MJ. The hapC gene of Aspergillus nidulans is involved in the expression of CCAAT-containing promoters. Mol. Gen. Genet. 1996;251:412–421. doi: 10.1007/BF02172369. [DOI] [PubMed] [Google Scholar]
- 79.Weidner G, d'E;nfert C, Koch A, Mol PC, Brakhage AA. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 1998;33:378–385. doi: 10.1007/s002940050350. [DOI] [PubMed] [Google Scholar]
- 80.Mabey Gilsenan JE, Atherton G, Bartholomew J, Giles PF, Attwood TK, Denning DW, Bowyer P. Aspergillus genomes and the Aspergillus cloud. Nucleic Acids Res. 2009;37:D509–D514. doi: 10.1093/nar/gkn876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.