Abstract
CpG dinucleotides are mutational hotspots associated with cancer and genetic diseases. Thymine DNA glycosylase (TDG) plays an integral role in CpG maintenance by excising mispaired thymine and uracil in a CpG context and also participates in transcriptional regulation via gene-specific CpG demethylation and functional interactions with the transcription machinery. Here, we report that protein kinase C α (PKCα) interacts with TDG and phosphorylates amino-terminal serine residues adjacent to lysines acetylated by CREB-binding protein (CBP) and p300 (CBP/p300). We establish that acetylation and phosphorylation are mutually exclusive, and their interplay dramatically alters the DNA mispair-processing functions of TDG. Remarkably, acetylation of the amino-terminal region abrogates high-affinity DNA binding and selectively prevents processing of G:T mispairs. In contrast, phosphorylation does not markedly alter DNA interactions, but may preserve G:T processing in vivo by preventing CBP-mediated acetylation. Mutational analysis suggests that the acetyl-acceptor lysines are not directly involved in contacting DNA, but may constitute a conformationally sensitive interface that modulates DNA interactions. These findings reveal opposing roles of CBP/p300 and PKCα in regulating the DNA repair functions of TDG and suggest that the interplay of these modifications in vivo may be critically important in the maintenance of CpG dinucleotides and epigenetic regulation.
INTRODUCTION
Cytosine methylation in vertebrates is an important epigenetic mechanism regulating gene expression (1) that also contributes to CpG dinucleotide instability by promoting spontaneous base damage and increased susceptibility to endogenous and environmental mutagens (2). Hydrolytic deamination of cytosine and 5-methylcytosine respectively generates uracil (U) and thymine (T) moieties mispaired with guanine (G) (3). If these G:T/U mispairs remain unrepaired, they give rise to C to T transition mutations associated with oncogenic transformation and genetic diseases (4). In fact, ∼25% of all somatic mutations in the p53 tumor suppressor gene in human tumors involve C to T transitions at CpG, and, in some tumors, this figure rises to almost 50% (5).
Thymine DNA glycosylase (TDG) and methyl-CpG binding domain protein 4 (MBD4) mediate excision of mispaired thymine (G:T) and uracil (G:U) in the CpG context (6–8) and also process various modified pyrimidines (9). Excision of the aberrant base generates a cytotoxic abasic site that is subsequently processed by the coordinated action of other base-excision repair (BER) enzymes, including apurinic/apyrimidinic endonuclease (APE) and DNA polymerase β (Polβ) (10). MBD4 localizes predominantly to transcriptionally inactive heterochromatin while TDG is mostly excluded from these regions and associates with sequence specific transcription factors and cofactors (11–15). These findings suggest that TDG is targeted to transcriptionally active regions of the genome and that BER may be coupled to transcription. Interestingly, overexpression of TDG was shown to reactivate a hormone-regulated transgene silenced by CpG methylation, suggesting a role for TDG in epigenetic regulation (16). Recent studies have demonstrated that recruitment of TDG, in concert with other BER enzymes and DNA methyltransferases 3a/b, to the promoter regions of estrogen-responsive genes is essential to establish cyclic methylation/demethylation patterns in transcriptionally active chromatin (17).
TDG contains a highly conserved central glycosylase domain flanked by divergent amino- and carboxy-terminal regions (18). The amino-terminal region of mammalian TDG contains a hydrophilic lysine-rich region (residues 70–118) that is acetylated by CREB-binding protein (CBP) and p300 (CBP/p300) while the carboxy-terminal region is modified by covalent conjugation of small ubiquitin-like modifier (SUMO) protein (15,19). The amino-terminal region is essential for nonspecific DNA interactions as well as tight binding to abasic sites and processing of G:T mispairs (18,20,21). The tight association of TDG to the abasic site following base excision prevents enzyme turnover, thereby limiting mispair-processing efficiency (21). TDG contains two separate SUMO-binding motifs located in the amino- and carboxy-terminal regions that mediate noncovalent binding to SUMO, which is required for translocation to PML oncogenic domains (20).
CBP/p300 and TDG form ternary complexes with DNA in vitro that retain the ability to mediate base excision and histone acetylation, suggesting that the recruitment of CBP/p300 in vivo may promote chromatin remodelling at the site of repair (15). Furthermore, TDG potentiates CBP-dependent transcription by means of intrinsic SUMO-binding activity (20). Covalent SUMO conjugation to a carboxy-terminal lysine residue effectively abrogates DNA binding and association with CBP, while acetylation of the amino-terminal region may regulate interactions with accessory factors (15,20,21). The reported effects of CBP/p300-mediated acetylation on the activities of BER enzymes (TDG, Polβ and flap endonuclease 1) (15,22,23) suggest an important role for CBP/p300 in coordinating BER.
In light of the critical role of the amino-terminus in G:T processing, we have undertaken to identify additional covalent modifications in this region with potential regulatory functions. Previous studies have shown that TDG is phosphorylated in living cells (24) and in silico analysis identified several putative protein kinase C (PKC) α/β/γ phosphorylation sites in the amino-terminal lysine-rich regulatory domain. PKC comprises a family of 11 related signalling proteins with different tissue distributions and cofactor requirements that participate in diverse cellular processes such as proliferation, apoptosis and differentiation (25). PKC signalling is activated by oxidative stress (26), and there is evidence that Polβ and mismatch repair proteins Msh2 and Msh6 are regulated by PKC phosphorylation (27,28). In this study, we identify a novel mechanism of crosstalk between CBP and PKCα that regulates the DNA mispair-processing functions of TDG through mutually exclusive covalent modification of the amino-terminal region. We demonstrate that acetylation of lysine residues not directly involved in DNA binding, selectively and potently abrogates G:T processing, whereas phosphorylation of adjacent serine residues by PKCα may preserve this function by preventing CBP-mediated acetylation. These findings highlight the importance of covalent modifications in regulating a DNA repair enzyme integral to the maintenance of CpG dinucleotides and epigenetic regulation.
MATERIALS AND METHODS
Plasmids
pCMX-based mammalian expression vectors for FLAG epitope-tagged TDG have been previously described (15). Amino acid substitution mutants of TDG were constructed using the Quickchange mutagenesis kit (Stratagene) according to the manufacturer’s directions and were verified by sequencing. A retroviral TDG expression vector was generated using the pLNCX vector (Clontech). Other expression vectors have been previously described (15,20,29).
Antibodies and peptides
TDG-specific rabbit polyclonal antibody has been previously described (20). Human PKCα-specific polyclonal antibodies (sc-208 or sc-208-G) were obtained from Santa Cruz Biotechnology. Antipolyhistidine (HS1) and anti-FLAG monoclonal antibodies (M2) were obtained from Sigma-Aldrich. TDG peptides obtained from GenScript Corporation were analysed by mass spectrometry and quantified by reverse phase HPLC.
Cell culture and metabolic labelling
NIH3T3 fibroblasts were maintained in Dulbecco’s minimal essential medium (D-MEM) containing penicillin/streptomycin and supplemented with 10% fetal calf serum. For subcellular localization experiments, cells were grown under serum-free conditions for 4 h and then treated with either 100 nM phorbol 12-myristate 13-acetate (PMA) or vehicle [dimethyl sulphoxide (DMSO)] for 15 min in serum-containing media. For metabolic-labelling experiments, 150 mm dishes of NIH3T3 or HEK 293T cells were transfected with TDG expression vectors using the Polyfect transfection reagent (Qiagen). Approximately 24 h later, the culture medium was replaced with serum and phosphate-free D-MEM, and the cells were incubated at 37°C for 2 h. Subsequently, 32P-orthophosphate (200 µCi/ml) was added to the media followed by a 2.5-h incubation at 37°C, which included treatment with PMA or vehicle (DMSO) during the final 30 min. Cells were then washed with phosphate-buffered saline (PBS), and whole-cell extracts were prepared for immunoprecipitations as described below. Immunoprecipitates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and incorporation of 32P was detected by phosphorimaging. Equal loading of TDG was verified by immunoblotting.
Preparation of whole-cell extracts and immunoprecipitations
Whole-cell extracts for immunoprecipitation were prepared from 100-mm dishes of NIH3T3 cells stably expressing FLAG–TDG and control cells transduced with the empty expression vector (pLNCX). Cells grown to 80% confluency were harvested by scraping, then pelleted by centrifugation and resuspended in 500 µl of Lysis buffer [50 mM Tris–HCl pH 7.9, 300 mM NaCl, 1 mM EDTA, 1 mM EGTA 10% glycerol, 0.5% Triton X-100, 1 mM dithiothreitol (DTT), 50 mM sodium fluoride, 200 µM sodium orthovanadate and proteinase inhibitors—20 μg/ml pepstatin A, 10 μg/ml aprotinin, 1 µg/ml leupeptin, 0.5 mM PMSF] and incubated on ice for 30 min. Subsequently, the cell lysate was diluted with 500 µl of dilution buffer (50 mM Tris–HCl pH 7.9, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1 mM DTT, 50 mM sodium fluoride, 200 µM sodium orthovanadate and proteinase inhibitors) and insoluble material removed by centrifugation. Whole-cell extracts were precleared twice with 50 µl (50% v/v) rabbit IgG-agarose (Sigma-Aldrich) for 30 min at 4°C and immunoprecipitated with 50 µl (50% v/v) anti-FLAG affinity resin (M2 agarose, Sigma-Aldrich). Immunoprecipitated proteins were separated by SDS–PAGE and detected by immunoblotting. In the case of immunoprecipitations of radiolabelled in vivo phosphorylated TDG (pTDG), detection following SDS–PAGE was carried out by phosphorimaging and equal loading of TDG was verified by immunoblotting.
2D PAGE
Cell lysates and (two-dimensional) 2D PAGE analysis were performed as previously described (30). Cells were grown to 80–90% confluency in 150-mm dishes and treated with PMA as described above. Approximately 150 µg of cell lysate was fractionated by isoelectric focusing using a 7-cm Immobiline DryStrip gel (pH 6.2–7.5; Amersham/Pharmacia). Subsequently, proteins on the Immobiline Dry strip were fractionated by SDS–PAGE, and TDG was detected by immunoblotting.
Protein purification and in vitro interaction assays
Protein purification, GST-based interaction assays, nickel-based pull-downs and immunoprecipitation procedures have been previously described (15,20).
Base excision and electrophoretic mobility shift assays
Base-excision assays and electrophoretic mobility shift assays (EMSA) were performed as previously described using identical oligonucleotide substrates (20). Either 25 or 12 ng of recombinant TDG was incubated at 30°C with 10 ng of radiolabelled duplex oligonucleotide (25 base pairs) containing a single G:U or G:T mispair in 20 µl of buffer (25 mM HEPES–KOH [pH 7.8], 1 mM EDTA, 0.1 mg/ml BSA and 1 mM DTT). Reactions were carried out for 30 min followed by the addition of 2.5 µl 50% glycerol, and 10 µl of the reaction was used directly for EMSA. EMSA samples were fractionated using 6% polyacrylamide gels cast in 0.25 × TBE and pre-run for 1.5 h in 0.5 × TBE. Protein–DNA complexes were detected by autoradiography or phosphorimaging. The remainder of the reaction was treated with alkali to cleave the abasic sites generated by base release, and subsequently the samples were analysed by denaturing polyacrylamide electrophoresis as described by Hardeland et al. (2002).
Avidin–biotin coupled DNA-binding assays
The avidin–biotin coupled DNA-binding (ABCD) assay was performed as described in ref. 15 using identical biotinylated duplex oligonucleotides. Briefly, streptavidin-coated paramagnetic beads (MagneSphere, Promega) were prepared by washing 0.6 ml (1 mg/ml) with ABCD assay buffer [50 mM Tris–HCl (pH 7.9), 150 mM NaCl, 10% glycerol, 5 mM MgCl2, 0.1% NP40 and 0.5 mM DTT] and resuspending in a final volume of 200 µl. Approximately 250–500 ng of annealed oligonucleotide was incubated for 30 min at room temperature with 10 µl of the streptavidin-coated paramagnetic bead slurry and 100–500 ng of purified bacterially expressed TDG in a total volume of 50 µl of ABCD buffer. Following incubation, beads were washed five times with 200 µl of ABCD buffer, and bound proteins were recovered by resuspending in 12 µl Laemmli buffer and incubation at 90°C for 5 min. Proteins were fractionated by SDS–PAGE and detected by immunoblotting with an antipolyhistidine antibody. In experiments addressing the displacement of TDG from abasic sites, we first incubated TDG with DNA at room temperature for 10 min, and subsequently GST–APE was added with further 30-min incubation.
Preparation of acetylated and phosphorylated TDG for biochemical studies
Acetylated TDG (acTDG) was prepared by incubating recombinant TDG (750 ng) with 1 µg recombinant CBP and 1.25 mM acetyl coenzyme A (AcCoA) in acetylation buffer (20 mM HEPES [pH 7.8], 1 mM EDTA, 1 mM DTT, 10 mM sodium butyrate and 10% glycerol) at 30°C for 90 min. Subsequently, acetylation reactions were dialysed for 60 min against Nickel-binding buffer (NiBB) (50 mM Tris–HCl [pH. 8.0], 100 mM NaCl, 5% glycerol, 0.1% NP40, supplemented with 200 µM sodium orthovanadate, 50 mM sodium fluoride and 10 mM sodium butyrate). NiBB (500 µl) and 60 µl (50% v/v) Ni–NTA agarose were then added to the dialysed reactions, and they were incubated at 4°C for 90 min. The Ni–NTA agarose was then washed three times with 200 µl NiBB, and TDG was eluted three times with 50 µl of NiBB containing 1 M imidazole. Eluted TDG was then dialysed against NETN (50 mM Tris [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.1% NP-40, 10% glycerol and 1 mM DTT) for 60 min. The final concentration of TDG was determined by immunoblotting. Phosphorylated TDG (pTDG) was prepared by incubating recombinant TDG (750 ng) with 50 mU recombinant PKCα (Calbiochem) and 1 mM ATP in 50 µl PKC phosphorylation buffer (20 mM HEPES, pH 7.2, 1 mM sodium orthovanadate, 25 mM glycerol-3-phosphate, 1 mM DTT, 1 mM CaCl2, 15 mM MgCl2, 10 mg/ml 1,2-dioleyl-sn-glycerol and 100 mg/ml phosphatidylserine) for 90 min at 30°C. Mock reactions were performed in the absence of ATP. Subsequently, TDG was purified as described above.
Protein acetylation and phosphorylation assays
Purified phosphorylated or mock-pTDG (100 ng) was incubated with ∼100 ng of purified, recombinant CBP in 30 µl of acetylation buffer in the presence of 1.5 µM 14C acetyl AcCoA and incubated for 30 min at 30°C prior to stopping of the reaction by addition of 30 µl of Laemmli buffer and incubation at 95°C for 5 min. Reaction components were then separated by SDS–PAGE. After separation, the gel was fixed in a 30% methanol, 10% acetic acid solution and treated with amplifying solution (Amersham) prior to imaging by autoradiography or phosphorimaging. Mock-acetylated or acTDG were incubated with 0.25 mU of recombinant PKCα in phosphorylation buffer with 1 µl γ32P-ATP. Reactions were incubated at 30°C for 30 min and then stopped by addition of Laemmli buffer and heated at 95°C for 5 min. Reaction products were then separated by SDS–PAGE. The gel was fixed with a 30% methanol and 10% acetic acid solution, and 32P incorporation was detected by autoradiography. To examine the effect of DNA on acetylation or phosphorylation, we preincubated TDG with DNA for 30 min on ice before proceeding with the reaction.
Immunostaining and microscopy
NIH3T3 cells were fixed for 15 min with 4% formaldehyde in PBS followed by a 10-min incubation with 0.1 M glycine in PBS. Cells were then permeabilized with 0.5% Triton X-100 in PBS for 10 min. Immunostaining was performed with the appropriate primary antibodies and fluorophore-conjugated Donkey secondary antibodies (CY3, FITC) (Jackson ImmunoResearch Laboratories). Epifluorescence imaging was performed on an Axiovert 200M inverted microscope equipped with an Apotome (Carl Zeiss) using appropriate fluorophore-specific filter sets. Z-series images (63 × magnification) were acquired at 0.5-µm intervals and processed with Axiovision software and Adobe Photoshop. Fluorescence intensity plots were obtained by performing a line scan bisecting the cell using Axiovision software.
RESULTS
Phorbol ester-stimulated phosphorylation of TDG in living cells
We have previously shown that TDG is acetylated by CBP/p300 at four lysine residues located within an amino-terminal region essential for DNA binding and G:T processing (Figure 1A) (15,20,21). In silico analysis, using the Scansite algorithm (31), revealed several potential PKC α/β/γ phosphoacceptor residues flanking acetyl-acceptor lysines, suggesting possible functional interplay between PKC and CBP/p300 signalling in TDG regulation. These residues are located within a short sequence motif (93SKKSGKS99) that is conserved in mouse, rat and human TDG. We initially investigated whether endogenous TDG is phosphorylated in living cells in response to treatment with PKC agonist PMA; serum-starved NIH3T3 mouse fibroblasts were treated for 15 min, and whole-cell lysates were subjected to 2D PAGE. Fractionated cellular proteins were immunoblotted with TDG-specific antibodies. Theoretically, each phosphorylation event leads to a 78 Da gain in molecular weight and a 0.11–0.14 decrease in isoelectric point. We observed increases in apparent molecular weight and discrete changes in isoelectric point in response to PMA treatment consistent with phosphorylation of TDG in vivo (Figure 1B). We next verified that TDG is phosphorylated in living cells by metabolic labelling of NIH3T3 cells transiently transfected with FLAG epitope-tagged TDG. Cells grown under serum-free conditions were labelled with 32P inorganic phosphate for 2.5 h, which included a 30-min treatment with either PMA or DMSO. TDG was immunoprecipitated from cell lysates with anti-FLAG resin and quantified by immunoblotting. Subsequently, comparable levels of TDG were analysed by SDS–PAGE, and incorporation of 32P was detected by phosphorimaging. We observed a basal level of phosphorylation that was enhanced by PMA treatment (Figure 1C, top panel). Pretreatment of cells with a PKCα/β-specific inhibitor (Gö6976) (32) prior to PMA stimulation abolished phosphorylation. Since the ubiquitous expression of PKCα (33) is more consistent with the wide tissue distribution of TDG, we focused our investigations on this isozyme. To obtain corroborating evidence for a link between PKCα and TDG, we examined the subcellular distribution of the endogenous proteins in NIH3T3 fibroblasts and undifferentiated P19 embryonal carcinoma cells by indirect immunofluorescence. In NIH3T3 cells cultured in the absence of serum, PKCα displayed predominantly cytoplasmic distribution while TDG was found almost exclusively in the nucleus (Figure 2A, panels I, III and V). Treatment with PMA for 15 min in the presence of serum, triggered nuclear translocation of PKCα as previously reported (34) and colocalization with TDG (Figure 2C, panels II, IV and VI). We observed mainly nuclear staining for PKCα (35) in untreated P19 cells and strong colocalization with TDG (Figure 2B). These findings are consistent with numerous studies demonstrating phosphorylation of nuclear proteins by PKCα (36).
Figure 1.
Phorbol ester-stimulated phosphorylation of TDG in living cells. (A) Illustration of the functional domains of mouse TDG and sites of posttranslational modification. The central-conserved glycosylase domain is sufficient for processing of G:U mispairs while a more divergent amino-terminal extension is required for tight DNA binding and G:T processing (20,21). Two SUMO-binding motifs (SBM1 and SBM2) and the sumoylation site (K341) are shown. A lysine-rich regulatory region located in the amino-terminus is acetylated by CBP/p300 at four distinct lysines (K70, K94, K95 and K98) (15). Putative protein kinase C (PKC) α/β/γ-phosphorylation sites (consensus [S/T-X-[R/K]) within a sequence (boxed) conserved in mouse, rat and human TDG are indicated by asterisks. Complete sequence alignments and accession numbers are found in Figure S1. (B) 2D PAGE analysis of cellular TDG demonstrates PMA-dependent alterations in apparent molecular weight and isoelectric point (pI). Cell lysates were prepared from NIH3T3 cells stimulated with PMA and then separated by 2D PAGE. TDG was detected by immunoblotting with a TDG-specific antibody. (B) In vivo metabolic labelling of transiently expressed TDG with 32P-orthophosphate. Transfected NIH3T3 fibroblasts were grown in serum-free media and metabolically labelled with or without PMA treatment. One population of transfected cells was pretreated with a PKCα/β inhibitor (Gö6976) before PMA stimulation. Immunoprecipitated TDG was fractionated by SDS–PAGE and analysed by phosphorimaging (upper panel) and immunoblotting (lower panel).
Figure 2.
Subcellular localization of TDG and PKCα in NIH3T3 fibroblast and P19 EC cells. (A) Subcellular localization of endogenous TDG and PKCα in NIH3T3 cells. Serum-starved cells were treated with PMA or DMSO and subsequently immunostained for TDG and PKCα. (B) Undifferentiated P19 embryonic carcinoma cells (not treated) were immunostained to detect endogenous TDG and PKCα. Representative optical sections generated by epifluorescence microscopy are shown. The fluorescence intensity plot illustrates the coincidence of peak fluorescence for TDG (CY3, red) and PKCα (FITC, green).
PKCα interacts directly with TDG and phosphorylates the amino-terminal region
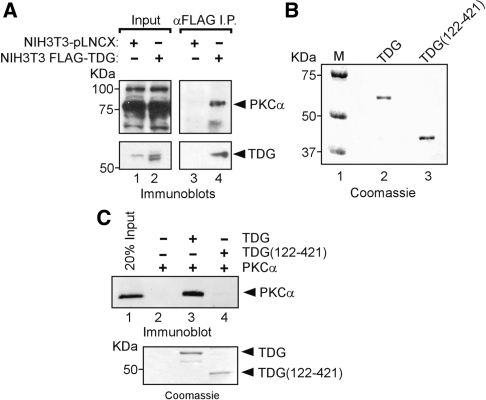
To determine whether TDG and PKCα associate in living cells, we carried out immunoprecipitations with anti-FLAG affinity resin on cell lysates derived from PMA-treated NIH3T3 fibroblasts stably expressing FLAG-tagged TDG. The amount of full-length FLAG-tagged TDG in these cells is comparable to that of endogenous TDG (Figure 3A, compare lanes 1 and 2). Immunoprecipitates were separated by SDS–PAGE and immunoblotted for TDG and PKCα. PKCα was only detected in immunoprecipitates derived from FLAG–TDG expressing cells but not from control cells, consistent with the association of these proteins in living cells (Figure 3A, compare lanes 3 and 4). Similar results were obtained using transiently expressed epitope-tagged proteins (Figure S2). To examine whether these proteins interact directly, we carried out in vitro interaction studies using commercially available recombinant PKCα and polyhistidine-tagged TDG or a truncated variant lacking the amino-terminal region (residues 1–121) (Figure 3B). Following incubation of these proteins, nickel-affinity resin was used to pull down TDG, and PKCα was detected by immunoblotting (Figure 3C). PKCα was retained on the nickel-affinity resin in the presence of full-length TDG but not with the amino terminal variant. Coomassie staining of an aliquot of the binding reaction confirmed that both TDG and TDG(122–421) were bound to the affinity resin. These findings establish a direct interaction between TDG and PKCα that is dependent on the amino-terminal region of TDG.
Figure 3.
PKCα associates directly with TDG. (A) PKCα coimmunoprecipitates with stably expressed TDG in NIH3T3 fibroblasts. Immunoprecipitations using anti-FLAG affinity resin were carried out on whole-cell extracts prepared from PMA-treated cells stably expressing FLAG–TDG or control cells transduced with the empty expression vector. Aliquots of the cell lysates and immunoprecipitated proteins were immunoblotted for PKCα and TDG. (B) Coomassie staining of polyhistidine-tagged TDG and TDG(122–421) used for in vitro protein interaction studies. (C) In vitro association of recombinant TDG and PKCα requires amino-terminal residues 1–121 of TDG. Approximately 1 µg polyhistidine-tagged TDG or TDG(121–421) were incubated with 10 ng of PKCα and subjected to pull down with nickel-affinity resin. As a control, PKCα was also incubated with beads alone. Bound proteins were subjected to SDS–PAGE and immunoblotting with a PKCα-specific antibody (upper panel). Binding of poly-histidine-tagged TDG and TDG(122–421) to the nickel-affinity beads was confirmed by Coomassie staining (lower panel).
We next assessed whether TDG is a direct substrate for PKCα phosphorylation in vitro, by incubating recombinant TDG or TDG(122–421) and PKCα in the presence of γ32P-ATP and essential cofactors; subsequently, the reaction products were fractionated by SDS–PAGE, and incorporation of 32P was measured by phosphorimaging (Figure 4A). Incorporation of radioactivity was observed only with full-length TDG, indicating that the amino-terminal region is essential for phosphorylation by PKCα. Considering that PKCα/β/γ consensus phosphorylation sites are located adjacent to acetyl-acceptor lysines 94, 95 and 98 (see Figure 1A), we employed two short peptides consisting of amino acid residues 68–91 and 91–107 in phosphorylation reactions to further delineate the location of phosphoacceptor serines or threonines. Analysis of the reaction products by SDS–PAGE and phosphorimaging revealed that the peptide containing residues 91–107 was robustly phosphorylated in vitro while the second peptide (residues 68–91) was not appreciably radiolabelled (Figure 4B). Phosphorylated residues were identified by examining the effect of alanine substitutions of potential phosphoacceptor residues in the context of the 91–107 peptide. This analysis indicated that substitution of serine 96 (S96) and serine 99 (S99) substantially decreased 32P incorporation, thereby identifying these residues as the principal phosphoacceptor sites (Figure 4C). In the context of full length bacterially expressed TDG, dual substitution of these residues with either alanine (S96–99A) or aspartate (S96–99D) resulted in ∼60 and 80% reduction in phosphorylation, respectively (Figure 4D). To assess whether these serines are phosphorylated in vivo, we transiently expressed FLAG-tagged wild-type TDG and TDG(S96–99D) in NIH3T3 fibroblasts and carried out metabolic labelling with radiolabelled inorganic phosphate with and without PMA stimulation. We observed that PMA treatment increased phosphorylation of wild-type TDG by ∼80% but had no detectable effects on the S96–99D mutant (Figure 4E, upper panel) confirming that these are the major phosphoacceptor sites. Immunoblotting analysis indicated the presence of comparable amounts of TDG in the different immunoprecipitates (Figure 4E, lower panel). These data confirm that phosphorylation of serine 96 and 99 of TDG is induced by phorbol ester stimulation in living cells.
Figure 4.
PKCα phosphorylates TDG on serines 96 and 99. (A) PKCα-mediated phosphorylation of TDG requires the amino-terminal region. In vitro phosphorylation reactions were performed in the presence of γ32P-ATP using 2 µg of TDG (lane 1) or TDG(122–421) (lane 2) and 0.3 ng of recombinant PKCα. Reaction products were fractionated by SDS–PAGE and incorporation of 32P was detected by phosphorimaging. (B) Delineation of the phosphorylated region using peptide probes. Equimolar amounts of TDG peptides (residues 68–91 and 91–107) and PKCα peptide substrate from glycogen synthetase (residues 1–8 and designated GS 1–8) along with the FLAG peptide were reacted with PKCα and analysed as indicated earlier. (C) Identification of phosphoacceptor residues by alanine substitution. In vitro phosphorylation of the TDG(91–107) (1 µg) peptide and alanine-substituted derivatives was performed and analysed by SDS–PAGE and phosphorimaging (top panel). Quantification of signal intensity is displayed in the bottom panel. Lysines acetylated by CBP/p300 are indicated with asterisks. (D) Dual alanine (lane 2) or aspartate (lane 3) substitutions of serine 96 and 99 reduces PKCα-mediated phosphorylation in full-length TDG. Recombinant TDG and the indicated substitution mutants (2 µg) were phosphorylated in vitro and analysed by SDS–PAGE and autoradiography. (E) In vivo metabolic labelling of transiently expressed TDG and S96-99D mutant with 32P-orthophosphate. Transfected NIH3T3 fibroblasts grown in serum-free media were metabolically labelled for 2.5 h, which includes treatment with either vehicle or PMA during the final 30 min of labelling. Proteins were immunoprecipitated with anti-FLAG resin and then fractionated by SDS–PAGE and phosphorimaged (upper panel). Aliquots of the immunoprecipitates were analysed by immunobloting to ensure equal loading of TDG (lower panel).
Amino-terminal acetylation and phosphorylation are mutually exclusive
The proximity of the acetylated residues to the PKCα phosphorylation sites (see Figure 1A) suggested that each modification may affect the other by altering the effective charge in this region. To investigate this, we established an assay whereby we initially acetylated or phosphorylated recombinant TDG in vitro and then used nickel-affinity chromatography to purify TDG from the modifying enzymes. Following quantification by immunoblotting, we assessed whether acTDG could be phosphorylated by PKCα and vice versa. We found that acTDG was not appreciably phosphorylated by PKCα (Figure 5A) and similarly, when pTDG was used as a substrate for CBP, acetylation was greatly reduced (Figure 5B). Quantification by phosphorimaging revealed a 10-fold and 3-fold reduction in phosphorylation and acetylation, respectively. The more moderate decrease in acetylation, following phosphorylation, is consistent with the presence of an additional acetyl-acceptor lysine (K70), which may not be affected by phosphorylation. To determine whether loss of positive charges at acetyl-acceptor lysines was responsible for inhibition of phosphorylation, we introduced charge neutralizing alanine substitutions at lysines 94, 95 and 98 (K94-95-98A triple mutant), which constitute the major acetylation sites in vitro and carried out in vitro phosphorylation (Figure 5C). Reduced levels of phosphorylation were observed with the mutant consistent with lysine residues being essential for optimal phosphorylation by PKCα. Serine to aspartate substitutions (S96 and S99), which mimic phosphorylation, also reduced acetylation consistent with the mutually exclusive relationship (Figure 5D). We next examined the relationship between these modifications in living cells by carrying out in vivo phosphorylation experiments in HEK 293T cells. This cell line was chosen due to the high-transfection efficiency that can be routinely achieved and the fact that TDG acetylation can be readily observed by metabolic labelling (data not shown). As with NIH3T3 fibroblast, we observed PMA-dependent phosphorylation of TDG, which could be attenuated by coexpression of CBP (60% reduction) (Figure 5E). Furthermore, the TDG mutant with alanine substitutions at acetyl-acceptor lysines was not phosphorylated in a PMA-dependent manner. These findings are in agreement with the in vitro data and consistent with a mutually exclusive relationship between acetylation by CBP/p300 and PKC-mediated phosphorylation. When we performed in vivo acetylation by metabolic labelling with 3H sodium acetate, we found surprisingly that both the S96-99D mutant and wild-type TDG were acetylated at comparable levels (data not shown). These findings are consistent with our unpublished data, indicating that TDG is acetylated by other acetylases in addition to CBP/p300.
Figure 5.
Acetylation and phosphorylation are mutually exclusive. (A) Acetylated TDG (acTDG) is refractory to phosphorylation by PKCα. Recombinant polyhistidine-tagged TDG was acetylated in vitro with CBP and then purified by nickel-affinity chromatography. acTDG was quantified by immunoblotting and ∼100 ng was used in phosphorylation reactions that included γ32P-ATP. Reaction products were analysed by SDS–PAGE and phosphorimaging. (B) Phosphorylated TDG (pTDG) is refractory to acetylation by CBP. pTDG was purified as indicated above and reacted with CBP and in the presence of 14C-acetyl coenzyme A (AcCoA). (C) Reduced in vitro phosphorylation of the K94-95-98A mutant. (D) Reduced in vitro acetylation of the S96-99D mutant. Recombinant proteins (1 µg) were phosphorylated or acetylated in vitro as described above. (E) Inhibition of in vivo TDG phosphorylation by coexpression of CBP or substitution of positively charged acetyl-acceptor lysines. HEK 293T cells were transfected with the indicated expression vector and metabolically labelled with and without stimulation with PMA.
Divergent effects of TDG acetylation and phosphorylation on DNA mispair processing
DNA binding has been reported to promote conformational changes in TDG (21). To address whether TDG could be acetylated or phosphorylated when bound to DNA, we carried out acetylation reactions with CBP in the presence of increasing amounts of either normally paired or G:T mispaired duplex oligonucleotides (Figure 6A). A considerable reduction in acetylation of TDG was observed in the presence of either oligonucleotide, while CBP autoacetylation was not significantly affected. These findings suggest that in vivo TDG bound to DNA is unlikely to be acetylated by CBP/p300. In contrast, the presence of DNA had little effect on the acetylation of GST-p53 and SET (TAF1β) proteins (Figure 6B). Additionally, the presence of duplex oligonucleotides caused a more moderate reduction in PKCα-mediated phosphorylation (Figure 6C), suggesting that phosphorylation of TDG may occur on DNA.
Figure 6.
DNA binding prevents CBP-mediated acetylation of TDG. (A) Dose-dependent inhibition of TDG acetylation by duplex oligonucleotides. TDG (150 ng) was preincubated with the indicated duplex oligonucleotides for 30 min on ice and then acetylated in vitro with CBP (100 ng) and 14C-AcCoA. Reaction products were analysed by SDS–PAGE and autoradiography. (B) DNA-dependent inhibition of CBP-mediated acetylation is specific to TDG. In vitro acetylation was performed with TDG (150 ng), GST-p53 (150 ng) and SET/TAF-1β (150 ng) recombinant proteins in the presence or absence of 200 ng of G:T mispaired oligonucleotide. (C) Phosphorylation of TDG in the presence of duplex oligonucleotides. In vitro phosphorylation reactions were performed as described in Figure 4 using TDG preincubated with 200 ng of the indicated oligonucleotides. (D) Alanine substitution of acetyl acceptor lysines enhances DNA binding. Electrophoretic mobility shift assay was carried out with radiolabelled duplex oligonucleotides bearing either a G:U or a G:T mispair with 25 ng of recombinant wild-type TDG or TDG(K94-95-98A).
The marked reduction in acetylation following DNA binding suggests that the acetyl-acceptor lysines may directly contact DNA. To investigate this possibility, we compared the DNA-binding activity of bacterially expressed recombinant wild-type TDG and the K94-95-98A mutant using the EMSA (37). We found that the alanine-substitution mutant displayed moderately enhanced binding to duplex oligonucleotides containing either G:U or G:T mispairs (Figure 6D). Interestingly, we found that the mutant bound to an abasic site was resistant to displacement by APE compared to wild-type TDG (Figure S3). These findings suggest that lysines 94, 95 and 98 are critical determinants of the DNA-binding properties of TDG and that these positively charged residues are not directly interacting with DNA but may be conformationally important.
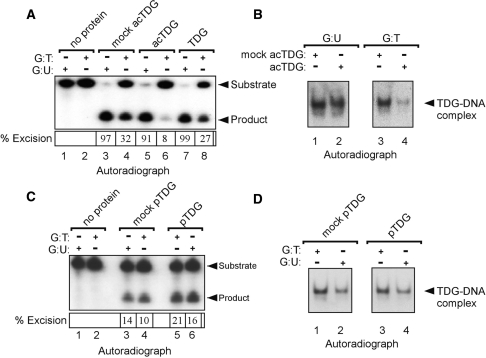
In light of the critical role of the amino-terminal region of TDG in G:T processing and the observed inhibition of CBP-mediated acetylation by DNA, we investigated whether acetylation or phosphorylation could modulate DNA interactions. To address this, we produced purified covalently modified (acetylated or phosphorylated) recombinant TDG in vitro using nickel-affinity chromatography (see Figure 5). As controls, we also carried out mock acetylation and phosphorylation reactions using heat-denatured CBP or by omission of ATP, respectively. We assayed in vitro modified TDG for DNA-binding and G:T/U processing activity using asymmetrically radiolabelled duplex oligonucleotides containing the indicated DNA mispairs. Remarkably, we observed that acTDG retained robust G:U processing activity but displayed severely reduced G:T processing (Figure 7A, compare lane 4 and 6). Consistent with these findings, we observed using both the ABCD-binding assay (data not shown) and EMSA (Figure 7B) that the binding of acTDG to G:U-mispaired oligonucleotides was comparable to that of mock acTDG, whereas binding to G:T mispaired DNA was substantially reduced. Therefore, these data indicate that acetylation of the amino-terminal region selectively abrogates the G:T processing functions of TDG. In contrast, the DNA binding and mispair-processing activities of pTDG were generally indistinguishable from mock phosphorylated protein (Figure 7C and D). To confirm that we achieved efficient phosphorylation of TDG in this experiment, we carried out an acetylation reaction with this material and observed a threefold reduction in incorporation of radiolabelled AcCoA (data not shown). Therefore, although PKCα-mediated phosphorylation does not appear to directly alter the processing functions of TDG it may prevent inhibition of G:T processing by preventing acetylation by CBP/p300.
Figure 7.
Divergent effects of acetylation and phosphorylation on DNA mispair processing. (A) Acetylation of TDG selectively abrogates G:T processing. Purified acetylated TDG (acTDG) (25 ng) was prepared as described in Figure 5, and base-excision assays were carried out using asymmetrically radiolabelled duplex oligonucleotides (10 ng) bearing either G:U or G:T mispairs. Reaction products were treated with alkali to cleave the abasic sites and analysed by denaturing PAGE and autoradiography. (B) acTDG does not stably bind oligonucleotides bearing a G:T mispair. Aliquots of base-excision reactions described above were subjected to electrophoretic mobility-shift assays to determine binding to G:U or G:T mispaired oligonucleotides. (C) Phosphorylation of TDG does not detectably alter G:T/U processing activity. Phosphorylated (pTDG) and mock pTDG (12 ng) were tested for ability to excise mispaired uracil and thymine. (D) DNA-binding analysis of phosphorylated and mock pTDG. Aliquots of the base-excision reactions were subjected to electrophoretic mobility-shift analysis. Supplementary Figures S4 and S5 show images of EMSA gels that include unbound DNA probe.
DISCUSSION
We have elucidated a unique interplay between acetylation and phosphorylation in regulating the DNA-repair functions of TDG. We show that these posttranslational modifications occur on adjacent residues in the amino-terminus and are mutually exclusive. Remarkably, acetylation by CBP/p300 selectively abrogates G:T processing while phosphorylation by PKCα may preserve this function in vivo by preventing CBP-mediated acetylation. Our findings suggest that the opposing regulatory roles of CBP/p300 and PKC may have profound effects on the functions of TDG in CpG maintenance and epigenetic regulation.
We investigated a regulatory role for PKC in TDG-mediated base excision on the basis of the proximity of putative PKCα/β/γ phosphorylation sites and acetyl-acceptor lysines (K94, K95 and K98). These residues are located within a sequence motif conserved in mouse, rat and human TDG (93SKKSGKS99). Our studies indicate that, in mouse fibroblasts, PKCα translocates to the nucleus in response to PMA stimulation and phosphorylates residues in the amino-terminal DNA-binding domain of TDG. Accordingly, we have shown that in NIH3T3 cells, stably expressed TDG coimmunoprecipitates with endogenous PKCα. Furthermore, 2D PAGE analysis and metabolic labelling indicate that TDG is phosphorylated in a PMA-dependent manner in vivo, and this effect was abrogated by a PKCα/β inhibitor. Using recombinant bacterially expressed proteins, we have shown that PKCα interacts directly with TDG, and this association requires the amino-terminal region of TDG (residues 1–122). The phosphorylation sites were mapped in vitro using peptides bearing alanine substitutions of potential phosphoacceptor residues. We identified two serine residues (S96 and S99) that when mutated in the context of full-length TDG, substantially reduced phosphorylation by PKCα. Consistent with this, in vivo PMA-dependent phosphorylation of transiently expressed TDG was substantially reduced when the phosphoacceptor serines were substituted with aspartate. Although we have focused on PKCα in our studies, these phosphoacceptor serines may be phosphorylated by other PKC subtypes. We have used PMA to induce nuclear translocation of PKCα; however, it is well known that in fibroblasts this is a normal physiological response to growth factor (e.g., PDGF and EGF) stimulation (38). Therefore, phosphorylation of TDG may be regulated by mitogenic signals. Interestingly, in undifferentiated P19 EC cells, PKCα is mostly nuclear and colocalizes with TDG, consistent with a role in the regulation of nuclear processes.
Considering the proximity of phosphoacceptor serines and acetyl-acceptor lysines and the different charge characteristics of these residues upon covalent modification, it should perhaps not be surprising that acetylation effectively prevents phosphorylation and vice versa. We found that acetylation of recombinant TDG decreases subsequent phosphorylation by 10-fold while phosphorylation decreases acetylation by at least threefold. Furthermore, substitution of phosphoacceptor serines with aspartate, which mimics phosphorylation, reduced acetylation in vitro while replacement of positively charged lysines with alanine-reduced phosphorylation. Consistent with in vitro studies, PMA-induced phosphorylation in vivo was abrogated by the removal of acetyl-acceptor lysines and reduced by CBP overexpression. Surprisingly, the phosphorylation mimic (S96–99D) was acetylated in vivo at similar levels to wild-type TDG. This is most likely attributable to acetylation of TDG in vivo at other lysine residues by other acetylases. The mutually exclusive nature of these adjacent modifications is reminiscent of the cross-talk observed on the amino-terminal KS dipeptide of histone H3; in this case, phosphorylation of S10 blocks both K9 acetylation and methylation while K9 dimethylation antagonizes S10 phosphorylation (39–42). Similar cross-talk has been suggested for nonhistone proteins (43,44).
In exploring the functional significance of these covalent modifications, we first established whether they could occur when TDG is bound to DNA as conformational changes in this context have been reported (19). We found that DNA-bound TDG was very resistant to acetylation while CBP autoacetylation was not altered. In contrast, phosphorylation by PKCα was much less affected by the presence of DNA. These findings suggest that in vivo PKCα may phosphorylate DNA-bound TDG while acetylation by CBP/p300 requires uncoupling from DNA. The observation that CBP/p300-mediated acetylation abrogates binding to duplex DNA and G:T processing may be relevant to both the CpG maintenance functions of TDG as well as its recently postulated role in gene-specific CpG demethylation (45). acTDG retains the ability to process G:U mispairs in vitro, consistent with previous reports indicating that this function does not require the amino-terminal region (20,21). However, the reduced ability of acTDG to bind DNA may interfere with the genome scanning functions attributed to DNA glycosylases, which could severely hinder detection of DNA mispairs and other DNA lesions in vivo. G:U and G:T mispairs at CpG dinucleotides are generated by hydrolytic deamination of cytosine and methyl cytosine, respectively. To restore cytosine methylation following repair of G:T mispairs, a mechanism is required to discern between mispairs arising from either the methylated or unmethylated cytosine. CBP/p300-mediated acetylation may provide a mechanism to discriminate between the two deamination products. Recent studies employing chromatin immunoprecipitation assays have shown that TDG transiently occupies the promoters of several estrogen responsive genes (11,17), and it has been postulated that TDG and other BER enzymes are essential for the cyclical CpG demethylation patterns observed on these genes during transcription. As the DNA-binding activity and G:T processing functions of TDG are dramatically reduced by CBP-mediated acetylation, this may serve as a powerful mechanism to regulate CpG demethylation and/or the release of TDG from promoters. The finding that CBP/p300-mediated acetylation of Polβ reduces end-trimming activity and impairs participation in BER (22) suggests that CBP/p300 may generally act as negative regulator of BER. This is also in agreement with the role of acetylation in reducing the nuclease activity of another BER enzyme, flap endonuclease 1 (Fen1) (23). A recent study provided evidence for the direct excision of 5-methylcytosine by MBD4, and this activity is stimulated by PKC phosphorylation (46). Therefore, PKC signalling likely plays a central role in regulating CpG demethylation by MBD4 and TDG.
CBP/p300 as well as other protein acetylases respond to DNA damage and other cellular stresses by acetylating cellular proteins such as tumor suppressor p53 (47–49). Covalent modification of key-cellular regulatory proteins is an integral signalling mechanism in DNA damage response that leads to cycle arrest, apoptosis and cellular senescence. Acetylation of TDG may serve to block DNA repair as part of an apoptotic response to cellular stresses such as excessive DNA damage. In this context, it is interesting to note that both phosphorylation by PKC and acetylation by p300 has been shown to inactivate Polβ, while, in this study, we demonstrate that phosphorylation of TDG by PKCα may preserve G:T processing by preventing acetylation by CBP/p300. Altogether, these findings suggest that the crosstalk between different signalling pathways could provide exquisite regulation of the different steps of BER in response to physiologic signals or stresses. Although, in this study, we have focused on the classic functions of TDG in processing G:T/U mispairs, it also processes other damaged bases and may be important in cellular responses to oxidative stress. Along these lines, since PKC isoenzymes are activated by reactive oxygen species (50,51), TDG may be a crucial downstream target that would also be subject to opposing regulation by CBP/p300.
The tight binding of TDG to abasic sites produced by base excision prevents enzyme turnover and limits processing efficiency (21). It has been postulated that sumoylation of mammalian TDG serves to promote release of the enzyme from abasic sites (19). Structural analysis of an amino- and carboxy-terminal deleted human TDG conjugated to SUMO revealed the presence of a protruding helix that interferes with DNA binding (52). However, analysis of sumoylated full-length human TDG by limited proteolysis suggested that sumoylation promotes conformational changes involving interaction of the carboxy- and amino-terminal regions (21). We have shown previously that mouse TDG contains two separate conserved amino-terminal and carboxy-terminal SUMO-binding motifs that interact intramolecularly with the conjugated SUMO and may account for the sumoylation-induced conformational changes (20). Interestingly, both sumoylation and acetylation by CBP/p300 abrogate DNA binding and processing of G:T mispairs (see Figure 8). However, in contrast to sumoylation, which can occur on DNA, we show that CBP-mediated acetylation requires the uncoupling of TDG from DNA. Furthermore, acTDG retains the ability to form stable complexes with abasic sites as evidenced by the stable binding observed following the processing of G:U mispairs. It is plausible that acetylation promotes limited conformational changes within the amino-terminus in contrast to the more extensive changes that are associated with sumoylation. Interestingly, substitution of lysines 94, 95 and 98 with alanines did not mimic the effects of acetylation on DNA binding, and this mutant was resistant to displacement by APE when bound to abasic sites (Figure S3). These findings suggest that these lysine residues play critical roles in DNA binding and mispair processing, and the effects of acetylation are not strictly due to loss of positive charges.
Figure 8.
Cross-talk between TDG posttranslational modifications. Previous studies have shown that sumoylation of human and mouse TDG induces a dramatic increase in G:U processing activity by promoting enzyme turnover (19,20). In contrast, sumoylation (19,20) or acetylation by CBP (this study) abrogate DNA binding and G:T processing (19,20). TDG sumoylation also drastically reduces interactions with CBP/p300, thereby preventing efficient acetylation (20). The present studies reveal that the phosphorylation of serine residues adjacent to acetyl-acceptor lysines by PKCα prevents acetylation by CBP and may preserve G:T processing in vivo.
We have provided biochemical evidence for the interplay of CBP/p300 and PKCα in modulating the DNA-repair functions of TDG. Our studies provide insights into the complex roles of posttranslational modifications in regulating genome maintenance and gene-expression pathways. The fact that both CBP/p300 and PKC signalling pathways are deregulated in oncogenesis (25,53,54) suggest that TDG may be a downstream target that may be functionally compromised and contribute to the genomic instability associated with cancer. Interestingly, TDG has recently been shown to efficiently excise 5-fluorouracil from DNA and plays a role in cellular responses to this commonly used chemotherapeutic agent (55,56). Our studies indicate it may be possible to alter the DNA damage processing functions of TDG in vivo by targeting the signalling pathways that mediate acetylation and phosphorylation of this enzyme. Future studies will establish the utility of our findings in this context and whether TDG is suitable target for cancer therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Cancer Society to M.T.; Ontario Graduate Scholarship and an award from the Canadian Institutes of Health Research Strategic Training Program to R.D.M.; Funding for open access charge: Canadian Cancer Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Rob Sladek for critical reading of the manuscript, Judy Lieberman for the SET/TAF1beta bacterial expression vector Chris Ward at London Regional Proteomics Centre for help with 2D–PAGE analysis and Yuhua Dong for technical assistance.
REFERENCES
- 1.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr. Top Microbiol. Immunol. 2006;301:259–281. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 4.Pfeifer GP. p53 mutational spectra and the role of methylated CpG sequences. Mutat. Res. 2000;450:155–166. doi: 10.1016/s0027-5107(00)00022-1. [DOI] [PubMed] [Google Scholar]
- 5.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 6.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 7.Neddermann P, Jiricny J. The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J. Biol. Chem. 1993;268:21218–21224. [PubMed] [Google Scholar]
- 8.Neddermann P, Gallinari P, Lettieri T, Schmid D, Truong O, Hsuan JJ, Wiebauer K, Jiricny J. Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 1996;271:12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- 9.Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst.) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 11.Chen D, Lucey MJ, Phoenix F, Lopez-Garcia J, Hart SM, Losson R, Buluwela L, Coombes RC, Chambon P, Schar P, et al. T:G mismatch-specific thymine-DNA glycosylase potentiates transcription of estrogen-regulated genes through direct interaction with estrogen receptor alpha. J. Biol. Chem. 2003;278:38586–38592. doi: 10.1074/jbc.M304286200. [DOI] [PubMed] [Google Scholar]
- 12.Chevray PM, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc. Natl Acad. Sci. USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucey MJ, Chen D, Lopez-Garcia J, Hart SM, Phoenix F, Al-Jehani R, Alao JP, White R, Kindle KB, Losson R, et al. T:G mismatch-specific thymine-DNA glycosylase (TDG) as a coregulator of transcription interacts with SRC1 family members through a novel tyrosine repeat motif. Nucleic Acids Res. 2005;33:6393–6404. doi: 10.1093/nar/gki940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Missero C, Pirro MT, Simeone S, Pischetola M, Di Lauro R. The DNA glycosylase T:G mismatch-specific thymine DNA glycosylase represses thyroid transcription factor-1-activated transcription. J. Biol. Chem. 2001;276:33569–33575. doi: 10.1074/jbc.M104963200. [DOI] [PubMed] [Google Scholar]
- 15.Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 16.Zhu B, Benjamin D, Zheng Y, Angliker H, Thiry S, Siegmann M, Jost JP. Overexpression of 5-methylcytosine DNA glycosylase in human embryonic kidney cells EcR293 demethylates the promoter of a hormone-regulated reporter gene. Proc. Natl Acad. Sci. USA. 2001;98:5031–5036. doi: 10.1073/pnas.091097298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 18.Gallinari P, Jiricny J. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature. 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 19.Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohan RD, Rao A, Gagliardi J, Tini M. SUMO-1-dependent allosteric regulation of thymine DNA glycosylase alters subnuclear localization and CBP/p300 recruitment. Mol. Cell. Biol. 2007;27:229–243. doi: 10.1128/MCB.00323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinacher R, Schar P. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr. Biol. 2005;15:616–623. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- 22.Hasan S, El-Andaloussi N, Hardeland U, Hassa PO, Burki C, Imhof R, Schar P, Hottiger MO. Acetylation regulates the DNA end-trimming activity of DNA polymerase beta. Mol. Cell. 2002;10:1213–1222. doi: 10.1016/s1097-2765(02)00745-1. [DOI] [PubMed] [Google Scholar]
- 23.Hasan S, Stucki M, Hassa PO, Imhof R, Gehrig P, Hunziker P, Hubscher U, Hottiger MO. Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol. Cell. 2001;7:1221–1231. doi: 10.1016/s1097-2765(01)00272-6. [DOI] [PubMed] [Google Scholar]
- 24.Um S, Harbers M, Benecke A, Pierrat B, Losson R, Chambon P. Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 1998;273:20728–20736. doi: 10.1074/jbc.273.33.20728. [DOI] [PubMed] [Google Scholar]
- 25.Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nat. Rev. Cancer. 2007;7:281–294. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 26.Gopalakrishna R, Jaken S. Protein kinase C signaling and oxidative stress. Free Radic. Biol. Med. 2000;28:1349–1361. doi: 10.1016/s0891-5849(00)00221-5. [DOI] [PubMed] [Google Scholar]
- 27.Christmann M, Tomicic MT, Kaina B. Phosphorylation of mismatch repair proteins MSH2 and MSH6 affecting MutSalpha mismatch-binding activity. Nucleic Acids Res. 2002;30:1959–1966. doi: 10.1093/nar/30.9.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tokui T, Inagaki M, Nishizawa K, Yatani R, Kusagawa M, Ajiro K, Nishimoto Y, Date T, Matsukage A. Inactivation of DNA polymerase beta by in vitro phosphorylation with protein kinase C. J. Biol. Chem. 1991;266:10820–10824. [PubMed] [Google Scholar]
- 29.Duguid JR, Eble JN, Wilson TM, Kelley MR. Differential cellular and subcellular expression of the human multifuncitonal apurinic/apyrimidinic endonuclease (APE/Ref-1) DNA repair enzyme. Cancer Res. 1995;55:6097–6102. [PubMed] [Google Scholar]
- 30.Duncan JS, Gyenis L, Lenehan J, Bretner M, Graves LM, Haystead TA, Litchfield DW. An unbiased evaluation of CK2 inhibitors by chemo-proteomics: characterization of inhibitor effects on CK2 and identification of novel inhibitor targets. Mol. Cell. Proteom. 2008;7:1077–1088. doi: 10.1074/mcp.M700559-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martiny-Baron G, Kazanietz MG, Mischak H, Blumberg PM, Kochs G, Hug H, Marme D, Schachtele C. Selective inhibition of protein kinase C isozymes by the indolocarbazole Go 6976. J. Biol. Chem. 1993;268:9194–9197. [PubMed] [Google Scholar]
- 33.Wetsel WC, Khan WA, Merchenthaler I, Rivera H, Halpern AE, Phung HM, Negro-Vilar A, Hannun YA. Tissue and cellular distribution of the extended family of protein kinase C isoenzymes. J. Cell. Biol. 1992;117:121–133. doi: 10.1083/jcb.117.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmalz D, Hucho F, Buchner K. Nuclear import of protein kinase C occurs by a mechanism distinct from the mechanism used by proteins with a classical nuclear localization signal. J. Cell. Sci. 1998;111(Pt 13):1823–1830. doi: 10.1242/jcs.111.13.1823. [DOI] [PubMed] [Google Scholar]
- 35.Xu FY, Fandrich RR, Nemer M, Kardami E, Hatch GM. The subcellular distribution of protein kinase Calpha, -epsilon, and -zeta isoforms during cardiac cell differentiation. Arch. Biochem. Biophys. 1999;367:17–25. doi: 10.1006/abbi.1999.1229. [DOI] [PubMed] [Google Scholar]
- 36.Martelli AM, Evangelisti C, Nyakern M, Manzoli FA. Nuclear protein kinase C. Biochim. Biophys. Acta. 2006;1761:542–551. doi: 10.1016/j.bbalip.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 37.Fried M, Crothers DM. Equilibrium and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakashima S. Protein kinase C alpha (PKC alpha): regulation and biological function. J. Biochem. 2002;132:669–675. doi: 10.1093/oxfordjournals.jbchem.a003272. [DOI] [PubMed] [Google Scholar]
- 39.Edmondson DG, Davie JK, Zhou J, Mirnikjoo B, Tatchell K, Dent SY. Site-specific loss of acetylation upon phosphorylation of histone H3. J. Biol. Chem. 2002;277:29496–29502. doi: 10.1074/jbc.M200651200. [DOI] [PubMed] [Google Scholar]
- 40.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 41.Lee DY, Northrop JP, Kuo MH, Stallcup MR. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J. Biol. Chem. 2006;281:8476–8485. doi: 10.1074/jbc.M511093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 43.Latham JA, Dent SY. Cross-regulation of histone modifications. Nat. Struct. Mol. Biol. 2007;14:1017–1024. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- 44.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 46.Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–1012. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- 47.Markham D, Munro S, Soloway J, O’Connor DP, La Thangue NB. DNA-damage–responsive acetylation of pRb regulates binding to E2F-1. EMBO Rep. 2006;7:192–198. doi: 10.1038/sj.embor.7400591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 50.Gopalakrishna R, Anderson WB. Ca2+- and phospholipid-independent activation of protein kinase C by selective oxidative modification of the regulatory domain. Proc. Natl Acad. Sci. USA. 1989;86:6758–6762. doi: 10.1073/pnas.86.17.6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konishi H, Tanaka M, Takemura Y, Matsuzaki H, Ono Y, Kikkawa U, Nishizuka Y. Activation of protein kinase C by tyrosine phosphorylation in response to H2O2. Proc. Natl Acad. Sci. USA. 1997;94:11233–11237. doi: 10.1073/pnas.94.21.11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435:979–982. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 53.Ionov Y, Matsui SI, Cowell JK. A role for p300/CREB binding protein genes in promoting cancer progression in colon cancer cell lines with microsatellite instability. Proc. Natl Acad. Sci. USA. 2004;101:1273–1278. doi: 10.1073/pnas.0307276101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giles RH, Peters DJM, Breuning MH. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 55.Fischer F, Baerenfaller K, Jiricny J. 5-Fluorouracil is efficiently removed from DNA by the base excision and mismatch repair systems. Gastroenterology. 2007;133:1858–1868. doi: 10.1053/j.gastro.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 56.Kunz C, Focke F, Saito Y, Schuermann D, Lettieri T, Selfridge J, Schar P. Base excision by thymine DNA glycosylase mediates DNA-directed cytotoxicity of 5-fluorouracil. PLoS Biol. 2009;7:e91. doi: 10.1371/journal.pbio.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.