Abstract
MicroRNAs (miRNAs) bind to Argonaute proteins, and together they form the RISC complex and regulate target mRNA translation and/or stability. Identification of mRNA targets is key to deciphering the physiological functions and mode of action of miRNAs. In mammals, miRNAs are generally poorly homologous to their target sequence, and target identification cannot be based solely on bioinformatics. Here, we describe a biochemical approach, based on tandem affinity purification, in which mRNA/miRNA complexes are sequentially pulled down, first via the Argonaute moiety and then via the miRNA. Our ‘TAP-Tar’ procedure allows the specific pull down of mRNA targets of miRNA. It is useful for validation of targets predicted in silico, and, potentially, for discovery of previously uncharacterized targets.
INTRODUCTION
MicroRNAs (miRNAs) are short (20–24 nt) non-coding RNAs transcribed from intergenic or intronic sequences as long precursors that are sequentially processed by the endonucleases Drosha and Dicer into short double-stranded sequences (1–3). They regulate gene expression at the post-transcriptional level: in the cytoplasm, miRNAs guide a complex of proteins that includes a member of the Argonaute family, together with which they form the RISC complex, toward a target messenger RNA (1–4). Dependent on the degree of homology between the target and the small RNA, the RISC complex will either cleave the target messenger or inhibit its translation (5,6).
MiRNAs are important gene regulators, both during development and in adults (7). Unraveling their mode of action and the cell pathways they control requires the identification of their mRNA targets. In mammals, miRNAs are in most cases poorly homologous to their targets. Various algorithms exist for prediction of miRNA targets, based on the ‘seed’, a sequence of 7–8 nt at the 5′ of the miRNA (from 2 to 10 nt) that is generally completely homologous to the target (4). Prediction programs are continually being optimized, and the most recent versions of these programs predict a high number of targets for each miRNA, several hundreds in many cases (4,8). These, however, are only predictions that need to be validated experimentally. Validation is generally based on reporter assays, in which the predicted miRNA target sequence is inserted into the 3′ UTR of the reporter gene, thereby conferring sensitivity to the miRNA (9). This reporter assay proves that a given mRNA is potentially a target for a miRNA. However, to directly prove the mRNA/miRNA interaction and its physiological relevance, an assay demonstrating that the mRNA is physically bound to the miRNA inside cells is needed.
Biochemical approaches have been designed to address this issue. In some cases, the Argonaute-containing complexes are pulled down, and associated miRNAs and mRNAs are identified using various procedures (10–12). Associated mRNAs are then subjected to in silico searches for the seed sequences of associated miRNAs. This approach limits the identification of targets to those containing a perfect or near-perfect seed match, which may or may not be the case in vivo (4,5,13–15). Another approach uses the miRNA as a primer for target mRNA amplification by qRT-PCR, but this has been reported only for C. elegans (16).
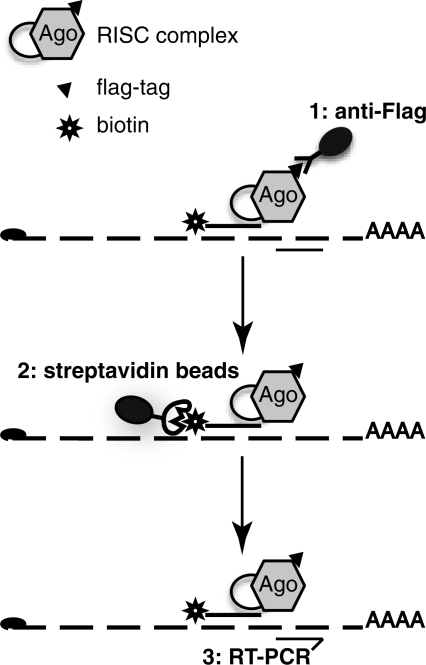
Another series of studies has attempted to pull down mRNAs specifically associated with a miRNA of interest. These biochemical assays are based on the introduction of biotinylated synthetic miRNAs into cells, followed by a pull-down on streptavidin beads (17). This technology has helped identify new targets for miRNAs (15,18), but its application remains limited. Indeed, one-step procedures for pull down are generally prone to high levels of background. Here, in order to circumvent this major concern, we developed a two-step procedure (Figure 1), in which the mRNA/miRNA complex is first pulled down with anti-FLAG antibodies and then purified on streptavidin beads. This procedure greatly reduced the background and allowed us to demonstrate unambiguously a physical association between miR-20a and its mRNA target E2F-1. Our Tandem Affinity Precipitation Target identification (TAP-Tar) thus provides an assay for direct validation of target mRNAs.
Figure 1.
Strategy for mRNA/miRNA pull-down. Extracts from cells expressing a tagged version of Argonaute protein and transfected with biotinylated miRNA are first immunoprecipitated using anti-FLAG antibodies, and then affinity purified on streptavidin beads. mRNAs are quantified by qRT-PCR.
MATERIALS AND METHODS
Cell culture and transfection
HeLa S3 (XLP) cells were cultured with Dulbecco’s modified Eagle’s medium (Invitrogen) containing 10% foetal calf serum. The HeLa S3 cell lines stably expressing Flag-HA-AGO1 and Flag-HA-AGO2 were obtained using retroviral vectors as previously described (19). A cell line transduced with the empty pREV vector was used as a control.
Synthetic miRNAs (15 nM) were transfected into cells using HiPerfect (Qiagen) according to the manufacturer’s instructions. Synthetic miRNA sequences were: (i) miR20a strand: UAAAGUGCUUAUAGUGCAGGUAG; (ii) miR20a* strand: ACUGCAUUAUGAGCACUUAAAGU; (iii) miR125b1 strand: UCCCUGAGACCCUAACUUGUGA; and (iv) miR125b1* strand: ACGGGUUAGGCUCUUGGGAGCU. Synthetic miRNAs were biotinylated at the 3′-end with a C10O4 spacer (purchased from Sigma-Aldrich). No other spacer was tested. Coupling the biotin to the 5′-end impaired miRNA efficiency (data not shown), and no other coupling site was tested.
Western blot
Cells were harvested 3 days after transfection and lysed using 300 mM NaCl, 50 mM Tris pH 7.5, 0.4% NP40 and 10 mM MgCl2. Alternatively, for TAP-Tar analysis, equivalent volumes of lysate or eluate were diluted in 20 mM Tris pH 8. Migration, transfer and staining were performed using standard procedures. Antibodies used were: mouse anti-HA, clone HA-7, Sigma, 1:1000; mouse anti-E2F1, KH95, Santa Cruz Biotechnology, 1:500; mouse anti-β-actin, clone AC-15, Sigma, 1:1000; peroxydase-anti-mouse Fab, A9917, Sigma, 1:5000.
qRT-PCR
For qRT-PCR, RNA was extracted using TRI reagent (Euromedex). When RNA levels were low RNA was purified using nucleospin RNA XS kits (Macherey-Nagel). In some cases, qRT-PCRs were performed directly on eluates, without prior RNA extraction; this procedure did not modify PCR efficiency (data not shown). One-step qRT-PCRs were performed using the QuantiTect SYBR Green RT-PCR kit (Qiagen) as recommended by the manufacturer on a LightCycler (Roche). Analyses were performed as advised by Roche. Primers used were as follows: cyclophilin A forward, GTCAACCCCACCGTGTTCTT; cyclophilin A reverse, CTGCTGTCTTTGGGACCTTGT; E2F1, commercially available Hs_E2F1_1_SG QuantiTect primer assay (Qiagen).
TAP-Tar
Cells were transfected with biotinylated miRNA (15 nM) as described above; the medium was replaced with fresh medium 6 h afterwards, and cells were harvested 2 h later. Cells were lysed at −80°C for 1 h in 10% glycerol, 20 mM Tris (pH 8), 0.2 mM EDTA, 0.5% NP-40, 0.5 M KCl, 1 mM DTT, supplemented with 2 × protease inhibitor (Roche) and 1 u/µl RNAse OUT (Invitrogen) (volume to volume with the cell pellet).
Extracts were submitted to sequential affinity purification, first using anti-Flag M2 agarose beads (A2220, Sigma; 20 µl for 100 µl of cell extract) followed by washes and elution with 1 mg/ml Flag peptide (sequence DYKDDDDK) in 10% glycerol, 20 mM Tris (pH 8), 0.2 mM EDTA, 0.5% NP-40, 0.1 M KCl, 1 mM DTT, and then complexed on M280 magnetic streptavidin beads (Invitrogen; 100 µl of beads for 50 µl of Flag eluate, concentration of beads adjusted to 10 mg/ml) in the same buffer. Streptavidin beads were washed 6 times with 10% glycerol, 20 mM Tris (pH 8), 0.2 mM EDTA, 0.5% NP-40, 0.35 M KCl, 1 mM DTT, and bound complexes were eluted in high salt—25 mM Tris HCl pH 7.4, 1 mM EDTA, 1 M KCl, 10% glycerol, 1 mM DTT, 2 × protease inhibitor (Roche) 1 u/µl RNAse OUT (Invitrogen). In some experiments, RNA was directly extracted from the beads without elution, using the RA1 reagent from the nucleospin RNA XS kit (Macherey–Nagel). Proteins were directly extracted in 4 × NuPAGE LDS sample buffer.
RESULTS AND DISCUSSION
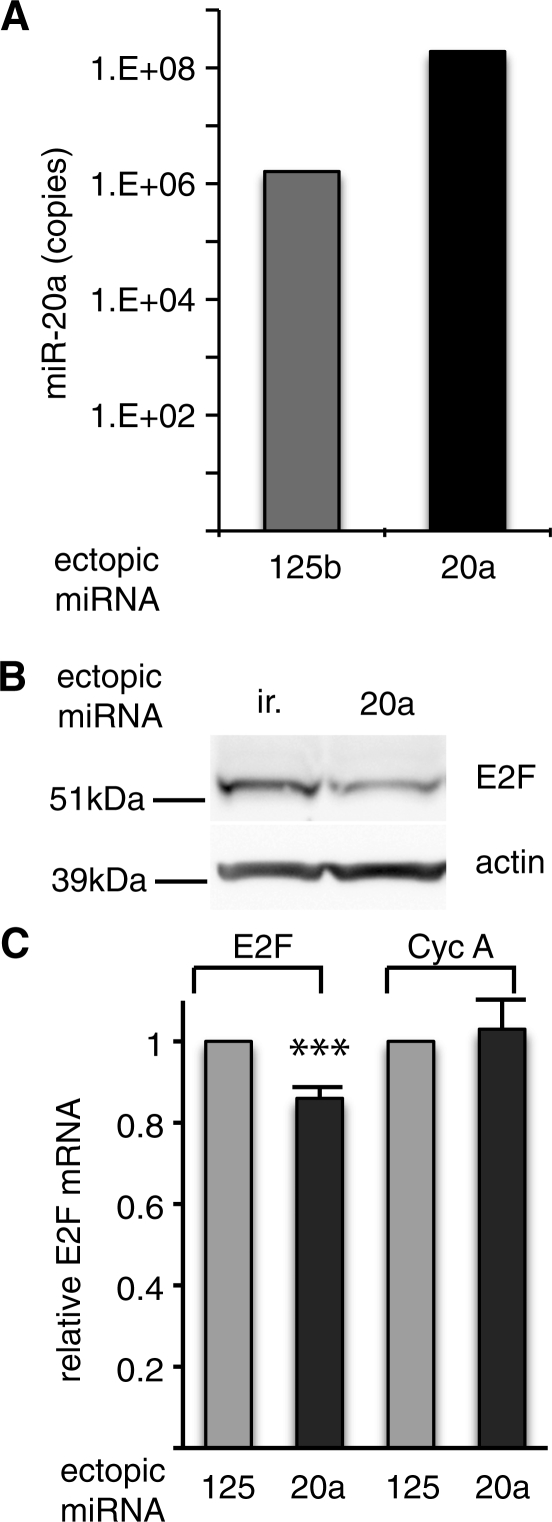
Mir-20a was used to set up the assay, because both mir-20a and its previously validated target E2F1 (20) are expressed in HeLa cells (Figure 2), even though E2F1 is a low-abundance mRNA. The HeLa-derived cell line expressing a tagged version of AGO1 was transfected with biotinylated miR-20a or with biotinylated miR-125b as a negative control. Direct quantification of the miRNA by qRT-PCR showed a vast excess of the ectopic miRNA over the endogenous species (Figure 2A for miR-20a, data not shown for miR-125b). Ectopic biotinylated miR-20a miRNA was active, as it was able to down-regulate the previously characterized target protein E2F1 (Figure 2B). Moreover, a small decay of E2F1 mRNA was also reproducibly observed in miR-20a-transfected cells (Figure 2C), as previously shown for other miRNA targets (8,21–24).
Figure 2.
Biotinylated miR-20a is expressed and active. Flag-HA-AGO1 cells were transfected with biotinylated miR-20a, miR-125b or an irrelevant sequence (ir.) as indicated. (A) Levels of miR-20a were quantified using qRT-PCR (Log scale); representative of two independent experiments. (B) E2F-1 protein, a previously characterized miR-20a target (20), monitored by western blot 3 days after transfection; representative of 3 independent experiments. (C) E2F-1 mRNA monitored by qRT-PCR. Standard deviation is from 10 independent experiments. ***P < 0.001.
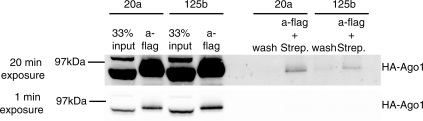
We first confirmed the association between the transfected miRNA and the Argonaute 1 protein complex. AGO1-transduced cells were transfected with biotinylated miR-20a, or biotinylated miR-125b as a control, and cell extracts analyzed after pull down with anti-FLAG followed by streptavidin. Around 2% of total AGO1 protein was pulled down using this procedure with both miRNAs (Figure 3). Maximum association, as judged after a one-step procedure on streptavidin beads, was observed as early as 8 h after transfection (Supplementary Figure S1, upper panel), similar to what was observed by Ohrt et al. (25). The association between biotinylated miR-20a and AGO1 slightly decreased thereafter, but remained detectable at least up to 48 h after transfection. AGO1 was not pulled down in the absence of biotinylated miRNA. Similar results were obtained with Flag-HA AGO2, although the percentage of precipitated complex was lower and a higher background was consistently observed (Supplementary Figure S1, lower panel). These results demonstrate a rapid and specific physical association between the FLAG-HA tagged Argonaute proteins and the biotinylated miRNA. The difference between AGO1 and AGO2 could be explained by a differential incorporation of miR-20a in AGO1 versus AGO2 complex. Alternatively, it could be due to the higher level of expression of Flag-AGO2 compared with Flag-AGO1 (data not shown).
Figure 3.
Interaction between biotinylated miR-20a and AGO proteins. Extracts of cells expressing tagged AGO1, and transfected with miR-20a or miR-125b as indicated, were immunoprecipitated with anti-Flag (a-Flag) followed by streptavidin (a-Flag + strep.) and analyzed by western blot with anti-HA antibodies; the final wash (wash) was also analyzed.
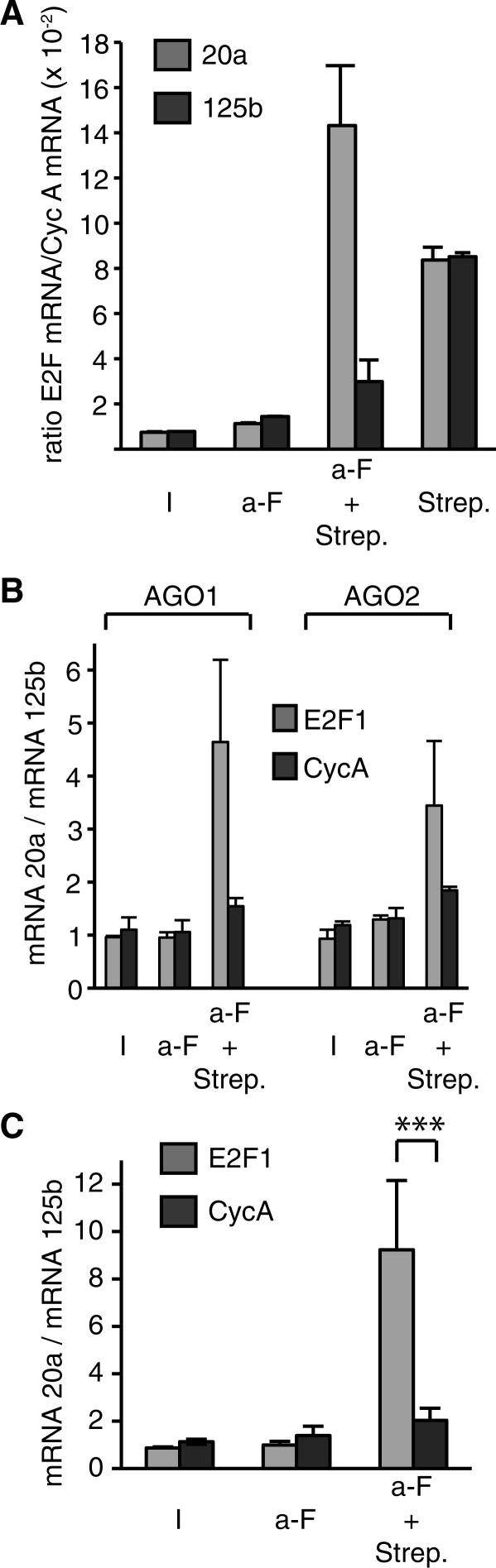
We next compared the efficiency of the tandem affinity purification procedure with that of the previously published one-step technology for target enrichment. Extracts from cells transfected with biotinylated miRNAs were submitted to sequential affinity purification as described in Figure 1 or to a simple pull-down with streptavidin. E2F1 mRNA and Cyclophilin A mRNA, a highly expressed housekeeping gene used as a control, were quantified using qRT-PCR. Precipitation of the complex via the miRNA with streptavidin resulted in a significant enrichment of E2F1 target (Figure 4A, Strep.). However, a similar enrichment was observed with biotinylated miR-125b, an irrelevant miRNA for which no similarity with E2F1 was found (http://www.targetscan.org/cgi-bin/vert_50/view_genetable.cgi?gs=E2F1&taxid=9606&members=miR-17-5p/20/93.mr/106/519.d&showcnc=1&shownc=1). Thus, this procedure is not optimal with regard to specificity. Indeed, one-step purification procedures are usually less specific than two-step protocols, each step providing enrichment and decreasing background. Moreover, endogenous biotinylated proteins, as well as free biotinylated miR-20a not incorporated into the RISC complex, are present in the total lysates and may bind to the streptavidin beads if not previously eliminated, competing with loading of RISC complexes. To overcome this problem, the complex was first purified via the Argonaute protein prior to precipitation via the biotinylated miR-20a miRNA moiety, thus reducing contaminants. Using this protocol, E2F1 mRNA enrichment using AGO1 protein (Figure 4A, a-F + strep) or AGO2 protein (data not shown) was greater. Most importantly, this enrichment was restricted to miR-20a, for which the ratio E2F1 mRNA : Cyclophilin A mRNA was increased by 15-fold as compared with the input, whereas such an enrichment was not observed with miR-125b, even though the two miRNAs were both bound to Argonaute proteins (Figure 3). Similar results were obtained with Flag-HA-AGO1 and Flag-HA-AGO2 (Figure 4B). However, enrichment was slightly less pronounced with AGO2, possibly due to the target mRNA cleaving activity of AGO2 (26) that impairs detection of the mRNA by RT-PCR. In any case, the procedure was more efficient with AGO1, and we thus focused on AGO1 complex.
Figure 4.
E2F1 mRNA is enriched in miR-20a complexes. (A) Extracts of cells expressing tagged AGO1 and transfected with biotinylated miR-20a or miR-125b as indicated, were submitted to affinity purification on streptavidin beads (Strep.), to immunoprecipitation with anti-Flag antibodies (a-F) or to tandem affinity purification (a-F + Strep.). E2F1 and Cyclophilin A mRNAs were quantified using qRT-PCR; results are presented as the ratio between E2F1 and Cyclophilin A mRNAs; representative of two experiments. (B) As in A except that cells expressed either AGO1 or AGO2 as indicated; results are presented as the ratio between miR-20a and miR-125b pull-downs for each mRNA, as indicated; representative of 10 independent experiments for AGO1 and 2 independent experiments for AGO2. (C) Extracts of cells expressing tagged AGO1 and transfected with biotinylated miR-20a or miR-125b, as indicated, were submitted to affinity purification on streptavidin beads (Strep.), to immunoprecipitation with anti-Flag antibodies (a-F) or to tandem affinity purification (a-F + Strep.). E2F1 and Cyclophilin A mRNAs were quantified using qRT-PCR; results are presented as the ratio between miR-20a and miR-125b pull-downs for each mRNA, as indicated; mean of 10 totally independent experiments; results are presented as the ratio between miR-20a and miR-125b pull-downs for each mRNA. The difference was significant. ***P < 0.001.
The biochemical yield of each step was in the expected range, as judged from the recovery of AGO1 proteins (∼50% after Flag immunoprecipitation, and 2% after streptavidin—final yield). The recovery of E2F1 mRNA was much lower (0.7% after Flag, and 0.4% after streptavidin, with a final yield of 0.01%). The low yield of the second step is partly due to the high avidity of biotin for streptavidin, which impairs elution; indeed, direct extraction of E2F1 resulted in much better recovery with a 4% yield. However, the difference between Argonaute protein recovery and E2F1 mRNA recovery most likely reflects a low proportion of E2F1 mRNA bound to the RISC complex in these cells. Thus, even with a low abundance mRNA, poorly bound to RISC, the procedure allowed us to validate the interaction between the miRNA and the mRNA.
The results with AGO1 were highly reproducible, as shown in the compilation of 10 experiments (Figure 4C). The association to the target mRNA was, at least for the most part, formed in cells, since the addition of biotinylated miR20a to cell extracts did not result in a pull-down of mRNA quantitatively as pronounced as that observed in transfected cells (Supplementary Figure S2).
Taken together, these results show that our tandem affinity procedure for mRNA target validation (TAP-Tar) is specific, robust, and provides a mean for direct target validation in cells. The major drawback of this approach is that test cells need to be transfected, similar to the standard reporter assay generally used to validate miRNA targets. Our assay, however, has the advantage of demonstrating a physical interaction between the target and the miRNA in a RISC complex, even though both Argonaute and the miRNA are of exogenous origin in these experiments. Moreover, provided that a transfectable cell can be used, our unbiased approach might be useful to discover previously uncharacterized new targets. Indeed, mRNAs that are pulled down using TAP-Tar can be globally analyzed using standard mRNA detection assays (arrays or deep sequencing). Coupled to bioinformatics analysis of the hits, similar to what has been done in several published studies (10–12,15,18), such a procedure might help to discover unpredicted targets and, ultimately, lead to refinement of target prediction programs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Commission Sixth Framework Programme Integrated Project SIROCCO contract number LSHG-CT-2006-037900. Funding for open access charge: Association pour la Recherche sur le Cancer (ARC; France) and not Governmental funds, as we had anticipated.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Linda Pritchard for critical reading of the manuscript and Thomas Tuschl for the kind gift of Argonaute plasmids.
REFERENCES
- 1.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 2.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 6.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 7.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 9.Kuhn DE, Martin MM, Feldman DS, Terry AV, Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 11.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc. Natl Acad. Sci. USA. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 15.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Andachi Y. A novel biochemical method to identify target genes of individual microRNAs: identification of a new Caenorhabditis elegans let-7 target. RNA. 2008;14:2440–2451. doi: 10.1261/rna.1139508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orom UA, Lund AH. Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods. 2007;43:162–165. doi: 10.1016/j.ymeth.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Hsu RJ, Yang HJ, Tsai HJ. Labeled microRNA pull-down assay system: an experimental approach for high-throughput identification of microRNA-target mRNAs. Nucleic Acids Res. 2009;37:e77. doi: 10.1093/nar/gkp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yahi H, Fritsch L, Philipot O, Guasconi V, Souidi M, Robin P, Polesskaya A, Losson R, Harel-Bellan A, Ait-Si-Ali S. Differential cooperation between heterochromatin protein HP1 isoforms and MyoD in myoblasts. J. Biol. Chem. 2008;283:23692–23700. doi: 10.1074/jbc.M802647200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 21.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 22.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 24.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohrt T, Mutze J, Staroske W, Weinmann L, Hock J, Crell K, Meister G, Schwille P. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008;36:6439–6449. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.