Abstract
Atomic force microscopy (AFM) can detect the adhesion or affinity force between a sample surface and cantilever, dynamically. This feature is useful as a method for the selection of aptamers that bind to their targets with very high affinity. Therefore, we propose the Systematic Evolution of Ligands by an EXponential enrichment (SELEX) method using AFM to obtain aptamers that have a strong affinity for target molecules. In this study, thrombin was chosen as the target molecule, and an ‘AFM-SELEX’ cycle was performed. As a result, selected cycles were completed with only three rounds, and many of the obtained aptamers had a higher affinity to thrombin than the conventional thrombin aptamer. Moreover, one type of obtained aptamer had a high affinity to thrombin as well as the anti-thrombin antibody. AFM-SELEX is, therefore, considered to be an available method for the selection of DNA aptamers that have a high affinity for their target molecules.
INTRODUCTION
Aptamers are rare functional nucleic acid motifs derived from libraries of nucleic acids by iterative rounds of selection and amplification using a process called Systematic Evolution of Ligands by EXponential enrichment (SELEX) (1–5). In the aptamer selection process, the oligonucleotide library is incubated with a target of interest and a buffer of choice at a given temperature. The bound oligonucleotides are then separated from the unbound oligonucleotides, either by filtration on nitrocellulose filters or by affinity processes such as covalent binding to a titer plate or streptavidin (SA)-coated beads. Aptamers have been selected for a wide variety of targets—for example, low-molecular compounds such as ethanolamine ATP (5–13) and proteins (14–17). Importantly, the isolated aptamers often show high specificity and affinity to their cognate targets. These properties expand the possible applications of aptamers, including their use in diagnosis (13,18–20), therapy and imaging processes (21,22).
Previously reported methods for aptamer selection have included affinity chromatography separation step (14–17). The successful selection of high-affinity aptamers from a library of nucleic acids depends mainly on the efficiency with which the unbound species can be separated from the bound sequences. In many cases, however, DNA aptamers selected by conventional SELEX strategy for adenosine triphosphate (KD: 1 × 10−6 order), hematoporphyrin (KD: 1 × 10−4 to 1 × 10−6) and thrombin (KD: 1 × 10−7 order), have a low affinity (7–17). Since the dissociation constant of antibody, often used for biosensing device, had been reported to be 1 × 10−8 to 1 × 10−10 M, the affinities of these reported DNA aptamers were weaker than that of antibody to antigen (23–27). When DNA aptamers are used with biosensing devices, a low affinity to the target molecule will result in unreliable results for detection sensitivity.
Atomic force microscopy (AFM) is a method that scans the imperceptible sample surface using a probe called a cantilever, detecting the weak force between sample surface and probe, and makes pictures of the sample surface. AFM can also dynamically detect the adhesion or affinity force between the sample surface and the cantilever (21,22,28,29). This notable feature is useful for measurement of the affinity force of biomolecule interactions using the biomolecule immobilized cantilever and the sample. This system is considered to be a selection method for aptamers. Therefore, we propose the SELEX method using AFM to obtain aptamers having a strong affinity to target molecules.
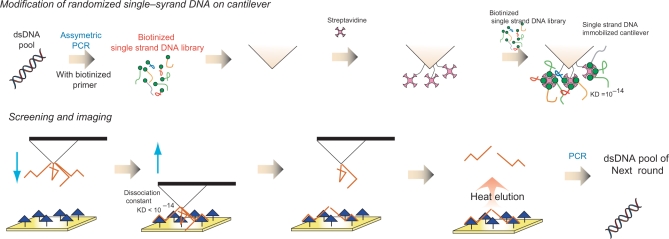
The selection scheme of this SELEX strategy is shown in Figure 1. DNA aptamers with a very strong affinity to target molecules were obtained by repeating this cycle. In this study, thrombin was chosen as the target molecule of the DNA aptamer, and the selection cycles were repeated for three rounds. As a result, the obtained DNA aptamers had a high affinity to thrombin than that of conventional thrombin aptamer. One type of DNA aptamer exhibited high affinity as well as that of an antibody.
Figure 1.
The scheme of a new SELEX strategy for functional oligonucleotide screening by AFM.
MATERIALS AND METHODS
Materials
Thrombin from human plasma and streptavidin from Streptomyces avidinii were purchased from Sigma (MO, USA). Anti-human thrombin antibody from sheep was purchased from Funakoshi (Tokyo, Japan). DNA polymerase was purchased from Roche Diagnostics (Basel, Switzerland). A microspin column was purchased from GE Healthcare (MO, USA). 3,3′-Dithiobis[sulfosuccinimidylpropionate] (DTSSP) was purchased from Pierce (MO, USA). The other chemicals used were analytical grade and were purchased from Nacalai Tesque (Kyoto, Japan).
Preparation of the cantilever as modified by single-strand DNA (ssDNA) library
In the primary round, for double-strand DNA library (dsDNA) production, a synthetic DNA oligonucleotide library (104-mer) with 60 random nucleotide sequences, 5′-TAGGGAATTCGTCGACGGATCC-N60-CTGCAGGTCGACGCATGCGCCG-3′, was amplified over 25 cycles of PCR (95°C, 15 s; 72°C, 30 s) using the following pair of primers: 5′-TAATACGACTCACTATAGGGAATTCGTCGACGGAT-3′ (P1) and 5′-CGGCGCATGCGTCGACCTG-3′ (P2). The ssDNA library was then obtained from the dsDNA by an additional 90 cycles of asymmetric PCR using 5′ biothinated P1 primer. The PCR product, ssDNA, was purified by microspin column.
To clear away the organic compounds that adhered on the cantilever, the cantilever was treated with UV for 2 h. The probe was then exposed to 100 µl of 2 mg/ml DTSSP solution in 20 mM acetate (pH 4.8) at room temperature for 30 min. After the reaction, the probe was dipped in 20 ml of ultra pure water to remove unreacted DTSSP. The succinimide immobilized probe was then doused at room temperature for 1 h with 100 µl of 1 mg/ml streptavidin solution in PBS, followed by washing with 20 ml of folding buffer. After immobilization of streptavidin on the cantilever, 100 µl of 5 µM biothinated ssDNA was dropped on the cantilever and incubated at room temperature for 30 min. Finally, the ssDNA immobilized probe was put in 20 ml of folding buffer containing 0.01% Tween 20 to remove unbound biothinated ssDNA.
Preparation of gold chip modified by thrombin
A gold chip was covered by 200 µl of 4 mg/ml DTSSP solution in 20 mM acetate at room temperature for 30 min. The chip was then washed with 10 ml of ultra pure water. After washing, the succinimide immobilized gold chip was covered with 200 µl of 6 µg/ml thrombin solution in PBS at room temperature for 1 h. Finally, the gold chip was washed with 10 ml of folding buffer.
SELEX strategy based on AFM
Force curve mapping was performed in the liquid cell of a SPA400-Nanonavi AFM unit (SII Nanotechnology Inc., Chiba, Japan) with the cantilever and gold chip described above. The force curve measurements were performed in folding buffer [50 mM Tris–HCl (pH 7.6), 300 mM NaCl, 30 mM KCl and 5 mM MgCl2]. Force curves were recorded at a velocity of 17 µm/s. Topographic images were captured at 64 × 64 pixels resolution with a scan size of 1 µm × 1 µm. The adhesion forces analysis between ssDNA immobilized on tip of cantilever and thrombin were measured at 4096 data points (64 × 64 point), and a histogram of these adhesion force was charted. According to the AFM-SELEX, scanning was carried out five times (4096 × 5 times) in round 1 and 2, and 4 times (4096 × 4 times) in round 3, respectively.
After the scanning and force curve mapping, the ssDNA-bound gold chip was washed with 10 ml folding buffer containing 0.01% Tween 20. Then, to elute ssDNA on the gold chip, the chip was dunked in 2 ml of 20 mM TE buffer containing 1% DMSO and incubated at 98°C for 10 min and on ice for 10 min. The eluted ssDNAs were precipitated by ethanol, dissolved in 100 µl of 20 mM TE buffer, and used for the next selection as a PCR template.
Cloning and sequencing
The dsDNAs obtained after the fourth rounds of selection were subcloned into pT7 blue vector, and then transformed into Escherichia coli (Nova blue). The plasmid DNA was isolated by the alkaline-extraction method. Twenty-two colonies were randomly selected after the fourth round, and these DNA sequences were determined by a dye-terminator method using CEQ 8000 (Beckman).
Affinity assay of obtained aptamers by AFM
For dsDNA production, obtained DNA was amplified over 15 cycles of PCR (95°C, 15 s; 72°C, 30 s) using P1 and P2 primer. The ssDNA was then obtained from the dsDNA by an additional 90 cycles of asymmetric PCR using 5′ biothinated P1 primer. The PCR product, ssDNA, was purified by microspin column.
The preparation of the cantilever immobilizing ssDNA and the gold chip immobilizing thrombin was followed by selection methods. The affinity force of the conventional thrombin aptamer and antibody to thrombin was analyzed as a positive control under the same conditions.
Fluorescence polarization measurement
The dissociation constant of TBA-1 was calculated by fluorescence polarization. Each of various concentrations of thrombin and FITC labeled TBA-1 (final concentration: 100 nM) was mixed in the folding buffer for 2 h, and fluorescence polarization measurement was carried out (EnVision, Perkin elmer, Wellesley, MA). As a negative control of thrombin, 100 nM streptavidin was used, and fluorescence polarization measurement was carried out under same condition. Each data was represented three independent experimental data, and each error bar was mean of standard deviation.
RESULTS
Selection of DNA aptamer using AFM
At first, the biotinated random ssDNA was immobilized on the cantilever through avidin, and the target molecules were immobilized on the gold chip. The prepared cantilever and gold chip were simultaneously applied to AFM as a DNA-modified probe and thrombin immobilized chip, respectively. When the cantilever approached the gold chip, the DNA aptamer that binds the target molecules would bind the target immobilized on the gold chip. If the affinity force with target is very strong, the avidin-biotin interaction is fractured, and DNA aptamer remains on the gold chip. The remaining DNA was recovered by heat elution amplified by PCR.
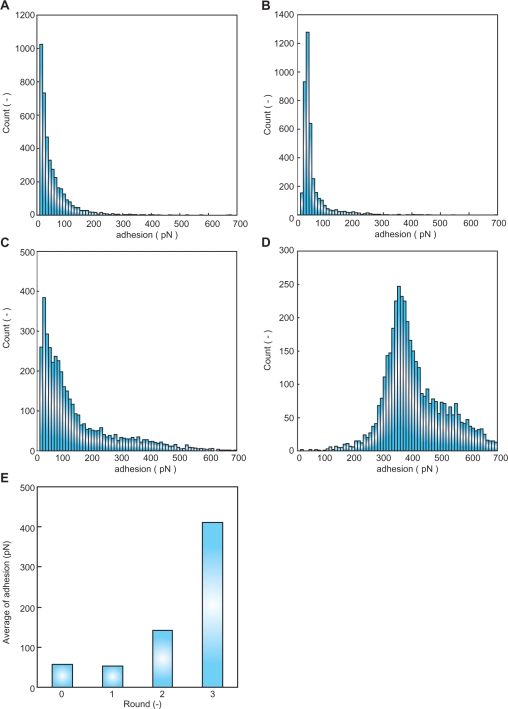
The selection cycle by AFM-SELEX was repeated for three rounds. The histogram of the affinity force between ssDNA and thrombin was developed from 4096 force curves from each round (Figure 2A–D). As a result, the affinity force between ssDNA and thrombin was gradually increased with repeated selection rounds. The initial ssDNA and round 1 elution pool had low affinity force to thrombin (Figure 2A and B). However, in the round 2 elution pool the force was higher, over 100 pN, and in the round 3 elution pool the histogram of affinity force had a peak at 320–329 pN. Using the histogram data, the average affinity force of each round was calculated (Figure 2E). The average force of the initial pool was ∼57.15 pN (Figure 2A and E). However, after only three repeated rounds of the AFM-SELEX method, the ssDNA pool had 411.52 pN of strong affinity to thrombin (Figure 2D and E).
Figure 2.
Histogram of adhesion force between ssDNA and protein for 4096 data points. (A) adhesion force between 0 pool ssDNA and thrombin, (B) adhesion force between round 1 elution pool and thrombin, (C) adhesion force between round 2 elution pool and thrombin, (D) adhesion force between round 3 elution pool and thrombin and (E) the average of affinity force between each round elution pool and thrombin.
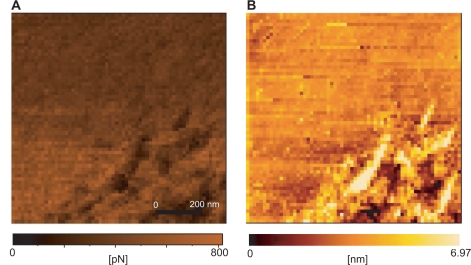
In Figure 3, the topography of the affinity force between the round 3 elution pool and thrombin is shown. The highest point of affinity force was concentrated late in the scanning (Figure 3A). Moreover, the force had no relationship with the roughness of the sample, since the higher force was not operating at the height of the surface roughness.
Figure 3.
Affinity image between round 3 elution pool and thrombin showing the affinity images (A) as well as the topography images (B).
Cloning and sequencing
The dsDNAs obtained after the third round of selection were subcloned into pT7 blue vector. Twenty-two colonies were randomly selected, and these DNA sequences were determined (Table 1). Many of the obtained DNAs had G-rich sequences. The sequence called TBA-1 had the largest share of obtained aptamers. Moreover, the sequences of TBA-1 and -2 had many ‘GGGGT’ motifs.
Table 1.
Sequence of obtained single-strand DNA
| Sequence name | Sequence of random region | Count |
|---|---|---|
| TBA1 | CCTAGTGTGCGTCGATGGGGTGGGGTGGGGCTGAGTTGGGGGGTGGGATCAATCAATCTGGTTT | 8 |
| TBA2 | CCTAGTGTGCGTCGATGGGGTGGGGTGGGGCTGAGTTGGGGGGTGGGATCAATCCATCTGGTCT | 2 |
| The other 1 | CCCACGGAGTCACCATTGATCACAACCCCAGCT | 1 |
| The other 2 | CCCACGGAGCCACCACCTTGATCACAACCCTCAGCT | 1 |
| The other 3 | CCCACGGAGTCACCCTTGATCACAACCTCAGCT | 2 |
| The other 4 | CCCACGGAGTCACCCTTGACCACAATCTCAGCT | 1 |
| The other 5 | CCCACGGAGCCACCCTTGATCACAGCCTCAGCT | 1 |
| The other 6 | CCCACGGAGTCACCCTTGATCACGAACCTCAGCT | 1 |
Binding assay using AFM
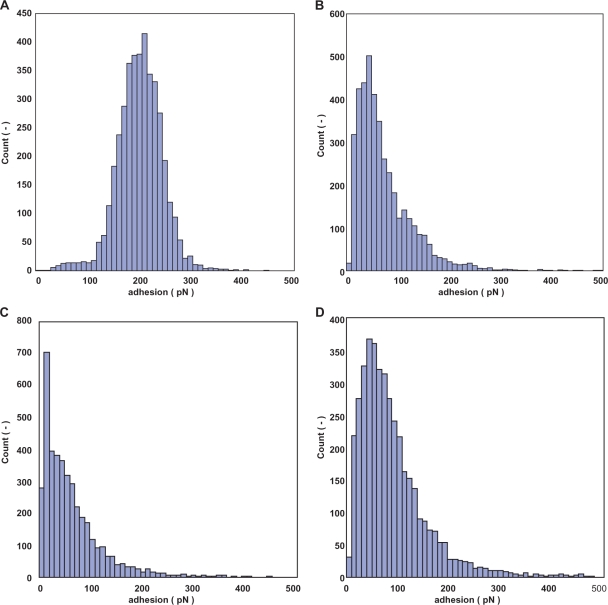
The immobilization method described above was followed by the SELEX protocol. The prepared cantilever and gold chip were applied to AFM to analyze the affinity force between ssDNA and thrombin. The resulting force histogram of TBA-1 had one peak at 210-219 pN (Figure 4A). The histogram of TBA-2 had two peaks at 40–49 and 110–119 (Figure 4B). The average affinity force between thrombin and the obtained aptamers was calculated. The obtained aptamers had a higher affinity for thrombin than N60 ssDNA and the conventional thrombin aptamer (Figure 4C and Table 2 ). TBA-1 had the highest affinity of the obtained DNA aptamers. The affinity force of the conventional DNA aptamer that has the ‘GGTTGGTGTGGTTGG’ sequence was also measured by AFM, and the resulting force average was 65.09 pN. The average affinity force was 205.52 pN, and the value was about three times higher than that of the conventional thrombin aptamer. The affinity force between anti-thrombin antibody and thrombin was then analyzed by AFM. The resulting average affinity force was 91.59 pN (Figure 4D and Table 2).
Figure 4.
Histogram of the affinity force between aptamer and thrombin for 4096 data points. (A) TBA-1, (B) TBA-2, (C) conventional thrombin aptamer and (D) anti-thrombin antibody.
Table 2.
The average of affinity force between obtained aptamer and thrombin
| Average of affinity force (pN) (mean ± SE) | Peak position (pN) | |
|---|---|---|
| N60 | 57.15 ± 0.96 | 10–19 |
| TBA-1 | 205.52 ± 0.70 | 210–219 |
| TBA-2 | 74.48 ± 0.90 | 40–49, 110–119 |
| Conventional thrombin aptamer | 65.09 ± 0.98 | 10–19 |
| Anti-thrombin antibody | 91.59 ± 1.09 | 50–59 |
Fluorescence polarization measurement
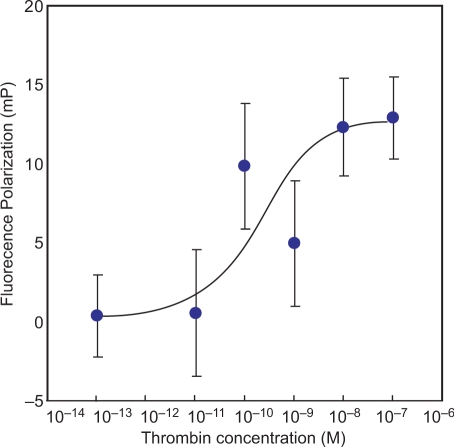
The fluorescence polarization measurement between TBA-1 and thrombin was carried out, and the recognition affinity obtained by AFM analysis was re-confirmed. The concentration of thrombin was changed from 10−13–10−7 M, and then polarization value was plotted against the concentration of thrombin (Figure 5). As a result, the fluorescence polarization value was increased with concentration dependency. On the other hand, polarization value of FITC-labeled TBA-1 against with 100 nM streptavidin was not increased significantly (Table 3).
Figure 5.
Binding curve of TBA-1 with thrombin. Each of thrombin solution (10−7–10−13 M) was mixed with TBA-1 (100 nM), and incubated for 2 h at room temperature. After incubation, the polarization was measured.
Table 3.
The fluorescence polarization value between TBA-1 and thrombin or streptavidin
| Polarization value (mP) | SE | |
|---|---|---|
| Thrombin | 12.9 | 2.60 |
| Avidin | 0.23 | 3.25 |
DISCUSSION
SELEX strategy using AFM
The selection of a DNA aptamer with AFM was performed using 60 random nucleotide sequences. After the first SELEX round, dsDNA was not amplified enough by 15 PCR cycles, since the amount of elution DNA was very small (data not shown). However, after the third SELEX round, the amount of dsDNA amplified by PCR was determined by PAGE to be large. Based on these results, selection by the AFM-SELEX strategy appeared to be very strong, and the ssDNA that binded strongly to thrombin was selected.
The selection cycle was repeated for three rounds. Figure 2 shows the histogram of adhesion force between ssDNA and thrombin. The initial ssDNA pool had a low adhesion force to thrombin, since the ssDNA had a low affinity for thrombin (Figure 2A). However, when the AFM-SELEX method was repeated for three rounds, the ssDNA had a strong affinity for thrombin (Figure 2D). Moreover, the third round elution pool did not have a large adhesion force to streptavidin, ∼67.14 pN (data not shown). This result shows that the third round elution pool bound specifically to thrombin. The affinity force between ssDNA and thrombin then gradually became stronger with repeated selection rounds (Figure 2E). Therefore, it is considered that ssDNA had an affinity for thrombin and was enriched by the repeating of the AFM-SELEX cycles.
In many articles on DNA aptamer, the selection cycle is repeated for more than eight rounds (8–12). However, in the selection method with AFM, the DNA aptamer with a high affinity for the target molecules was obtained by repeating only three rounds. Therefore, the AFM-SELEX strategy is a very useful method to rapidly obtain a DNA aptamer. In this new SELEX strategy using AFM, the oligonucleotide that could be bound to thrombin was only remained on gold chip. Therefore, compared with conventional SELEX strategy, it is considered that the non-specific bound of DNA was assumed to be reduced on gold chip surface, and the DNA aptamer that have high affinity to thrombin could be rapidly obtained.
The topography between round 3-elution pool and thrombin was analyzed (Figure 3B). In commonly, at the point of large roughness, the large force was occurred because of impact between cantilever and surface roughness. However, the high affinity forces, caused by molecular interaction between selected DNA aptamers and thrombin, were observed in every place, and this phenomenon was not related to the result of topology analysis (Figure 3A). Together these results, it was concluded that the specific affinity originated by molecular interaction was not related on surface roughness, and the specific adhesion force by affinity between round-3-elution pool and thrombin could be measured. Furthermore, the SELEX cycle number was suitable with three round since the affinity of TBA-1 to thrombin is significantly larger than that of anti-thrombin antibody at three rounds.
Sequence analysis
The third-round elution DNA that suggested the strongest binding to thrombin was subcloned to pT7 blue vector, and the sequence was determined. Many of the resultant obtained DNA sequences had a G-rich sequence (Table 1). Previously, thrombin aptamer possessing a G-rich sequence had been reported (14). According to this previous reported result, thus, it was assumed that the oligonucleotide sequence called TBA-1 could be functioned as a DNA aptamer against with thrombin.
Binding assay and determination of dissociation constant
The histogram of the affinity force between obtained aptamers called TBA-1 and -2 and thrombin was depicted from 4096 points of force curves (Figure 4A and B). The force histogram of TBA-1 had one peak at 210–219 pN (Figure 4A). These results suggest that there is a single binding site of TBA-1 to thrombin. On the other hand, the histogram of TBA-2 has three peaks at 40–49 and 110–119 pN (Figure 4B). The force value of the second peak is twice as large as that of the first peak. For these reasons, TBA-2 is considered to have two binding sites to thrombin.
The average affinity force between aptamer and thrombin was calculated from 4096 point force curve data (Table 2). The force average of TBA-1 was 203.52 pN, and the affinity of the N60 pool to thrombin was 57.14 pN. These results indicate that obtained ssDNAs have a high affinity to thrombin.
On the other hand, the conventional thrombin aptamers that have the ‘GGTTGGTGTGGTTGG’ sequence have the affinity force of 65.09 pN (Figure 4C and Table 2). Therefore, the affinity of obtained DNA aptamers to thrombin is stronger than that of the conventional thrombin aptamer. In previously report, the average of affinity force between conventional selected thrombin aptamer and thrombin is 4.45 pN. However, in this study, the affinity force was estimated to be 65.09 pN. It was considered that this difference was depended on the immobilization method and the DNA aptamer molecule number involved in immobilization. In addition, the affinity force between anti-thrombin antibody and thrombin is 91.59 pN (Figure 4D and Table 2). Therefore, the affinity of the TBA-1 aptamer to thrombin is stronger than that of antibody to thrombin.
For determination of dissociation constant (KD), the fluorescence polarization analysis was carried out. As a result, binding curve was increased with thrombin concentration dependency (Figure 5). However, in the case of streptavidin, the polarization value of FITC-labeled TBA-1 was not increased significantly (Table 3). Therefore, it is considered that the binding affinity of TBA to thrombin is specifically. By fluorescence polarization analysis, the KD value of TBA-1 against thrombin is estimated to be 200 pM (2 × 10−10 M). Although KD value of streptavidin–biotin interaction was 1 × 10−14 order, however, as a result, the binding affinity of TBA-1 to thrombin is 10 000-fold lower than that of streptavisdin–biotin interaction. Generally, free streptavidin exhibited a high affinity interaction described above. However, in the case of immobilized streptavidin, the KD value to biothin was decreased to 1 × 10−8 to 1 × 10−10 M order by conformation alternation (30). In AFM SELEX analysis, the streptavidin was covalently immobilized on the tip of surface of cantilever. Therefore, it was considered that KD value of streptavidin was decreased by immobilization, and as a result, the DNA aptamers with comparable lower affinity were selected.
As mentioned above, the KD value of isolated DNA aptamer TBA-1 was 1000 times lower than conventional aptamer having 200 nM of dissociation constant to thrombin (14). Moreover, this result was strongly supported to AFM analysis of TBA-1 affinity. Moreover, by AFM analysis, there was significant difference between the affinity of TBA-1 and the affinity of antibody to thrombin, and affinity force of TBA-1 was strongly compared with that of anti-thrombin antibody. Therefore, it was considered that the dissociation constant of TBA-1 to thrombin is <10−8 – 10−10 M, since the dissociation constant of antibody to antigen is ∼10−8 –10−10 M (23–27). However, there was no significant difference of dissociation constant between TBA-1 and antibody to thrombin. Therefore, it was considered that TBA-1 was optimized on AFM analysis.
In many reports, the dissociation constant of aptamers to their target molecules is 10 – 0.01 µM, and the aptamers that have low affinity are also selected (7–17). This is because the affinity force of obtained aptamers to their target cannot be controlled when using a conventional SELEX method. This AFM-SELEX strategy permit us to not only selecting quickly, but also obtaining high affinity DNA aptamer to the target as well as that of antibody to antigen. Therefore, this method may have a practical advantage for obtaining aptamers that have a strong affinity to targets.
Concluding remarks
In this study, a DNA aptamer that binds to thrombin with very high affinity and specificity was selected using AFM. The affinity force between ssDNA and thrombin grew gradually stronger upon repeating selection rounds. In addition, the sequences of obtained aptamers have many G-rich regions. One type of obtained aptamers, called TBA-1, has a strong affinity to thrombin compared with the conventional thrombin aptamer. This result suggests that DNA aptamers that bind to their targets with high affinity can be selected by AFM-SELEX. In addition, it suggests that DNA aptamers could be selected using fewer rounds, as compared with a conventional SELEX strategy. Considered together, these results indicate that this new SELEX strategy could be a viable candidate for the screening for various DNA aptamers with high affinities.
FUNDING
Funding for open access charge: Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan (No. 19021017 to C.O.), the foundation from New Energy and Industrial Technology Development Organization (NEDO) of Japan (No. 06B44019 to C.O.) and the Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe), MEXT, Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bass L, Cech R. Specific interaction between the self-splicing RNA of Tetrahymena and its guanosine substrate: implications for biological catalysis by RNA. Nature. 1984;308:820–826. doi: 10.1038/308820a0. [DOI] [PubMed] [Google Scholar]
- 2.Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 3.Osborne S, Ellington A. Nucleic acid selection and the challenge of combinatorial chemistry. Chem. Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- 4.Wilson DS, Szostak Z. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:647–661. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 5.Misono TS, Kumar KR. Selection of RNA aptamers against human influenza virus hemagglutinin using surface plasmon resonance. Anal. Biochem. 2005;342:312–317. doi: 10.1016/j.ab.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Davis JH, Szostak JW. Isolation of high-affinity GTP aptamers from partially structured RNA libraries. Proc. Natl Acad. Sci. USA. 2002;99:11616–11621. doi: 10.1073/pnas.182095699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 8.Huuizengen DE, Szostak JW. A DNA aptamer that’s binds adenosine and ATP. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 9.Miyachi Y, Shimizu N, Ogino C, Fukuda H, Kondo A. Selection of a DNA aptamer that binds 8-OHdG using GMP-agarose. Bioorg. Med. Chem. Lett. 2009;19:3619–3622. doi: 10.1016/j.bmcl.2009.04.130. [DOI] [PubMed] [Google Scholar]
- 10.Okazawa A, Maeda H, Fukusaki E, Katakura Y, Kobayashi A. In vitro selection of hematoporphyrin binding DNA aptamers. Bioorg. Med. Chem. Lett. 2000;10:2653–2656. doi: 10.1016/s0960-894x(00)00540-0. [DOI] [PubMed] [Google Scholar]
- 11.Niazi JH, Lee SJ, Kim YS, Gu MB. ssDNA aptamers that selectively bind oxytetracycline. Bioorg. Med. Chem. 2008;16:1254–1261. doi: 10.1016/j.bmc.2007.10.073. [DOI] [PubMed] [Google Scholar]
- 12.Mann D, Reinemann C, Stoltenburg R, Strehlitz B. In vitro selection of DNA aptamers binding ethanolamine. Biochem. Biophys. Res. Commun. 2005;338:1928–1934. doi: 10.1016/j.bbrc.2005.10.172. [DOI] [PubMed] [Google Scholar]
- 13.Kima YS, Jung HS, Matsuura T, Lee HY, Kawai T, Gua MB. Electrochemical detection of 17β-estradiol using DNA aptamer immobilized gold electrode chip. Biosens. Bioelectron. 2007;22:2525–2531. doi: 10.1016/j.bios.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Bock L, Griffin L, Latham J, Vernaas E, Toole J. Selection of single-strand DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 15.Hasegawa H, Sode K, Ikebukuro K. Selection of DNA aptamers against VEGF165 using a protein competitor and the aptamer blotting method. Biotechnol. Lett. 2008;30:829–834. doi: 10.1007/s10529-007-9629-6. [DOI] [PubMed] [Google Scholar]
- 16.Ogasawara D, Hasegawa H, Kaneko K, Sode K, Ikebukuro K. Screening of DNA aptamer against mouse prion protein by competitive selection. Prion. 2007;1:248–254. doi: 10.4161/pri.1.4.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang J, Yu T, Guo L, Xie J, Shao N, He Z. In vitro selection of DNA aptamer against abrin toxin and aptamer-based abrin direct detection. Biosens. Bioelectron. 2007;22:2456–2463. doi: 10.1016/j.bios.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 18.Guthrie JW, Hamula LA, Zhang H, Le XC. Assays for cytokines using aptamers. Methods. 2006;38:324–330. doi: 10.1016/j.ymeth.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Ikebukuro K, Yoshida W, Sode K. Aptameric enzyme subunit for homogeneous DNA sensing. Biotechnol. Lett. 2008;30:243–252. doi: 10.1007/s10529-007-9526-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bini A, Minunni M, Tombelli S, Centi S, Mascini M. Analytical performances of aptamer-based sensing for thrombin detection. Anal. Chem. 2007;79:3016–3019. doi: 10.1021/ac070096g. [DOI] [PubMed] [Google Scholar]
- 21.Basnar B, Elnathan R, Willner I. Following aptamer-thrombin binding by force measurements. Anal. Chem. 2006;78:3638–3642. doi: 10.1021/ac052289e. [DOI] [PubMed] [Google Scholar]
- 22.Lin L, Wang H, Liu Y, Yan H, Lindsay S. Recognition imaging with a DNA aptamer. Biophys. J. 2006;90:4236–4238. doi: 10.1529/biophysj.105.079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aixing D, Weiming T, Suping H, Wei L, Tiegui N, Zhaohu L, Baomin W, Qing X. Monoclonal antibody-based enzyme linked immunosorbent assay for the analysis of Jasmonates in plants. J. Integr. Plant Biol. 2008;50:1046–1052. doi: 10.1111/j.1744-7909.2008.00715.x. [DOI] [PubMed] [Google Scholar]
- 24.Larvor M, Ohaniance L, Nall B, Goldberg M. Measurement of the dissociation rate constant of antigen/antibody complexes in solution by enzyme-linked immunosorbent assay. J. Immunol. Methods. 1994;170:167–175. doi: 10.1016/0022-1759(94)90392-1. [DOI] [PubMed] [Google Scholar]
- 25.Dill K, Fraser C, Blomdahl J, Olson J. Determination of dissociation constant and concentration of an anti-DNA antibody by using the light-addressable potentiometric sensor. J. Biochem. Biophys. Methods. 1996;31:17–21. doi: 10.1016/0165-022x(95)00023-k. [DOI] [PubMed] [Google Scholar]
- 26.Krämer P, Gouzy M, Keß M, Kleinschmidt U, Kremmer E. Development and characterization of new rat monoclonal antibodies for procalcitonin. Anal. Bioanal. Chem. 2008;392:727–736. doi: 10.1007/s00216-008-2321-4. [DOI] [PubMed] [Google Scholar]
- 27.Hoylaerts M, Bollen A, De Broe M. The application of enzyme kinetics to the determination of dissociation constants for antigen-antibody interactions in solution. J. Immunol. Methods. 1990;126:253–261. doi: 10.1016/0022-1759(90)90158-r. [DOI] [PubMed] [Google Scholar]
- 28.Agnihotri A, Somana P, Siedlecki CA. AFM measurements of interactions between the platelet integrin receptor GPIIbIIIa and fibrinogen. Colloid Surf. B-Biointerfaces. 2009;71:138–147. doi: 10.1016/j.colsurfb.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 29.Yoshimura SH, Takahashi H, Otsuka S, Takeyasu K. Development of glutathione-coupled cantilever for the single-molecule force measurement by scanning force microscopy. FEBS Lett. 2006;580:3961–3965. doi: 10.1016/j.febslet.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 30.Huang SC, Stump MD, Weiss R, Caldwell KD. Binding of biothylated DNA to streptavidin-coated polystyrene latex: effects of chain length and particle size. Anal. Biochem. 1996;237:115–122. doi: 10.1006/abio.1996.0208. [DOI] [PubMed] [Google Scholar]