Abstract
The human Werner and Bloom syndromes (WS and BS) are caused by deficiencies in the WRN and BLM RecQ helicases, respectively. WRN, BLM and their Saccharomyces cerevisiae homologue Sgs1, are particularly active in vitro in unwinding G-quadruplex DNA (G4-DNA), a family of non-canonical nucleic acid structures formed by certain G-rich sequences. Recently, mRNA levels from loci containing potential G-quadruplex-forming sequences (PQS) were found to be preferentially altered in sgs1Δ mutants, suggesting that G4-DNA targeting by Sgs1 directly affects gene expression. Here, we extend these findings to human cells. Using microarrays to measure mRNAs obtained from human fibroblasts deficient for various RecQ family helicases, we observe significant associations between loci that are upregulated in WS or BS cells and loci that have PQS. No such PQS associations were observed for control expression datasets, however. Furthermore, upregulated genes in WS and BS showed no or dramatically reduced associations with sequences similar to PQS but that have considerably reduced potential to form intramolecular G4-DNA. These findings indicate that, like Sgs1, WRN and BLM can regulate transcription globally by targeting G4-DNA.
INTRODUCTION
In humans, deficiencies in the RecQ-family DNA helicases WRN or BLM cause Werner or Bloom syndromes, respectively. Werner syndrome (WS) is marked by premature features of aging, including graying and loss of hair, skin atrophy, cataracts, arteriosclerosis, osteoporosis and an increased incidence of certain cancers (1). Indeed, expression profiling of human fibroblasts has revealed that loss of WRN results in global transcriptional changes that resemble normal aging (2). Bloom syndrome (BS) is marked by an increased incidence of nearly all types of cancer, in addition to immune and developmental abnormalities (3). WRN and BLM function in multiple facets of DNA metabolism, including replication, repair, recombination and telomere maintenance (4,5). However, the understanding of how abrogation of WRN or BLM function is translated into the pathologies of WS or BS remains incomplete.
In yeast, genes that are repressed in sgs1Δ RecQ helicase mutants are significantly associated with transcription units containing sequences with the potential to form intramolecular G-quadruplexes (6). G-quadruplexes are a family of DNA (G4-DNA) or RNA (G4-RNA) structures that form when guanine residues from one or more nucleic acid strands form planar arrangements of four guanines (G-quartets) via Hoogsteen hydrogen bonding. These structures then stack to form G-quadruplexes, which can be highly stable under physiological temperature, pH and salt conditions, and which can differ from one another based on strand number and polarity, glycosidic bond angles, and loop sequence and topology (7). The distribution of potential intramolecular G-quadruplex-forming sequences (here referred to as PQS) in the human genome has recently been described (8–13). In addition, the formation of G-quadruplexes in vivo has been clearly demonstrated both at telomeres in S. lemnae and in human immunoglobulin class switch recombination regions that have been transcribed in Escherichia coli (14,15).
Yeast Sgs1 is highly active in binding and unwinding G4-DNA in vitro, with a clear preference for this substrate (16). Therefore, the preferentially altered expression of PQS-containing genes observed in sgs1Δ mutants suggests that Sgs1 modulates gene expression via direct effects on G4-DNA. WRN and BLM share with Sgs1 the propensity to unwind G4-DNA (16–18). All three of the proteins possess a conserved RQC domain, which binds G4-DNA with high affinity (Kd ∼ 5 nM) and thus confers G4-selectivity to the helicases (19).
To test the hypothesis that altered gene expression in WS and BS occurs preferentially at loci containing PQS, as observed for sgs1Δ yeast, we compared expression profiles of cultured normal human fibroblasts with those having mutations in WRN, BLM or RECQ4, the latter encoding a third human RecQ helicase homolog, mutation of which causes Rothmund–Thompson syndrome (RTS) (20,21). RECQ4 is an active DNA helicase (22), but lacks an RQC domain and therefore is not expected to target G4-DNA. In addition, if RECQ4 were to interact with G4-DNA, it would presumably do so in a fashion different from WRN and BLM. No gene expression studies have been described for RTS or BS. In the current study, we find that gene expression is considerably altered in WS and BS cells as compared with normal and RTS cells. Moreover, upregulated genes in WS and BS cells are significantly enriched for PQS. Importantly, there is no enrichment of PQS among genes with increased expression in RTS cells. These results represent the first demonstration that loss of WRN and BLM directly affects expression of PQS-containing loci genome-wide, and further suggest that this effect is directly mediated by G4-DNA in vivo.
MATERIALS AND METHODS
Cell culture conditions and media
Human fibroblast cell strains (WS: AG05229, AG12795, AG12797; BS: GM02932, GM03402, GM16891; RTS: AG18371, AG18375, AG05013; Normal/Wild-type: AG04054, AG06310, AG09975) were obtained from the Coriell Repository (Camden, NJ, USA), from donors matched for gender and of similar ages, and were at similar passage levels (Supplementary Table S1). Cells were cultured in MEM supplemented with Earle’s salts, 20% fetal bovine serum, 1x penicillin/streptomycin, and 1x fungizone in 3% O2 at 37°C and harvested for RNA extraction during active growth and at ∼80% confluence.
GeneChip microarray expression
Total RNA from the 12 fibroblast cell strains was isolated by extraction with TRIzol (Invitrogen) and purified using the RNeasy system (Qiagen). Total RNA was amplified by in vitro transcription using the Ovation RNA Amplification System V2 (NuGen). The resultant cDNA was fragmented and labeled using the FL-Ovation cDNA Biotin Module V2 (NuGen), and then purified using QIAquick columns (Qiagen), as specified by the Ovation System manual. Labeled probe was hybridized to Affymetrix U133A 2.0 GeneChips, and ultimately scanned using an Axon GenePix array scanner. The complete dataset is available on the Gene Expression Omnibus (GEO) database http://www.ncbi.nlm.nih.gov/geo/.
Statistical analysis of microarray expression experiment
The output files were normalized by Robust Multiarray Average (RMA), using the R package GCRMA (23) and gene expression levels were log2-transformed. The R/Bioconductor package limma (24) was applied to rank genes in order of evidence for differential expression of WS, BS, and RTS versus wild-type simultaneously using a P-value cut-off of 0.001. Hierarchical clustering (complete linkage/Euclidean distance) and Principle Component Analysis were applied to RMA-adjusted log2 expression levels to examine similarities between samples.
Identification of PQS and overlap with differentially expressed genes
Human genomic sequences (Release 18; without repeat-masking) were downloaded from the UCSC Genome Browser and scanned for PQS motifs, exactly as described (8). Specifically, intramolecular PQS was defined as a string matching the regular expression G3+N1–7G3+N1−7G3+N1−7G3+, where N1−7 represents a loop region of any nucleotide sequence of length 1–7. These motifs were then classified by the subgenic region in which their centers reside, according to the Release 18 RefSeq annotations: (i) 5′ flank (1 kb upstream of the transcription start), (ii) 5′ UTR, (iii) introns 1, 2 and 3, (iv) 3′ UTR and (v) 3′ flank (1 kb downstream of the transcript). When appropriate, motifs were categorized into multiple subgenic regions (e.g. in the case of overlapping transcripts). In addition, motifs were further categorized based on whether they are present on the sense or anti-sense strands. Overlapping motifs were considered to comprise a single motif, such that motifs describe tracts of at least 4 G-runs separated by loops of length no greater than 7.
For each subgenic region, the motif density was computed as the total number of motifs in the region divided by the size of the region in kilo base pairs. These densities were then averaged across all genes to give a mean density for that region type. The genomic density was computed as the number of motifs occurring in the genome divided by the genome size in kilo base pairs.
PQS was analyzed using the R/Bioconductor package GOstats (25) to determine the significance of their associations with differentially regulated probe sets, selected using a P-value cutoff of 0.01. Significance was determined by Fisher’s exact test, using a P-value cutoff of 10−4.
Overlap of pPQS control motifs with differentially expressed genes
In order to control for non G4-DNA-related sequence features such as GC-richness, we performed the above expression-correlation analysis with two other types of sequence motifs, neither of which by themselves are predicted to form G-quadruplexes. The first is the pattern G3+N1–7G3+N1–7G3+ with the constraint that an additional G3+ may not appear within eight nucleotides on either side; this represents a putative G-quadruplex-forming sequence with one G-run missing (pPQS). The second control pattern matches sequences that have an edit distance of up to two from PQS motifs. This corresponds to a sequence that differs from the putative G-quadruplex-forming pattern by having up to two nucleotide changes within the G-runs (pPQS-mut2). In particular, pPQS-mut2 conforms to the pattern G3+N1–7G3+N1;7G3+N1–7G3+, where at least one, and maximally two, G-runs must be GHG or where one G-run must be GHHG (where H is A, C or T).
RESULTS
Exploratory analysis of expression profiles of fibroblasts deficient for RecQ family proteins
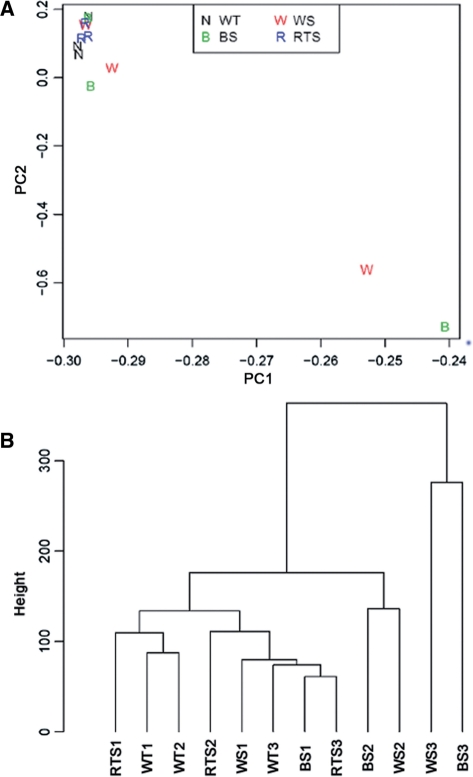
To examine differences in gene expression profiles among samples, we made use of principal component analysis (PCA) (Figure 1A) and hierarchical clustering (Figure 1B) (26) to analyze 12 expression datasets, including three samples each for normal, WS, BS and RTS (samples described in Supplementary Table S1). Specifically, PCA was performed by using an orthogonal linear transformation to simplify each complex expression dataset to two simple numerical values that could be graphically represented on a two-dimensional coordinate graph such that the greatest variance lies on the first axis and the next greatest variance on the second axis. Similarly, hierarchical clustering was achieved by partitioning the datasets into subsets (clusters) as a function of shared characteristics, here determined by Euclidean distance measures over all probe sets. We found that signal variations within the WS and BS groups were each larger than those within the normal and RTS samples. Furthermore, the normal and RTS samples clustered together with one WS and one BS sample, while the remaining four samples from the WS and BS groups lay relatively far apart by both PCA and clustering analyses. These findings indicate that (i) the expression profiles of cells from normal and RTS individuals are both similar to one another and relatively reproducible, whereas (ii) loss of WRN or BLM helicase function can produce more gene expression changes as compared with loss of RECQ4.
Figure 1.
Multivariate analyses of gene expression data. (A) Principle component analyses. Two-dimensional visualization of samples by the first two principal components (PC1 and PC2). (B) Average linkage clustering of samples by Euclidean distance. The vertical axis (Height) indicates the similarity between the clusters.
Differentially expressed genes in WS, BS and RTS cells
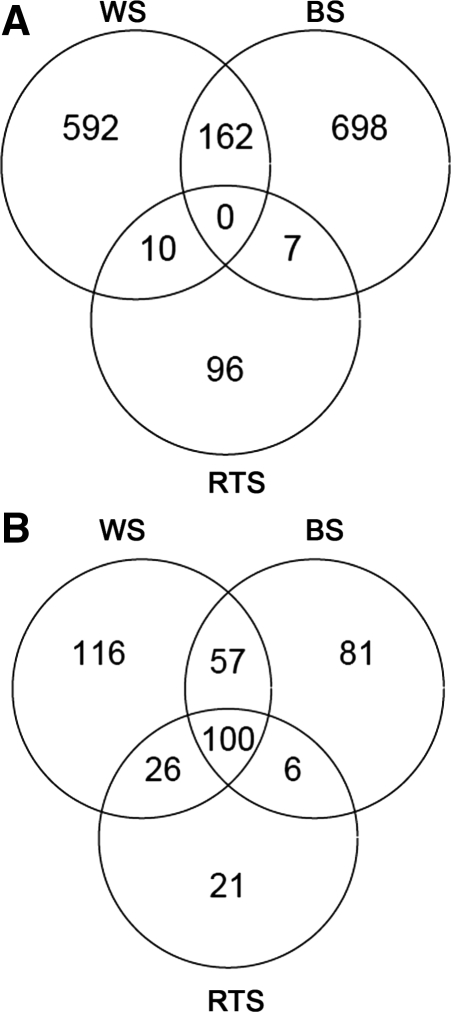
To further assess gene expression changes, we determined the number of gene probes differentially expressed in each of the three mutant groups (WS, BS and RTS) as compared with normal cells (Figure 2). These analyses revealed that considerably more gene probes were differentially expressed in the WS (up, n = 764; down, n = 299; total, n = 1063) or BS (up, n = 867; down, n = 244; total, n = 1111) than in the RTS cells (up, n = 113; down, n = 153; total, n = 264). In addition, more probe sets were upregulated than downregulated in both the WS and BS cells (72% and 75% upregulated, respectively), while this bias was not observed in RTS cells (42% upregulated). Furthermore, many differentially expressed probe sets were specific to WS only (n = 689) or BS only (n = 747). Taken together, these results suggest that, with respect to changes in gene expression in fibroblasts, deficiencies of WRN, BLM and RTS might have biological effects that are both complex and distinct from one another. For example, genes commonly affected in all three syndromes are downregulated, though those specifically dysregulated in WS and/or BS, but not RTS, tend to be upregulated. Based on the lack of an RQC domain in the RECQ4 helicase, a potential explanation for the different biological effects of these protein deficiencies on gene expression is that loss of WRN or BLM function causes upregulation of gene expression through dysregulation of G4-DNA, while a G4-DNA-independent mechanism that is altered similarly in all three diseases leads to the downregulation of certain genes.
Figure 2.
Differentially expressed gene probes in RecQ helicase-deficient fibroblast cell strains. (A) upregulated genes and (B) downregulated genes. Indicated values are subsets of a total 22277 gene probes.
Gene expression and potential intramolecular G-quadruplex forming sequences (PQS)
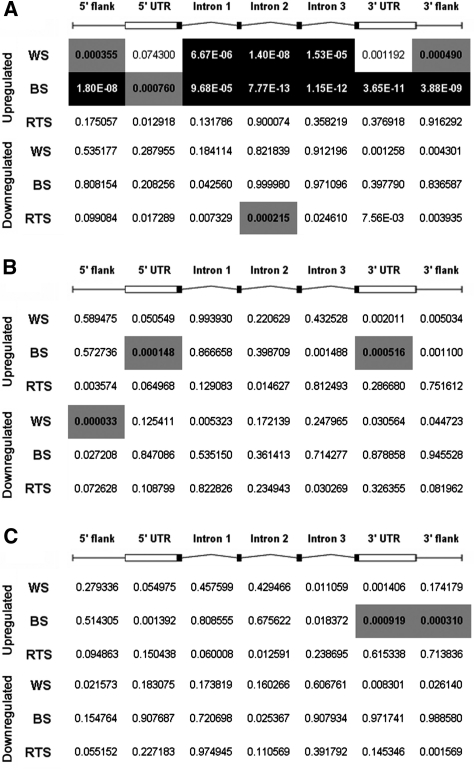
To test whether the upregulation of genes observed in WS or BS cells is related to PQS, a data set of human genes containing PQS within different subgenic regions was compared with the sets of differentially expressed genes (Figure 3A). The likelihood that the association between PQS and altered gene expression is due to random chance is indicated by a P-value based on the Fisher’s exact test. Intramolecular PQS was defined as G3+N1-7G3+N1-7G3+N1-7G3+, exactly as described (8). PQS predictions were performed for sequences on either the sense or anti-sense strands and within individual subgenic regions, including 1 kb regions upstream and downstream from transcription units, 5′ and 3′ UTRs, and introns. The 1 kb upstream region, here referred to as ‘5′ flank’, corresponds to the region of high PQS density previously identified within the promoters of human genes (9). PQS density differs according to subgenic region (8–10,27) (Supplementary Table S3). However, we chose to examine multiple regions because PQS-related regulation by WRN or BLM need not correlate with regions having the highest PQS density. Indeed, it was reported recently that upon expression of a selective G4-DNA-binding single chain antibody in human cells, there is preferential regulation of loci possessing PQS near their transcription end sites (TESs), even though PQS density around TESs is generally low (28). Remarkably, we found that genes that are upregulated in WS and BS cells correlate well with genes that are predicted to form intramolecular G-quadruplexes (P < 10−4 for the majority of subgenic regions). No such association was observed for downregulated genes in WS or BS cells. Further, there was no significant enrichment of PQS in or near genes that are upregulated in RTS samples, although there was a weak association with the second intronic region for downregulated genes. These results are consistent with the fact that the WRN and BLM, but not RECQ4, helicases have G-quadruplex-binding RQC domains. In addition, PQS was compared with altered gene expression in cultured fibroblasts from individuals with a genetic disorder unrelated to WS and BS, Setleis syndrome (GEO ID number GSE16524) (29). As expected, there was no association between PQS in the majority of subgenic regions and gene expression changes in Setleis syndrome, and only weak associations for downregulated genes and PQS in the third intron and 3′ UTR (P ∼ 10−4; data not shown).
Figure 3.
Combined analysis of overlap significance in the indicated subgenic locations between differentially expressed genes and genes with PQS or PQS-related motifs having much lower G4-DNA forming potential (pPQS or pPQS-mut2). Values are given for associations with PQS (A), pPQS (B) and pPQS-mut2 (C). Significance is computed by one-sided Fisher’s test. Categorized by region, P-values are colored to highlight significant overlaps: white text within a black background indicates a P-value lower than 10−4. Black text within a gray background indicates a P-value of intermediate significance, between 10−4 and 10−3.
To control for the possibility that the association of upregulated gene expression in WS and BS fibroblasts might simply be a function of a sequence feature associated with PQS, such as GC-richness, and not intramolecular G4-DNA forming potential per se, we performed the above expression-correlation analysis with two other types of sequence motifs, neither of which are predicted to form stable intramolecular G-quadruplexes. First, we tested for associations with loci having only partial PQS (pPQS) motifs conforming to the pattern G3+N1−7G3+N1−7G3+ and lacking a fourth G3+ sequence within eight nucleotides of the pattern. Thus, pPQS are highly similar to PQS, but are less likely to form intramolecular G4-DNA. Gene expression was compared with pPQS and while most pPQS-containing regions showed no associations with upregulated loci, pPQS within the 5′ and 3′ UTRs was weakly associated with upregulated genes in BS (Figure 3B). However, we note that, with respect to the 3′ UTR, the association of BS upregulated genes with bona fide G-quadruplex-forming sequences (PQS) was much more significant than the association with pPQS control sequences (10−11 versus 10−4). Gene expression was also compared with a second control data set comprising genes containing sequences that differ from the PQS pattern by up to two nucleotides (pPQS-mut2) (Figure 3C). Similar to the results for pPQS, the majority of gene regions containing pPQS-mut2 showed no significant associations with upregulated loci in WS or BS cells, while weak associations were observed for upregulated genes in BS and control sequences in the 3′ UTR and 3′ flank. As above, actual PQS sequences in these regions show much more significant associations with BS upregulated loci as compared with pPQS-mut2 control sequences (3′ UTR, 10−11 versus 10−4; 3′ flank, 10−9 versus 10−4). As an additional control, we also compared differentially expressed genes in WS and BS with genes containing simple tetranucleotide repeat sequences that are predicted not to form stable G-quadruplexes ([AAAB]5 or 9, [AAGG]5 or 9, [AGAT]5 or 9 and [ATCC]5 or 9) (30). As expected, no significant associations were found (all P-values > 10−3; data not shown).
Previous analyses of PQS in the human genome have revealed strand asymmetries within various subgenic locations (8,10,12,14). We therefore investigated whether the PQS associated with differentially expressed genes identified in the current study demonstrate strand bias. Towards this end, we classified PQS motifs according to strand, as well as subgenic region, and assessed strand-specific PQS associations with altered gene expression using the Fisher’s exact test. Interestingly, the PQS associations are similar for the anti-sense and sense strands (Supplementary Table S2). This observed lack of strand bias is also true for similar analyses involving pPQS and pPQS-mut2 control sequences (Supplementary Tables S4 and S5, respectively).
The similar patterns of altered gene expression in WS and BS cells raised the question of how many differentially expressed genes are shared by these two syndromes. When all genes are considered, whether they contain PQS or not, ∼20% of all upregulated genes in WS and BS are shared (Supplementary Table S6). Interestingly, when the analysis is restricted to targets with PQS, the overlap improves for both the 3′ flank (WS) and 3′ UTR (WS and BS). These data, therefore, provide further evidence of the relationship between G4-DNA and altered gene expression in WS and BS cells.
DISCUSSION
We have found highly significant associations between genes that have intramolecular PQS and those from which transcripts accumulate in primary cultured fibroblasts from individuals with WS or BS. Further, the current study is the first report of PQS-associated changes in global gene expression in the context of human genetic disease. The preferential upregulation of gene expression at loci with PQS in WS and BS cells, together with the preference of the WRN and BLM helicases to unwind G4-DNA in vitro, argues that the observed gene expression changes involve G4-DNA. Furthermore, there was no such preferential regulation of PQS loci in RTS cells, which have defects in the RECQ4 helicase. Because RECQ4 lacks the RQC domain that enables WRN and BLM to bind G4-DNA, there is no reason to expect RECQ4 to have a preference for G4-DNA substrates, consistent with our findings and supporting the specificity of the PQS association in WS and BS. Although RQC domains mediate the interaction of RecQ-family helicases with G4-DNA, they are not generally required for all helicases to unwind G-quadruplexes. For example, the FANCJ helicase unwinds G4-DNA but does not possess an RQC domain (31). So, while it is expected that RECQ4, as a RecQ helicase, would require an RQC domain for interaction with G4-DNA, we cannot completely rule out a role for the protein at these structures. Nevertheless, if RECQ4 were to interact with G4-DNA, it would do so in a RQC-independent manner, suggesting that it might recognize different G4 targets than WRN or BLM, or interact in a different fashion with the same targets. Additional studies will be required to determine why RTS cells lack significant PQS-associated changes in gene expression, but the simplest prediction is that RECQ4 lacks the same selectivity for G4-DNA as WRN and BLM. Concerning the association of gene expression changes and PQS in WS and BS cells, an alternative to the proposed G4-DNA-dependent mechanism is that these syndromes are associated with the altered activity of transcription factors that bind duplex DNA targets that, when present in tandem arrays, happen to conform to the PQS pattern. Such an explanation, however, is inconsistent with the lack of correlation between gene expression changes and the partial PQS patterns, both of which (pPQS and pPQS-mut2) would still be predicted to bind ∼75% as many copies of such transcription factors (as only one of four G-runs are ablated in the partial PQS sequences). In addition, there are also no known transcription factors that both bind G-rich sequences and have altered activity in WS or BS.
Using the same or similar algorithms to the one we employed to identify PQS, previous studies have found enrichment of PQS in the upstream promoter regions, 5′ UTRs and first introns of genes (9,10,12). Our findings are consistent with these results, but genes differentially expressed in WS and BS cells do not show greater enrichment within these subgenic locations, as compared with others. A possible explanation for this observation is that WRN and BLM target only genes with particular kinds of G4-DNA structures (e.g. with particular loop sequences or topologies), which are then dysregulated in their absence. It is also possible that only a subset of the G4 structures targeted by WRN and BLM affect gene expression and the distribution of such structures is not identical to the distribution of PQS overall. Such a case would be similar to the situation in yeast, where the most PQS lies upstream of promoters, but the strongest association with altered gene expression in sgs1Δ mutants is with PQS within the transcription unit (6). In particular, we find that genes upregulated in WS and BS cells are enriched for PQS in essentially all subgenic regions, including upstream promoter regions, introns and downstream regions flanking the TES (transcription end site) (Figure 3A and Supplementary Table S2). While it is unclear how modulation of G4 structures in these particular regions results in differential gene expression, we speculate that G4-DNA in different subgenic regions can have different mechanistic effects on transcriptional regulation. More specifically, it is possible that G4-DNA in intronic regions and regions proximal to the TES can affect the efficiency of transcriptional elongation and/or termination, whereas similar structures within promoter-proximal regions affect transcriptional initiation (9,10). As noted earlier, the intersection of differentially expressed genes in WS and BS shows enrichment for genes with PQS in the 3′ flank and 3′ UTR (Supplementary Table S6), suggesting that genes with G4-DNA near the TES are regulated by a similar mechanism in both WS and BS cells.
We further find that while there is some overlap between genes that are dysregulated in WS and BS, the majority of differentially expressed genes are unique to one or the other syndrome. For PQS loci, this difference might be explained by the WRN and BLM helicases acting on different types of G4-DNA structures located in different genes, or a differential activity at the same structures. An intriguing possibility is that such differences might contribute to the unique pathologies of these disorders, but additional studies will be required to test this speculation.
Our findings also indicate that the PQS-related mechanism likely operates via transcriptional regulation, rather than by altered mRNA stability, as PQS on sense and anti-sense strands have similar associations with upregulated mRNA levels in WS and BS cells. Furthermore, WRN and BLM, despite being able to unwind RNA–DNA and DNA–DNA duplexes, have no known RNA–RNA unwinding activity (32,33). As a result, these helicases are unlikely to process G4-RNA, indicating that the simplest mechanism by which gene expression changes are engendered in WS and BS cells is through modulation of G4-DNA in genes, themselves, and not altered stability of the resulting transcripts. However, as our studies only examined mRNA abundance, they do not rule out the possibility of additional effects of PQS on RNA processing or translation in WS and BS.
We found previously that Saccharomyces cerevisiae cells lacking the Sgs1 RecQ helicase, which possesses an RQC domain and actively unwinds G4-DNA, also have quadruplex-related changes in gene expression. Although our new findings extend the association from yeast to human cells, there are important differences between the two sets of findings. sgs1Δ mutants downregulate expression of loci having quadruplex-forming potential within their transcription units, whereas WS and BS cells upregulate loci with PQS, and particularly in the case of BS, this association extends to several subgenic regions including those upstream and downstream of transcription units. There are several possible explanations for these differences. One is that transcriptional regulatory factors that bind or are excluded by G4-DNA could vary between yeast and human cells, and thus the regulatory consequences of G4-DNA formation would depend upon the particular milieu of such factors. Another is that instead of global differences in regulatory factors, the local contexts in which G4-DNA can form might vary between organisms. For example, G-quadruplex forming sequences targeted by WRN and BLM in humans might tend to lie within regions where repressors (e.g. nucleosomes) bind duplex DNA and are excluded by G4-DNA formation leading to increased transcription, whereas those targeted by Sgs1 in yeast might instead reside at locations that impede transcription upon G4-DNA formation. A third possibility is that the spectrum of particular G4-DNA structures formed might also vary between organisms, perhaps also distinguishing how loci with quadruplex-forming potential could be differentially regulated in yeast and man. Additional studies will be required to determine if the repression of gene expression in sgs1Δ mutants versus the activation of gene expression in WS and BS cells reflects different regulatory functions of G4-DNA or different effects of the yeast and human RecQ proteins.
Previously published studies have found sequences with the capacity to form G4-DNA in several human cancer-related genes (hif1-α, ret, vegf, k-ras, c-kit, c-myc, bcl2, pdgfa) (34–44). In our datasets, the majority of these factors are not differentially expressed upon loss of WRN or BLM helicase activity (not shown). However, expression of vegf, c-kit, c-myc and pdgfa was found to be downregulated in WS and BS cells (data not shown), consistent with previous reports that stabilization of G4 structures in these genes results in inhibition of their expression (34,38,40–44), and notable given that G4-DNA recognized by WRN or BLM in wild-type cells is likely stabilized in WS and BS cells. That expression of the majority of the indicated genes is unchanged in WS and BS cells suggests that not all G4-DNA elements are affected by WRN or BLM. In addition, while both our data and previous studies suggest that vegf, c-kit, c-myc and pdgfa transcription is inhibited by stabilization of G4-DNA, the majority of PQS-associated expression changes in WS and BS cells comprise upregulation events, indicating that the G4-DNA-dependent inhibition of gene expression observed for the specific examples listed above is not, in this case, representative of the global trend following gross stabilization of G4-DNA (i.e. upregulation). We also note that previous findings support both upregulation and downregulation of gene expression via G4-DNA related mechanisms. For example, in human cells following treatment with the G4-binding ligand TmPyP4 or expression of a single-chain antibody specific for G4-DNA, expression from PQS loci was altered preferentially, with some loci upregulated and others downregulated (28,45).
As mentioned above, owing to the fact that the WRN and BLM helicases have been shown to bind and unwind G-quadruplexes, it is likely that G4-DNA levels are increased in their absence. Although we consider it less likely, we note that if their helicase activities are inhibited by other factors (e.g. p53) (46), it is possible that WRN and BLM might instead promote G4-DNA formation and thus levels could diminish in their absence. Altered levels of G4-DNA might affect transcription directly via changes in chromatin. For example, in yeast and human cells, reduced levels of nucleosome occupancy are associated with PQS (6,47,48), and thus higher levels of G4-DNA could be associated with elevated levels of transcription in WS or BS cells. Alternatively, increased G4-DNA levels in WS or BS cells might result in the creation binding sites for transcriptional activators that bind G4-DNA (49–52) or the destruction of binding sites for repressors that bind duplex DNA. Yet another possibility is that the lack of binding of WRN or BLM to G4-DNA simply relieves occlusion of transcriptional activators, without a concomitant change in G4-DNA levels. Future studies will be aimed at discriminating between these possibilities. Regardless of the specific mechanism being employed, however, it is clear that the transcriptional program is altered in cells lacking WRN and BLM and that this alteration is associated, in part, with DNA sequences having intramolecular G-quadruplex forming potential.
Beyond any significance our findings might have for the pathogenesis of WS and BS, they provide important data supporting previous observations indicating roles for G4-DNA in transcriptional regulation in human cells. Earlier studies have relied on mutagenesis of PQS elements to indicate the importance of the element in regulation or on the activity of small molecule ligands that bind and stabilize G4-DNA selectively to impact regulation via PQS elements (28,34,35,40,42,45,53). In the first set of cases, it is difficult to completely exclude the possibility that mutagenesis not only perturbs G4-DNA forming potential but also the association of canonical duplex-DNA binding transcription factors, whereas in the second set it is possible that the ligands induce G4-DNA that would not otherwise form naturally, thus altering expression via perturbation of natural mechanisms. In the case of PQS-associated upregulation of gene expression in WS and BS, the changes reflect the loss of G4-DNA processing helicases, thus providing support for the idea that G4-DNA does function in natural gene regulation, at least in the setting of disease. Furthermore, because our analyses were performed genome-wide, they allowed for the repeated testing of the association for each of the thousands of PQS-containing genes, thus providing statistical support of the connection between PQS and altered gene expression in WS and BS. It will be a challenge for future studies to dissect the precise mechanisms underlying the associations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Pennsylvania Institute on Aging Pilot Award (to L.-S.W. and F.B.J.); and P01-AG031862 and 1R01AG021521 (to F.B.J.), including a supplement to J.E.J. (National Institute on Aging). Funding for open access charge: Department of Pathology and Laboratory Medicine, University of Pennsylvania.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the members of the Johnson and Wang labs for helpful discussions, Don Baldwin and the Penn Microarray Facility for advice and performing the microarray studies.
REFERENCES
- 1.Martin GM. Genetic syndromes in man with potential relevance to the pathobiology of aging. Birth Defects Orig. Artic. Ser. 1978;14:5–39. [PubMed] [Google Scholar]
- 2.Kyng KJ, May A, Kolvraa S, Bohr VA. Gene expression profiling in Werner syndrome closely resembles that of normal aging. Proc. Natl Acad. Sci. USA. 2003;100:12259–12264. doi: 10.1073/pnas.2130723100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis NA, German J. Molecular genetics of Bloom's; syndrome. Hum. Mol. Genet. 1996;5(Spec No):1457–1463. doi: 10.1093/hmg/5.supplement_1.1457. [DOI] [PubMed] [Google Scholar]
- 4.Brosh RM, Jr, Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007;35:7527–7544. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell. Mol. Life Sci. 2007;64:2306–2322. doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hershman SG, Chen Q, Lee JY, Kozak ML, Yue P, Wang LS, Johnson FB. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shirude PS, Okumus B, Ying L, Ha T, Balasubramanian S. Single-molecule conformational analysis of G-quadruplex formation in the promoter DNA duplex of the proto-oncogene c-kit. J. Am. Chem. Soc. 2007;129:7484–7485. doi: 10.1021/ja070497d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 18.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom's; and Werner's; syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber MD, Duquette ML, Shiels JC, Maizels N. A conserved G4 DNA binding domain in RecQ family helicases. J. Mol. Biol. 2006;358:1071–1080. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 20.Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 21.Vennos EM, James WD. Rothmund–Thomson syndrome. Dermatol. Clin. 1995;13:143–150. [PubMed] [Google Scholar]
- 22.Xu X, Liu Y. Dual DNA unwinding activities of the Rothmund–Thomson syndrome protein, RECQ4. EMBO J. 2009;28:568–577. doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909. [Google Scholar]
- 24.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 25.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 26.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd edn. New York: Springer; 2009. [Google Scholar]
- 27.Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 28.Fernando H, Sewitz S, Darot J, Tavare S, Huppert JL, Balasubramanian S. Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGaughran J, Aftimos S. Setleis syndrome: three new cases and a review of the literature. Am. J. Med. Genet. 2002;111:376–380. doi: 10.1002/ajmg.10632. [DOI] [PubMed] [Google Scholar]
- 30.Bacolla A, Larson JE, Collins JR, Li J, Milosavljevic A, Stenson PD, Cooper DN, Wells RD. Abundance and length of simple repeats in vertebrate genomes are determined by their structural properties. Genome Res. 2008;18:1545–1553. doi: 10.1101/gr.078303.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, Hickson ID, Vindigni A. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J. Biol. Chem. 2008;283:17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki N, Shimamoto A, Imamura O, Kuromitsu J, Kitao S, Goto M, Furuichi Y. DNA helicase activity in Werner's; syndrome gene product synthesized in a baculovirus system. Nucleic Acids Res. 1997;25:2973–2978. doi: 10.1093/nar/25.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bejugam M, Sewitz S, Shirude PS, Rodriguez R, Shahid R, Balasubramanian S. Trisubstituted isoalloxazines as a new class of G-quadruplex binding ligands: small molecule regulation of c-kit oncogene expression. J. Am. Chem. Soc. 2007;129:12926–12927. doi: 10.1021/ja075881p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai J, Chen D, Jones RA, Hurley LH, Yang D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 38.Grand CL, Han H, Munoz RM, Weitman S, Von Hoff DD, Hurley LH, Bearss DJ. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol. Cancer Ther. 2002;1:565–573. [PubMed] [Google Scholar]
- 39.Guo K, Pourpak A, Beetz-Rogers K, Gokhale V, Sun D, Hurley LH. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 2007;129:10220–10228. doi: 10.1021/ja072185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun D, Liu WJ, Guo K, Rusche JJ, Ebbinghaus S, Gokhale V, Hurley LH. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol. Cancer Ther. 2008;7:880–889. doi: 10.1158/1535-7163.MCT-07-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verma A, Yadav VK, Basundra R, Kumar A, Chowdhury S. Evidence of genome-wide G4 DNA-mediated gene expression in human cancer cells. Nucleic Acids Res. 2009;37:4194–4204. doi: 10.1093/nar/gkn1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Q, Zhang R, Wang XW, Spillare EA, Linke SP, Subramanian D, Griffith JD, Li JL, Hickson ID, Shen JC, et al. The processing of Holliday junctions by BLM and WRN helicases is regulated by p53. J. Biol. Chem. 2002;277:31980–31987. doi: 10.1074/jbc.M204111200. [DOI] [PubMed] [Google Scholar]
- 47.Wong HM, Huppert JL. Stable G-quadruplexes are found outside nucleosome-bound regions. Mol. Biosyst. 2009 doi: 10.1039/b905848f. 10.1039/b905848f. [DOI] [PubMed] [Google Scholar]
- 48.Halder K, Halder R, Chowdhury S. Genome-wide analysis predicts DNA structural motifs as nucleosome exclusion signals. Mol. Biosyst. 2009 doi: 10.1039/b905132e. 10.1039/b905132e. [DOI] [PubMed] [Google Scholar]
- 49.Etzioni S, Yafe A, Khateb S, Weisman-Shomer P, Bengal E, Fry M. Homodimeric MyoD preferentially binds tetraplex structures of regulatory sequences of muscle-specific genes. J. Biol. Chem. 2005;280:26805–26812. doi: 10.1074/jbc.M500820200. [DOI] [PubMed] [Google Scholar]
- 50.Palumbo SL, Memmott RM, Uribe DJ, Krotova-Khan Y, Hurley LH, Ebbinghaus SW. A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 2008;36:1755–1769. doi: 10.1093/nar/gkm1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yafe A, Shklover J, Weisman-Shomer P, Bengal E, Fry M. Differential binding of quadruplex structures of muscle-specific genes regulatory sequences by MyoD, MRF4 and myogenin. Nucleic Acids Res. 2008;36:3916–3925. doi: 10.1093/nar/gkn340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shklover J, Etzioni S, Weisman-Shomer P, Yafe A, Bengal E, Fry M. MyoD uses overlapping but distinct elements to bind E-box and tetraplex structures of regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2007;35:7087–7095. doi: 10.1093/nar/gkm746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simonsson T, Henriksson M. c-myc suppression in Burkitt's; lymphoma cells. Biochem. Biophys. Res. Commun. 2002;290:11–15. doi: 10.1006/bbrc.2001.6096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.