Abstract
We have determined diversities exceeding 1012 different sequences in an annealing and melting assay using synthetic randomized oligonucleotides as a standard. For such high diversities, the annealing kinetics differ from those observed for low diversities, favouring the remelting curve after annealing as the best indicator of complexity. Direct comparisons of nucleic acid pools obtained from an aptamer selection demonstrate that even highly complex populations can be evaluated by using DiStRO, without the need of complicated calculations.
INTRODUCTION
Recent technological developments have employed nucleic acid libraries of staggering diversity. In contrast to arrayed libraries, random combinatorial libraries allow simple and inexpensive handling of greater than a billion different sequences in less than a millilitre. Nowadays the most diverse libraries are used in Systematic Evolution of Ligands by EXponential enrichment (SELEX) experiments (1,2) where highly variable initial nucleic acid pools of up to 1015 individual sequences are employed (3).
Despite the progress made towards the sequencing of up to 107 clones at a time by second generation techniques (4), to date, no method allows the reliable assessment of diversity of larger nucleic acid libraries.
A simple assay to estimate the complexity of DNA has been described earlier (5). The so-called C0t analysis is usually performed to measure the size of genomes by observing the absorbance at 260 nm during annealing of complementary DNA fragments of variable lengths. This assay has been adjusted to be applicable to PCR fragments generated from SELEX experiments using the same principles while reducing ambiguities due to fragment length and primer sequences (6).
A further adjustment to modern real-time PCR equipment, termed AmpliCot, and its application to the measurement of T-cell repertoire diversity has been introduced more recently (7). AmpliCot makes use of the double stranded DNA (dsDNA) binding dye SYBR green to measure the annealing kinetics of even small quantities of DNA. In addition to the C0t1/2 value (5) commonly used as an indicator of DNA complexity, a linear correlation of the inverse slopes of the kinetic plots and template diversity was described. AmpliCot was evaluated by mixtures of 96 individual clones and the results were used to assign diversities of up to 106 by extrapolation. However, apart from the use of such small pools of individual sequences no attempt has been made to validate calculated diversities by calibration of the diversity assays.
To monitor the course of nucleic acid library selections and predict a positive outcome, it is important to reliably determine the quality of libraries and the dynamics of DNA populations during the selection experiments. We sought to develop a general and reliable DNA standard that can be used for the calibration of diversities covering the broad range from single clones up to 1012 unique molecules.
MATERIALS AND METHODS
Preparation of dsDNA standard
All randomized synthetic oligonucleotides were synthesized by Purimex RNA/DNA Oligonucleotides (Grebenstein, Germany), using standard phosphoamidite chemistry with equally pre-mixed bases at random positions, and were purified via HPLC. Diversity standard oligonucleotide templates were converted to dsDNA by PCR in PTC200-cycler (Biozym Scientific, Hess. Oldendorf, Germany). Briefly, 100 µl reactions with 2 µM primers Ri5 and PO4Ri3 (obtained from Operon, Cologne, Germany with 20 pmol standard oligonucleotide template, 1.5 mM MgCl2, (Promega, USA), 5 U GoTaq®-polymerase (Promega) using four cycles and an annealing temperature of 55°C, yielding an 80 bp PCR product. Residual single-stranded DNA and primers were removed by S1-nuclease (Roboklon, Berlin, Germany) treatment for 15 min at 55°C in S1-buffer (Roboklon). Resulting dsDNA PCR-products were purified by binding to sbeadex® magnetic particles (AGOWA, Berlin, Germany) according to manufacturers’ protocol. DNA yield was determined spectrophotometrically (Nanodrop).
Fluorescent diversity assay
The assay was performed using the SYBR Green PCR Core Kit (Applied Biosystems, USA) in 96 well plates with a Biorad Lightcycler device. Each well contained 1.5 µg of dsDNA sample diluted in 50µl buffer. Reannealing measurements were initiated by denaturation for 2 min at 95°C, and subsequent annealing for 180 min at the respective constant annealing temperature taking one measurement per minute. Remelting was performed starting at 20°C with incremental steps of 0.5°C for 7 s until the final temperature of 98°C was reached.
RESULTS
Design of the diversity standard
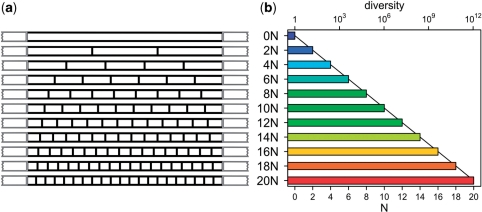
Chemical synthesis of oligonucleotides allows the incorporation of random nucleotides at predefined positions. One randomized nucleotide N yields four different sequences and any further randomized nucleotide adds to the diversity exponentially with D = 4N, where N is the number of randomized nucleotides. An even distribution of randomized bases was chosen over a cluster, in order to mimic naturally occurring situations such as somatic hypermutation in the hypervariable regions of T-cell receptors. We thought that the stringency for small sequence differences is stronger in such an arrangement than in consecutively randomized areas. By this method we designed several oligonucleotide libraries with varying numbers of random nucleotides denoted 2–20 N, based on a single template sequence flanked by conserved primers (Figure 1 and Table 1). The synthetic oligonucleotides were converted to dsDNA by four cycles of PCR and treated by S1 nuclease to remove any unpaired ssDNA. We confirmed the introduction of all four nucleotides at the randomized positions by batch sequencing the oligonucleotides with the conventional Sanger method. Other randomized oligonucleotides obtained from the same supplier, synthesized by the same method, have been subjected to next generation sequencing. Those displayed less than 3.2% deviation from ideal 25% distribution for any base in more than 700 000 sequenced bases in randomized regions (data not shown).
Figure 1.
(a) Schematic diagram of diversity standard sequences. Variable positions marked as vertical black bars, flanking primer regions as grey open areas. (b) Variability and resulting diversity of standard DNA fragments.
Table 1.
List of all oligonucleotide sequences used in this study
| Name | Application | Sequence |
|---|---|---|
| Ri5 | Primer | 5′-GGGAATTCGAGCTCGGTACC |
| PO4Ri3 | Primer | PO4-5′-CCAAGCTTGCATGCCTGCAG |
| Div0N | Diversity-Standard | 5′-TTCGAGCTCGGTACCGGTAATGT |
| GTAAATCTACTTCCTTCTCAGCGCCCCCGTG | ||
| GCTGCAGGCATGCAAG | ||
| Div2N | Diversity-Standard | 5′-TTCGAGCTCGGTACCGGTAATGT |
| GTAAANCTACTTCCTTCTCAGNGCCCCCGTG | ||
| GCTGCAGGCATGCAAG | ||
| Div4N | Diversity-Standard | 5′-TTCGAGCTCGGTACCGGTAATGN |
| GTAAATCTNCTTCCTTCNCAGCGCCCNCGTG | ||
| GCTGCAGGCATGCAAG | ||
| Div6N | Diversity-Standard | 5′-TTCGAGCTCGGTACCGGNAATGT |
| GNAAATCTNCTTCCTNCTCAGCNCCCCCGNG | ||
| GCTGCAGGCATGCAAG | ||
| Div8N | Diversity-Standard | 5′-TTCGAGCTCGGTACCGGNAATGN |
| GTAANTCTANTTCCNTCTCNGCGCNCCCGNG | ||
| GCTGCAGGCATGCAAG | ||
| Div10N | Diversity-Standard | 5′-TTCGAGCTCGGTACCGNTAANGT |
| GNAAANCTANTTCNTTCNCAGNGCCNCCGNG | ||
| GCTGCAGGCATGCAAG | ||
| Div12N | Diversity-Standard | 5′-TTCGAGCTCGGTACCNGTNATNT |
| GNAAATCNACNTCNTTNTCAGNGCNCCNGTN | ||
| GCTGCAGGCATGCAAG | ||
| Div14N | Diversity-Standard | 5′-TTCGAGCTCGGTACCNGNAATNT |
| NTAANTNTACNTNCTTNTNAGCNCNCCCNTN | ||
| GCTGCAGGCATGCAAG | ||
| Div16N | Diversity-Standard | 5′-TTCGAGCTCGGTACCNGNANTNT |
| GTNANTNTACNTNCNTCTNANCNCCCNCNTN | ||
| GCTGCAGGCATGCAAG | ||
| Div18N | Diversity-Standard | 5′-TTCGAGCTCGGTACCNGNANTNT |
| NTNAATNTNCNTNCNTNTCANCNCNCNCNTN | ||
| GCTGCAGGCATGCAAG | ||
| Div20N | Diversity-Standard | 5′-TTCGAGCTCGGTACCNGNANTNTN |
| TNANTNTNCNTNCNTNTNANCNCNCNCNTN | ||
| GCTGCAGGCATGCAAG |
Re-annealing kinetics
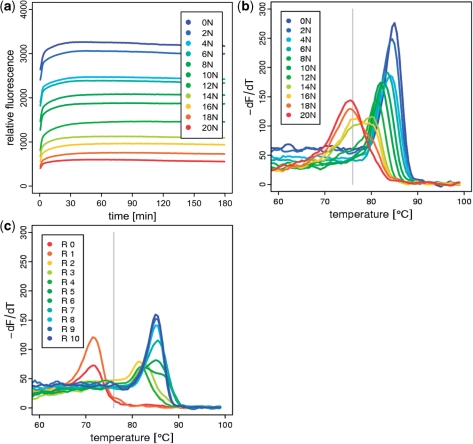
As expected we observed that the annealing kinetics varied with the diversity of the used oligonucleotide library. However, despite prolonged annealing times of up to 6 h, the SYBR green fluorescence of 10N and higher stayed well below 50% of its initial value, which made the determination of a C0t1/2 value impossible for libraries with diversities higher than 106 (Figure 2a). Triplicate experiments were variable at low diversities (Supplementary Figure 1) and the observed kinetics did not fit the calculations proposed by Baum and McCune (7), especially when diversities exceeded 4 × 103.
Figure 2.
(a) Reannealing kinetics observed for the diversity standard at 76°C, (b) remelting curves for diversity standard after reannealing at 76°C, (c) remelting curves for daunomycin aptamer selection rounds (R 0–R 10) after reanneling at 76°C. Grey lines indicate initial annealing temperature.
Remelting profiles and annealing temperature dependence
We observed that the remelting curves following an annealing step were much more reproducible in terms of melting temperature (Supplementary Figure 2) and characteristic for each of the assayed diversities (Figure 2a and b). Differences are more obvious in the remelting assay when compared with the profiles obtained in the preceding reannealing assay. On an increase in diversity, a decrease in melting temperature from 86°C towards 75°C was detected, denoting the amount of imperfectly formed heteroduplex. We noticed that the threshold of this shift was partly dependent on the applied annealing temperature. An annealing temperature close to the initial melting point of 80°C leads to a shift towards the heteroduplex population for comparatively low diversities (8 N, 105); higher annealing temperatures yielded curves that followed the same pattern as the unmelted, perfectly hybridized samples as described earlier (7,8). Using the lower annealing temperatures of 78°C and 76°C, the shift occurs gradually in higher diversities of 10 N (106) and 12 N (107), respectively (Supplementary Figure 3). The choice of the annealing temperature will determine the resolution in a given diversity range. Generally, this data demonstrates that there is no continuous linear correlation that determines the annealing kinetics, especially for higher diversities.
Uneven distribution of diversities
As most naturally occurring pools of nucleic acids consist of unevenly distributed sequences, we attempted to simulate this situation by admixtures of varying diversities. The retrieved remelting profiles indicate that by greatly increasing diversity, the pool assumes the kinetics of the lower diversity. This differs from the data provided by Baum and McCune (7), where one clone was mixed to a final concentration of 50% with a pool of 96 different sequences. Thus strong disparities in clonal distribution may yield misleading values by all melting and annealing kinetics methods.
Evaluation of a SELEX experiment
We applied our new diversity standard to analyze the population dynamics of an aptamer selection against daunomycin (9). We compared PCR aliquots from each selection round (Figure 2c) with our diversity standard (Figure 2b) from the relative distribution of peaks and amplitudes in the remelting assay. The initial population quickly collapses from the initial 1015 (25 N) pool to 107 (12 N) after just two rounds of selection. The relative plateau of gradual decrease from 106 (10 N) to 4 × 104 (6 N) correlates with the emergence of moderate binders in the fifth selection round (10). The final diversity of the tenth round was 1 in 16 (2 N) matching the results obtained by cloning and sequencing (9).
DISCUSSION
The diversity standard was easily synthesized and was shown to perform as anticipated in melting studies. We have been able to distinguish DNA populations ranging from single clones to 1012 different sequences by using our diversity standard. Previous attempts to describe population diversities by calculation of melting curves lacked necessary internal calibrations and therefore could provide misleading data in some cases. We decided to use the remelting curve as a more characteristic indicator for the analyzed diversity over the usual reannealing profiles. The newly discovered discontinuity may be attributed to the distribution and length of fully annealed regions and corresponding changes in kinetics. Another explanation could be that the exceedingly small concentration of exactly complementary strands diluted into a huge excess of highly similar sequences could result in the formation of kinetically trapped heteroduplexes. Such heteroduplexes may become prevalent beyond 10 N (106), thus the previously assumed co-linearity of diversities and the annealing process is disrupted.
Other well-established sensitive biochemical assays like ELISA and RT–PCR have already made use of an internal standard for calibration as even small changes may dramatically affect the results. We suggest that a synthetic standard adapted to the sequences of interest can be used to inexpensively and quickly determine diversities of unknown pools. Alternatively, as most nucleic acid libraries are of variable length and consist of unevenly distributed sequences, our DiStRO may serve as a gold standard. Independently performed assays referenced by DiStRO should allow direct comparison of libraries. As naturally derived libraries would display more pronounced differences in clonal copy numbers, our described method using synthetic oligonucleotides will remain partially empirical. However, the defined nature of such oligonucleotides permits reproducible generation and application of the standard. The monitoring of selection experiments of highly complex libraries like in SELEX or other in vitro selection techniques (11,12) including library generation (13) is one of the most direct applications. Additionally, calibrated diversity assays could also be useful for the evaluation of DNA pools and provide an economical setup in the rapidly evolving technology of next generation sequencing (14,15); especially paired end sequencing techniques which generate fragments of defined lengths that should facilitate the design and application of a dedicated DiStRO (16).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union through the EFRE program (ProFIT grant number 10139409). Funding for open access charge: Max Planck Society for the Advancement of Science.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Stoltenburg R, Reinemann C, Strehlitz B. SELEX—a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Mardis ER. Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 5.Britten RJ, Kohne DE. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968;161:529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- 6.Charlton J, Smith D. Estimation of SELEX pool size by measurement of DNA renaturation rates. RNA. 1999;5:1326–1332. doi: 10.1017/s1355838299991021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baum PD, McCune JM. Direct measurement of T-cell receptor repertoire diversity with AmpliCot. Nat. Methods. 2006;3:895–901. doi: 10.1038/NMETH949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 9.Wochner A, Cech B, Menger M, Erdmann VA, Glökler J. Semi-automated selection of DNA aptamers using magnetic particle handling. BioTechniques. 2007;43:344. doi: 10.2144/000112532. 346, 348. [DOI] [PubMed] [Google Scholar]
- 10.Wochner A, Glökler J. Nonradioactive fluorescence microtiter plate assay monitoring aptamer selections. BioTechniques. 2007;42:578. doi: 10.2144/000112472. 580, 582. [DOI] [PubMed] [Google Scholar]
- 11.Gupta RD, Tawfik DS. Directed enzyme evolution via small and effective neutral drift libraries. Nat. Methods. 2008;5:939–942. doi: 10.1038/nmeth.1262. [DOI] [PubMed] [Google Scholar]
- 12.Lynch SA, Gallivan JP. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Res. 2009;37:184–192. doi: 10.1093/nar/gkn924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein V, Hollfelder F. An efficient method to assemble linear DNA templates for in vitro screening and selection systems. Nucleic Acids Res. 2009;37:e122. doi: 10.1093/nar/gkp589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lister R, Gregory BD, Ecker JR. Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond. Curr. Opin. Plant Biol. 2009;12:107–118. doi: 10.1016/j.pbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 16.Fullwood MJ, Wei C, Liu ET, Ruan Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19:521–532. doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.