Abstract
Ribosomal protein (rp)S5 belongs to the family of the highly conserved rp’s that contains rpS7 from prokaryotes and rpS5 from eukaryotes. Alignment of rpS5/rpS7 from metazoans (Homo sapiens), fungi (Saccharomyces cerevisiae) and bacteria (Escherichia coli) shows that the proteins contain a conserved central/C-terminal core region and possess variable N-terminal regions. Yeast rpS5 is 69 amino acids (aa) longer than the E. coli rpS7 protein; and human rpS5 is 48 aa longer than the rpS7, respectively. To investigate the function of the yeast rpS5 and in particular the role of its N-terminal region, we obtained and characterized yeast strains in which the wild-type yeast rpS5 was replaced by its truncated variants, lacking 13, 24, 30 and 46 N-terminal amino acids, respectively. All mutant yeast strains were viable and displayed only moderately reduced growth rates, with the exception of the strain lacking 46 N-terminal amino acids, which had a doubling time of about 3 h. Biochemical analysis of the mutant yeast strains suggests that the N-terminal part of the eukaryotic and, in particular, yeast rpS5 may impact the ability of 40S subunits to function properly in translation and affect the efficiency of initiation, specifically the recruitment of initiation factors eIF3 and eIF2.
INTRODUCTION
Despite a large body of information provided by X-ray analysis of prokaryotic ribosomes, the role of many prokaryotic ribosomal proteins (rp’s) remains rather obscure. Even less is known about the functions of eukaryotic rp’s. Thirty-four rp families are present in all the domains of life, and 33 additional families are specific to Archaea and Eucarya (1,2). Apparently, these proteins have evolved to play distinct roles in archaeal and eukaryotic ribosome biogenesis, structure and function (3). Many rp’s (like eukaryotic rp L9; homolog of prokaryotic L6) show an extremely high degree of conservation and display very little variation in size and amino acid (aa) composition (1–4). Others possess less pronounced similarity (1–4). Interestingly, the proportion of universally conserved rp’s is higher in the small ribosomal subunit (two-thirds are conserved), whereas only 50% of the rp’s are conserved in the large ribosomal subunit (1,2). This high degree of conservation of rp’s from the small ribosomal subunit has been attributed to the higher degree of conservation of the small subunit ribosomal RNA (rRNA) and the rRNA regions with which they interact (1). Yet, many details of the evolution of sequence and structure of rp’s from both small and large ribosomal subunits are unclear and remain to be established.
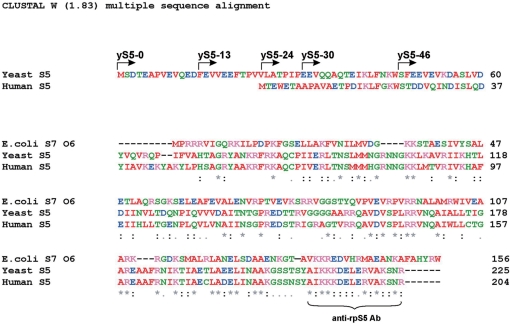
rpS5 belongs to a family of conserved rp’s that includes bacterial rpS7 (1). rpS5/7 proteins share about 30% identity at the aa level and possess a conserved central/C-terminal region and variable N-terminal ends (4–6). The extreme carboxy-terminal 16 aa of the rpS5/S7 proteins are extremely conserved in all organisms spanning several kingdoms (4–6). This suggests that this region of the rpS5 and rpS7 proteins serves an important function. The protein forms part of the exit (E) site on the 30S/40S ribosomal subunits and contributes to the formation of the mRNA exit channel (7,8). The high degree of sequence similarity between rpS5 and rpS7 and their location suggest conservation of function(s) of this protein in the ribosome. Indeed, mutations in rpS7/S5 are detrimental for the cell function and/or can significantly perturb the translation process, leading to an increased capacity for frameshifting and read-through (6,9). In contrast to the high degree of conservation of the C-terminal region, the rpS5/S7 proteins show variability in aa sequence composition and length at the N-terminal end. It is evident that many fungal and insect (fly) rpS5 proteins are the longest and prokaryotic rpS7 proteins are the shortest members of this family (1,4,5). Interestingly, Saccharomyces cerevisiae rpS5 is 69 aa longer than the Escherichia coli rpS7 (strain O6) protein; and human rpS5 is 48 aa longer than the E. coli rpS7, respectively (Figure 1). The reason why eukaryotes and in particular fungi have evolved a longer rpS5 protein is not clear.
Figure 1.
Alignments of amino acid sequences of ribosomal proteins S7/S5. E. coli strain O6:H1/CFT073 (NP_755978), S. cerevisiae rpS5 (NP_012657) and H. sapiens rpS5 (NP_001000) have been used. Arrows indicate the start sites of the truncated protein variants prepared and used in this study. The peptide used to elicit anti-rpS5 antibodies is underlined.
We have previously obtained and characterized a yeast strain in which yeast rpS5 was replaced by its 21 aa shorter human rpS5 homolog (6). We concluded that the negatively charged N-terminal extension of yeast rpS5 might affect the ribosomal recruitment of specific mRNAs as well as play important roles in ensuring the efficiency and fidelity of elongation (6). Although human rpS5 is 67% identical and 79% similar to S. cerevisiae rpS5 (6), it nevertheless could not be excluded that the observed differences between the wild-type (wt) and humanized yeast strains were due to the overall dissimilarity of the yeast and human rpS5 proteins, rather than to the dissimilarity of their N-terminal regions.
To further investigate the function of the yeast rpS5 and to better understand the role of the N-terminal region of the eukaryotic protein, we obtained and characterized yeast strains in which the wt yeast rpS5 was replaced by its truncated variants, lacking 13, 24, 30 and 46 N-terminal amino acids, respectively. Our data further corroborated our previous observations, suggesting that the N-terminal part of the yeast S5 plays important roles in ensuring the efficiency and accuracy of elongation as well as initiation processes and may in addition play important roles in recruiting certain initiation factors to the 40S ribosome.
MATERIALS AND METHODS
Yeast strains and growth methods
Yeast strains used in this study are: Research Genetics (Huntsville, AL, USA) WT haploid BY4741 (MATa, his3-1, leu2-0, met15-0, ura3-0), diploid BY4743 ([4741/4742] MATa/MATα, his3-1/his3-1, leu2-0/leu2-0, lys2-0/+, met15-0/+, ura3-0/ura3-0, RPS5/rps5::kanMX). The rpS5 yeast strains expressing full-length and truncated version of the RPS5 gene from the plasmid: BY47yS5-0 Mata his3-1, leu2-0, ura3-0, rps5::kanMX, <yrps5; LEU2, 2μ> BY47yS5-13 Mata his3-1, leu2-0, ura3-0, rps5::kanMX, <yrps5-13; LEU2, 2μ>, BY47yS5-24 Mata his3-1, leu2-0, ura3-0, rps5::kanMX, <yrps5-24; LEU2, 2μ>, BY47yS5-30 Mata his3-1, leu2-0, ura3-0, rps5::kanMX, <yrps5-30; LEU2, 2μ>, BY47yS5-46 Mata his3-1, leu2-0, ura3-0, rps5::kanMX, <yrps5-13; LEU2, 2μ> were obtained as follows: cDNA of yeast rp S5 was amplified by polymerase chain reaction (PCR) using 5′-AAAAAGGATCCACCGACCATTCCAAAGATGTCTGA-3′ (BY47yS5-0), 5′-TTATAGGATCCATGTTCGAAGTTGTTGAAGAA-3′ (BY47yS5-13), 5′-TATGGGATCCATGTTGGCTACTCCAATTCCAGAAGAAG-3′ (BY47yS5-24), 5′-AAAAAGGATCCATGGAAGAAGTCCAACAAGCTCAAACCG-3′ (BY47yS5-30), 5′-AAAAAGGATCCATGTCTTTTGAAGAAGTTGAAGTTAAGGATGC-3′ (BY47yS5-46) as forward primers (initiation codon is in bold) and 5-AAAAAGTCGACAGCTTCTTAACGGTTAGACTTGGCAAC-3′ as reverse primer (termination codon is in bold) and yeast high molecular DNA as a template. The PCR fragments were further digested with BamHI and SalI and cloned into p425TEF (2 µ, LEU2) vector (10) digested with the same enzymes. The resultant pTEF_yS5 plasmids were transformed into the heterozygous diploid. Transformants were allowed to sporulate [using standard protocols (11)] and tetrads were dissected. Haploid clones able to grow on yeast extract, peptone and dextrose (YPED) medium, resistant to G418 sulfate and expressing human rpS5 were selected. An rps5::kanMX genotype of these clones was further verified by PCR using 5′-CATAATACCAAGAAAAGAGACTAGAAATAACCG-3′ and 5′-GAAAAACATATGTAATATTGAAAATCTTTCACTTTTTTTAG-3′ primers.
Yeast cultures were grown as indicated using either synthetic media containing 0.67% Difco yeast nitrogen base, 1% ammonium sulfate, 2% glucose (or galactose) and supplemented with the appropriate amino acids or YEPD medium (11). Transformation was done using the lithium acetate method (12). For polysome analysis, yeast cells were grown in YEPD medium with 2% glucose.
Reporter plasmids
The p281 plasmid containing lacZ under GAL1/10 promoter has been described previously (13). The p281-4-URE2 vector has also been previously described (14,15). Programmed −1 and +1 frameshifting test reporters containing L-A, Ty1 or Ty3 frameshift signals, respectively, between the Renilla and Firefly luciferase genes and nonsense suppression test reporters containing three different stop codons (UAA, UAG or UGA) in the firefly luciferase gene (16,17) were provided by Dr Jonathan D. Dinman (University of Maryland). All luciferase reporter plasmids were transformed into wt and BY47yS5 strains and grown on the minimal YNB medium.
Fractionation of polyribosomes and 40S and 60S subunits
Fractionation of polyribosomes was done essentially as described (6). All procedures were performed at 4°C except where indicated. Yeast cells from 50 ml of log phase culture were pelleted, treated for 10 min with 100 µg/ml cycloheximide and repelleted. Lysates were made by glass bead cell disruption (three to five cycles of 1 min each), with intermittent cooling on ice, in buffer which contained 100 mM KCl, 2.5 mM magnesium acetate, 20 mM HEPES·KOH, pH 7.4, 14.4 mM β-mercaptoethanol, 100 µg/ml cycloheximide. Cell debris was removed by centrifugation at 7000 r.p.m. for 8 min. Polyribosomes, ribosomes and subunits were resolved in either 7–50% or 5–30% (17 000 r.p.m., 18 h) and (20 000 r.p.m., 18 h), respectively, sucrose gradients containing 100 mM KCl, 5 mM MgCl2, 20 mM HEPES·KOH, pH 7.4 and 2 mM dithiothreitol using a Beckman SW32.1 rotor. Gradients were collected using the ISCO Programmable Density Gradient System with continuous monitoring at 254 nm using an ISCO UA-6 absorbance detector. Analysis of the amounts of 40S and 60S subunits was done by centrifugation on low-Mg2+ sucrose density gradients as described by Hinnebusch and co-authors (18) with minor modifications. Subunits were resolved in 5–30% low Mg2+ sucrose gradients at 20 000 r.p.m. for 18 h using SW32.1 rotor. Analysis of ratios of 80S monosomes to polyribosome as well as 40S to 60S subunits was done essentially as before (19).
Western blotting
Western blotting was performed following standard procedures (20). Western blots were decorated with rabbit polyclonal anti-rpS5 or anti-rpS2/rpL30 antibodies followed by incubation with goat anti-rabbit HRP-conjugated antibodies. The anti-S2/L30 antibodies able to simultaneously recognize both S2 and L30 proteins were kindly provided by Dr Jonathan Warner (Albert Einstein College of Medicine, New York). The blots were then detected with an enhanced chemiluminescence detection kit (ECLTM, GE Healthcare, Piscataway, NJ, USA). The anti-eIF2α (21 ) and anti-eIF3b (PRT1) (22) antibodies used for western blotting were kindly provided by Drs Tom Dever (National Institutes of Health, Bethesda, MD, USA) and Alan Hinnebusch (National Institutes of Health), respectively.
rRNA analysis
Yeast strains were grown at 30°C in complete medium till mid-logarithmic phase. rRNA was extracted and subjected to denaturing gel electrophoresis as described (14,15). Gels were stained with ethidium bromide and scanned using a Typhoon imaging scanner. Northern blotting was done following standard procedures (23) with a probe 5′-CATGGCTTAATCTTTGAGAC-3′ complementary to nucleotides +34 to +53 within 18S rRNA and capable of recognizing both the 18S rRNA and the 20S pre-rRNA. The probe was end-labeled with the use of gamma-32P-ATP (MP Biomedicals, Solon, OH, USA) and used for hybridization.
Miscellaneous
Molecular cloning was performed following the general procedures described in Sambrook et al. (24). DNA sequencing was accomplished by the DNA Sequencing Core facility at Cleveland Clinic. Sodium dodecyl sulfate polyacrylamide gel electrophoresis was performed according to Laemmli (25). Yeast genomic DNA was isolated using the DNA-Pure™ Yeast Genomic Kit (PureBiotech, Middlesex, NJ, USA) and following the manufacturer’s protocol. β-Galactosidase activity was measured following the protocol described in the Clontech Yeast Protocols Handbook with o-nitrophenyl β-d-galactopyranoside as a substrate. Cell extracts were prepared by subsequent cycles of cell-freezing in liquid nitrogen and thawing at 37°C. Luciferase activities were measured using a dual luciferase assay kit (Promega Madison, WI, USA) as described by Dinman and co-authors (16,17,26).
RESULTS
Yeast rpS5 proteins lacking 13, 24, 30 and 46 aa residues from the N-terminal end substitute for the full-length protein in vivo
Many eukaryotic (and in particular yeast) rp’s are encoded by two genes; however, yeast rpS5 is encoded by a single and essential gene (5). To determine whether truncated rpS5 proteins can substitute for the full-length yeast rpS5 protein we have used an approach that ensures that truncated rpS5 proteins in the mutant strains will be expressed at levels equal to that of the full-length yeast rpS5 in the wt strain (6). In eukaryotic cells, and, particularly in yeast, balanced expression of rp’s is achieved at the post-transcriptional level, through the regulated turnover of rp’s (27–29). Rp’s that are present in excess are rapidly degraded until they reach levels identical to other rp’s (27–29). The coding sequences of rpS5 variants lacking 13, 24, 30 and 46 aa residues from the N-terminal end (Figure 1) were therefore cloned into the high copy number 2µ vector to ensure high levels of protein expression. An initiation (ATG) codon was added to all of the truncated gene variants to insure that translation of all the mRNA would be initiated properly. All rpS5 variants were placed under the control of the constitutive (TEF) promoter (derived from the elongation factor 1A gene), because the TEF gene belongs to the cluster of rp genes and its expression is tightly co-regulated with the expression of rp’s in yeast (30). We also cloned the full-length yeast rpS5 protein into the same plasmid to insure that the full-length protein would be expressed in the same context.
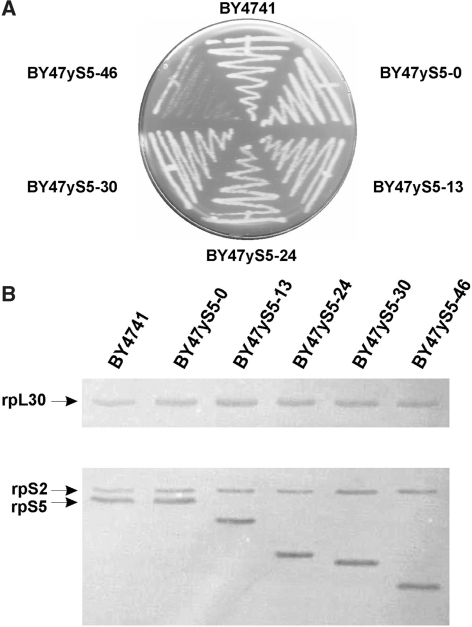
The resulting pTEF_S5s (2µ, LEU2) plasmids were transformed into a diploid heterozygous yeast strain with a single disrupted copy of the RPS5 gene. Transformants were allowed to sporulate and tetrads were dissected. Tetrad dissection analysis revealed that in the case of yeast strains carrying either the full-length rpS5, or gene/protein variants lacking 13, 24, 30 and 46 aa residues all four spores were viable; however, further truncation of rpS5 protein (by 50 aa) resulted in two inviable spores (data not shown) suggesting that an rpS5 gene variant encoding rpS5 lacking 50 N-terminal aa is not able to complement the full-length gene. PCR analysis using chromosomal DNA isolated from yeast clones showed that two of four viable spores contained a disrupted copy of the yeast RPS5 gene (data not shown). Thus, rpS5 variants expressed from the plasmid were the sole source of rpS5 in these strains, and we therefore concluded that they substitute for the full-length protein in vivo. The BY4743 strain transformed with the empty p425TEF plasmid (2 µ, LEU2) gave rise to only two viable colonies after sporulation and tetrad dissection, confirming that rpS5 is essential (5) for yeast viability (data not shown). Isogenic haploid strains with the genotype (MATa his3-1, leu2-0, ura3-0, rps5::kanMX, <rps5 full-length or variants; LEU2, 2μ>) were used for subsequent analysis. It should be noted that the haploid BY47yS5 strain carrying the full-length copy of the RPS5 gene expressed from the plasmid displayed growth rates equal to that of the haploid BY4741 strain; however, the BY47yS5-13, BY47yS5-24, BY47yS5-30 strains displayed moderately reduced (15–25%) growth rates in comparison with the BY4741 and BY47yS5 yeast strains (Figure 2A). The BY47yS5-46 strain revealed substantial reduction in growth rates, when grown on either rich YEPD medium (Figure 2A) or minimal SD medium (data not shown) in comparison with the BY47yS5 yeast strain. The doubling time for this strain at 30°C in liquid YEPD glucose medium was ∼3–3.2 h, whereas for all other strains it is ∼1.5–1.7 h.
Figure 2.
Cell growth and expression levels of the rpS5s in the wt and the mutant yeast strains. (A) Yeast cell growth. Cells were grown for 36 h on a solid YEPD agar medium containing 2% glucose. (B) Western blot analysis of the rpS5, rpS2 and rpL30 expression levels. Anti-rpS5 antibody derived against the AIKKKDELERVAKSNRC C-terminally conserved rpS5 peptide is capable of recognizing all the rpS5 protein variants.
The expression levels of yrpS5 proteins in the wt and all the mutant yeast strains were determined by western blotting using an antibody directed against the AIKKKDELERVAKSNR C-terminally conserved rpS5 peptide (Figure 1). We also used an antibody directed against the yeast rpS2, which also recognizes the yeast L30 (31) protein formerly known as L32 (31,32). This analysis showed equal levels of expression of the rpS5 proteins in all the strains, relative to S2 and L30 (Figure 2B). For further characterization, we chose the isogenic BY47yS5-0, BY47yS5-13, BY47yS5-24, BY47yS5-30 and BY47yS5-46 strains.
Cap-dependent but not the internal ribosome entry site-mediated translation initiation is severely impaired in yeast strain carrying rpS5 lacking 30 aa residues from the N-terminal end
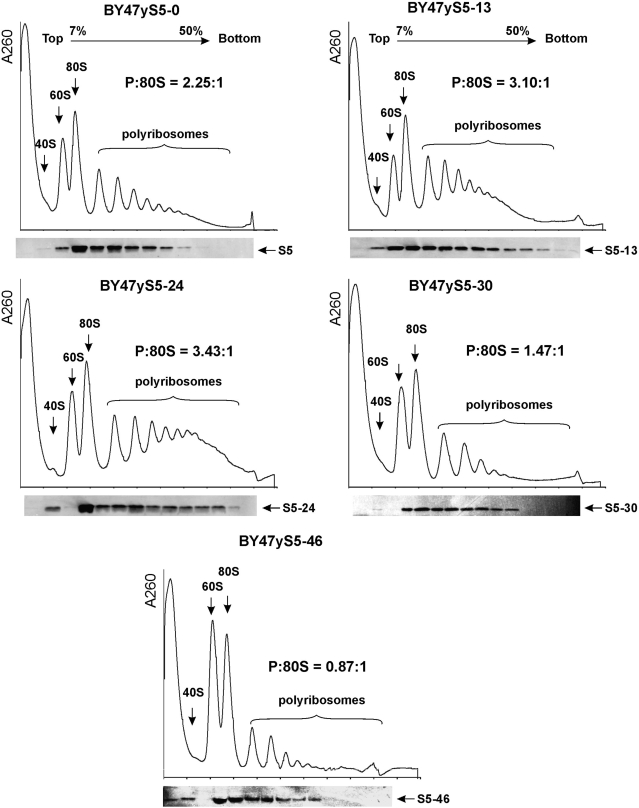
Polyribosome analysis is an efficient way to characterize the translational status of cells. The polyribosome: 80S monosome ratio is indicative of the efficiency of translation initiation. Sucrose density-gradient centrifugation analysis of cytoplasmic extracts prepared from the BY47yS5-0, BY47yS5-13, BY47yS5-24, BY47yS5-30 and BY47yS5-46 strains showed that protein synthesis is not severely perturbed in BY47yS5-13 and BY47yS5-24 strains in comparison with the BY47yS5-0 strain carrying the full-length copy of rpS5 (Figure 3). The polysome (P) to monosome (80S) ratios were 2.25 : 1, 3.10 : 1 and 3.43 : 1 for the BY47yS5-0, BY47yS5-13 and BY47yS5-24 strains, respectively. Both the BY47yS5-13 and BY47yS5-24 strains displayed a moderate increase in the heavy polyribosomal components similar to what we observed previously for the strain carrying a copy of the human rpS5 protein, which is 21 aa shorter (6). Further truncation of yeast rpS5 by 9 aa (strain BY47yS5-30) resulted in a reduction in the polysome: 80 monosome ratio (1.47 : 1) characteristic of a reduced rate of translation initiation. However, truncation of the protein by 46 aa resulted in more severe changes in polysome : 80 monosome ratio (0.87 : 1 in the BY47yS5-46 strain). Western blot analysis showed that all the rpS5 variants (including the rpS5-46 protein) were present in fractions containing 40S subunits, 80S ribosomes and polyribosomes (Figure 3); thus, confirming that the truncated proteins are stable components of mutant ribosomes and are actively involved in protein synthesis. These data suggest that truncation of rpS5 by 30 and 46 aa residues might significantly impair translation initiation.
Figure 3.
Ribosome profiles of the wt and mutant yeast strains containing truncated versions of the rpS5 protein and rpS5 western blot analyses. Extracts from isogenic wt (BY47yS5-0) and rpS5 mutant (BY47yS5-13, BY47yS5-24, BY47yS5-30 and BY47yS5-46) strains were resolved by velocity sedimentation on 7–50% sucrose gradients. Fractions were collected while scanning at A254. The positions of different ribosomal species are indicated. The ratio of the area under the polysomal (P) and 80S peaks is shown (P:80S). The data recorded by the PeakTrak program (ISCO gradient density gradient fractionation system) were exported to ASCII format and analyzed by UVProbe 2.10 Shimadzu software to assess the 80S and polysomal peak areas. Western blot analysis of individual fractions was done using an antibody directed against the AIKKKDELERVAKSNR C-terminally conserved rpS5 peptide.
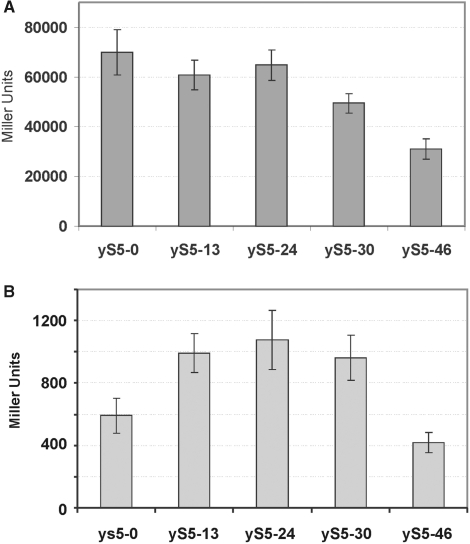
Additional evidence supporting this conclusion came from experiments utilizing a reporter plasmid carrying the lacZ gene under control of the GAL1 promoter (13). These experiments showed that LacZ expression in the mutant strains expressing rpS5-13 and rpS5-24 proteins was only mildly reduced (by ∼10–15%) compared to the strain expressing the full-length protein (Figure 6A). However, LacZ expression in the strain expressing the rpS5-30 protein was reduced by ∼30%, and in the strain expressing the rpS5-46 protein was reduced by more than 50%. These results corroborate the data from polyribosome analysis and suggested that cap-dependent translation is impaired in the strains expressing rpS5 lacking 30 and 46 amino N-terminal acid residues. Interestingly, expression mediated by the URE2 internal ribosome entry site (IRES) in the mutant BY47yS5-13, BY47yS5-24 and BY47yS5-30 strains was about 50% higher than in the BY47yS5-0 strain (Figure 6B). URE2 IRES-mediated expression was measured using the previously described Ure2p-lacZ reporter vector, in which the URE2 IRES was inserted in frame in front of the lacZ reporter gene but behind a stable hairpin structure (ΔG > −30 kcal/mol), which abolished cap-dependent lacZ expression (14,15). This suggests that, particularly in the BY47yS5-30 strain, it is cap-dependent initiation which becomes impaired. However, further truncation of rpS5 in BY47yS5-46 also results in reduction of IRES-mediated translation.
Figure 6.
Analysis of the cap-dependent and URE2 IRES-mediated translation in the wt mutant yeast strains. (A) Expression of reporter lacZ construct (13) under the control of the GAL1/10 promoter transformed into wt and mutant yeast strains. β-Galactosidase activity (relative units). (B) Expression of the monocistronic URE2 IRES p281-4-URE2 reporter lacZ construct (14,15) under the control of the GAL1/10 promoter transformed into wt and mutant yeast strains. β-Galactosidase activity was determined after a 20-h induction in 2% galactose at 30°C. Mean efficiencies determined from at least nine independent experiments are plotted and bars represent the corresponding standard error.
Ribosome biogenesis is not severely affected in the mutant yeast strains carrying rpS5 proteins lacking 13, 24, 30 and 46 aa residues from the N-terminal end
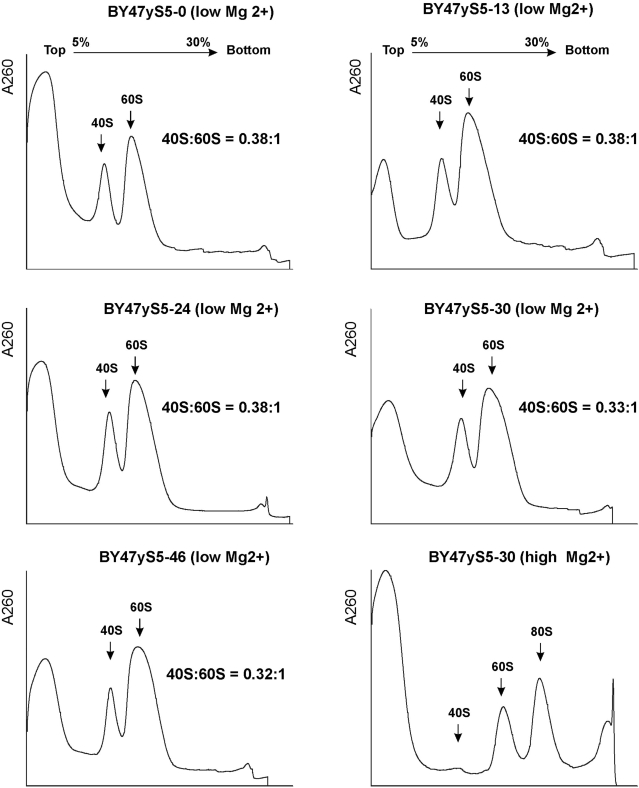
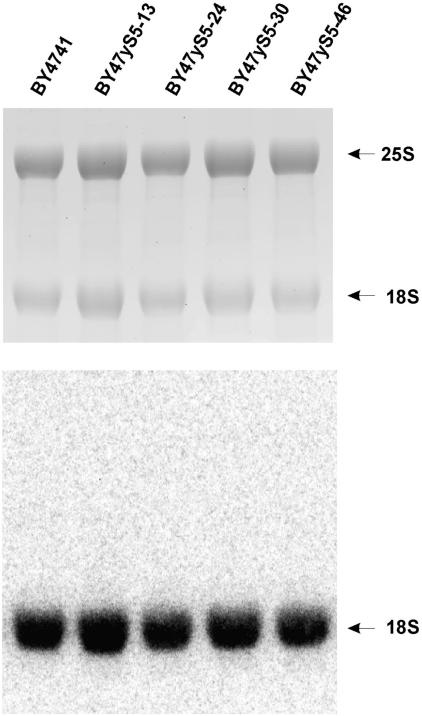
Rp’s that maintain the structural integrity of mature ribosomal subunits including rpS5 have been suggested to play important roles in ribosome biogenesis and intracellular transport (33–35). Therefore, it could not be completely excluded that the observed defects in translation initiation, particularly in the BY47yS5-30 and BY47yS5-46 strains, might in part be related to the affected biogenesis of 40S ribosomal subunits leading to their reduced amounts in the mutant yeast cells. However, analysis of the ratios of 40S–60S subunits done under conditions of low Mg2+, when polyribosomes and 80S ribosomes dissociate into free ribosomal subunits, does not support this suggestion. The isogenic BY47yS5-0, BY47yS5-13, BY47yS5-24, BY47yS5-30 and BY47yS5-46 strains had very similar amounts of 40S and 60S subunits (Figure 4). This suggests that the observed drop in polyribosome fractions and increase in 80S ribosomes, particularly in the BY47yS5-46, strain could most probably be attributed to the inability of mutant 40S subunits (containing truncated rpS5 protein lacking 46 N-terminal amino acids) to function properly in initiation, rather than to the reduced amounts of 40S subunits due to affected 40S biogenesis. Additional evidence supporting this observation came from the analysis of rRNA (Figure 5). We have not observed any substantial changes in 25S/18S ratios between the strains (Figure 5) and/or the appearance of the unprocessed 20S pre-rRNA species that would be indicative of affected 40S subunit biogenesis (34,35). We therefore conclude that the progressive truncations of the N-terminus of rpS5 described here do not seem to significantly affect the biogenesis of 40S subunits.
Figure 4.
Analysis of the amounts of 40S and 60S subunits in wt and mutant yeast strains by centrifugation on low-Mg2+ sucrose density gradients. Extracts from isogenic wt (BY47yS5-0) and rpS5 mutant (BY47yS5-13, BY47yS5-24, BY47yS5-30 and BY47yS5-46) strains were resolved by velocity sedimentation on 5–30% sucrose gradients under low Mg2+ as described (18). The positions of different ribosomal species are indicated. Right bottom panel shows ribosomal fractions from the BY47yS5-30 strain resolved under high Mg2+ concentration (2.5 mM) to indicate the position of the 80S peak on the same gradient. The ratio of the area under the 40S and 60 S peaks is shown (40S : 60S).
Figure 5.
rRNA analysis from wt and mutant yeast strains. Seven micrograms of total yeast RNA was separated on a denaturing agarose gel and subjected to hybridization with 18S and 20S pre-rRNA specific probe (18S-A) (33). Ethidium bromide stained gel (top). Northern blot (bottom). The positions of 25S and 18S rRNA species are indicated.
Fidelity of translation in the mutant rpS5 yeast strains
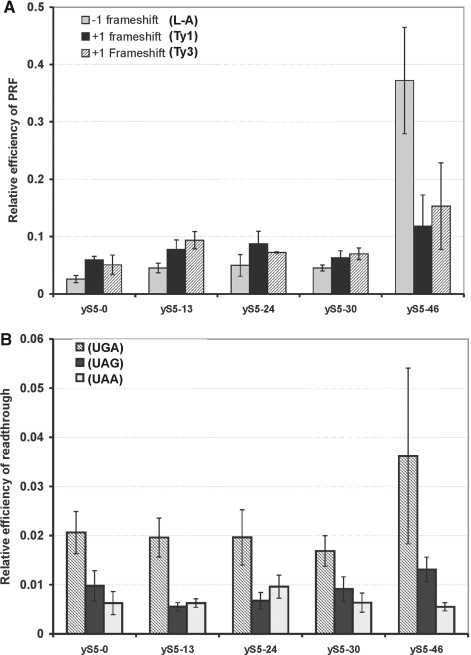
Rp S5/S7 forms part of the E-site, which plays an important role in maintaining the accuracy of translation (6,9). Modifications of the E-site in the prokaryotic ribosome and in particular of the rpS7 protein yielded more error-prone ribosomes (9). However, so far, few data are available concerning the function of the eukaryotic and in particular of yeast rpS5 (6). We therefore determined whether translation fidelity was compromised in the mutant yeast strains. To monitor translation fidelity, we used a bicistronic dual-luciferase reporter system, in which the frameshift signals derived from L-A virus, or Ty1 or Ty3 retrotransposons were inserted between the Renilla and firefly luciferase genes so that firefly luciferase can only be produced in the event of a frameshift (16,17). Interestingly, substitution of yrpS5 with its truncated versions resulted in only a moderate increase in programmed frameshifting (∼1.5–2.0-fold) for all three signals (Figure 7A) in the BY47yS5-0, BY47yS5-13, BY47yS5-24 and BY47yS5-30 strains and in a substantial increase in programmed frameshifting in the BY47yS5-46 strain (>10-fold in case of −1 frameshifting).
Figure 7.
Translation fidelity in the wt and mutant rpS5s yeast strains. (A) Programmed frameshifting efficiency. WT and mutant rpS5’s strain were transformed with the control, L-A, Ty1 and Ty3 frameshift reporter plasmids. Dual-luciferase assays were performed and programmed frameshifting (PRF) efficiencies were calculated as described (6,16,17,23). Mean efficiencies determined from at least six independent experiments are plotted and bars represent the corresponding standard error. (B) Termination codon read-through efficiency in wild-type and mutant yeast strains. The wt and human rpS5 mutant yeast strains expressing human rpS5 were transformed with control, UAA, UAG and UGA reporter plasmids and read-through efficiencies at each terminator were determined as described (6,14,15,21). Mean efficiencies determined from at least six independent experiments are plotted and bars represent the corresponding standard error. The significance of differences in signals between mutant and wt strain in all the experiments (P) above was at least <0.05, with the exception of UGA read-through signal in BY47yS5-46 strain.
We have further investigated whether nonsense suppression is affected in the mutant strains. To this end, we employed similar dual-luciferase reporters each containing one of the stop codons (UGA, UAG or UAA) inserted into the firefly luciferase gene so that firefly luciferase can only be produced as a result of a nonsense suppression (16,17). We have found that replacement of rpS5 by truncated versions thereof only moderately affected the ability of the mutant strains to suppress stop codons. Interestingly, there was almost no change in recognition of the UGA codon in the BY47yS5-0, BY47yS5-13, BY47yS5-24 and BY47yS5-30 strains and about a 2-fold (although statistically insignificant) increase in its read-through in the BY47yS5-46 strain (Figure 7B). The mutant BY47yS5-13, BY47yS5-24 strains exhibited hyperaccurate recognition of the UAG stop codon (∼1.75–2-fold enhancement), and no significant change in the recognition of the UAG or UGA codons (Figure 7B). Interestingly, there was almost no change in the efficiency of recognition of the UAA codon. Therefore, we concluded that the process of recognition of the STOP codons is less sensitive to the truncation of the N-terminal part of rpS5 than the elongation and initiation processes.
Association of eIF2 and eIF3 with mutant 40S ribosomal subunits containing rpS5 lacking 30 and 46 N-terminal amino acids
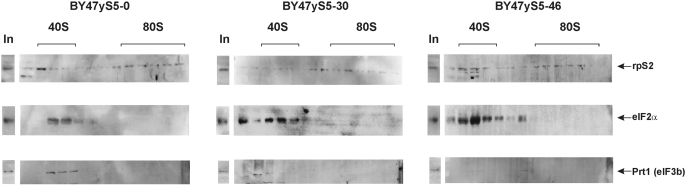
To gain further insights into why the initiation step is impaired in BY47yS5-30 and BY47yS5-46 strains, we assessed the association of initiation factors eIF2 and eIF3 with the 40S subunits in isogenic BY47yS5-0, BY47yS5-30 and BY47yS5-46 strains by western blotting using antibodies against the α-subunit of eIF2 (21) and the core eIF3b (PRT1) subunit of eIF3 (22). eIF3 is a multisubunit, multifunctional factor that interacts directly with the 40S subunit (36). eIF2 is a heterotrimer composed of α-, β- and γ-subunits, which is responsible for the attachment of initiator Met-tRNAiMet to the 40S subunit as the eIF2-GTP-Met-tRNAiMet ternary complex (37). Attachment of the eIF2-ternary complex to 40S subunits is promoted by eIF3 and also by eIF1 and eIF1A. The resulting 43S preinitiation complex containing eIF2, eIF3, eIF1 and eIF1A attaches to the eIF4F-associated cap-proximal region of mRNA (a step, which is likely stimulated by the eIF3/eIF4F interaction), and then scans downstream to the initiation codon where it forms the 48S initiation complex (37).
We have found that association of the mutant 40S subunits containing rpS5 lacking 30 and 46 aa residues from the N-terminal end (BY47yS5-30 and BY47yS5-46 strains) with eIF3 was reduced, whereas their association with eIF2 instead increased compared to the 40S subunit in the wt strain (Figure 8). The observed effect was more prominent in BY47yS5-46 strain than in the BY47yS5-30 strain.
Figure 8.
Association of initiation factors eIF2 and eIF3 with 40S ribosomal subunits in wt and mutant (BY47yS5-30 and BY47yS5-46) yeast strains. Extracts from isogenic wt (BY47yS5-0) and rpS5 mutant (BY47yS5-30 and BY47yS5-46) strains were resolved by velocity sedimentation on 5–30% sucrose gradients as described (15). Western blot analyses of individual fractions were done using antibodies directed against the initiation factors eIF2α and eIF3b (PRT1) and the ribosomal protein S2 respectively. The positions of 40S subunits and 80S ribosomes are indicated. ‘In’ for input, represents a 7% portion of each gradient fraction.
DISCUSSION
It is now evident that the mechanism of the catalysis of peptide bond formation by the ribosome is extremely conserved in all organisms (38–40), and this is reflected in the conserved structure of the peptidyl transferase active site composed of RNA (7,38). While catalysis of peptide bond formation does not seem to require rp’s, other steps in the translation process do involve rp’s, which participate in a variety of activities such as recruitment of tRNAs and/or translation factors (1–4). In addition, the additional rp’s present in the eukaryotic ribosome were suggested to stabilize eukaryote-specific rRNA expansion segments, participate in ribosomal biogenesis and play an important role in translational control of gene expression (3). Interestingly, the 34 rp families that are present in all the domains of life are not identical and exhibit varying degrees of similarity (1,2). Many eukaryotic proteins have evolved additional segments, the function(s) of which are not known, but may be related to differences in the translation apparatus between the different kingdoms (1,2).
To gain further insights into the function of eukaryotic (S. cerevisiae) rpS5/S7, and, in particular its N-terminal extension, which is absent in prokaryotes (E. coli), we characterized yeast strains in which the wt yeast rpS5 was replaced by truncated variants, lacking 13, 24, 30 or 46 N-terminal amino acids, respectively (Figures 1 and 2). The mutant yeast strains were all viable and their growth rates were only moderately (∼15–25%) reduced, with the exception of the strain expressing the rpS5-46 truncation mutant, which displayed a more than 2-fold reduction in growth rate (Figure 2A). This result suggests that the N-terminal ∼25 aa of yeast rpS5 may play only a modulatory role in the function of the yeast ribosome, whereas more distal amino acid residues play a considerably more important role in yeast cell physiology, likely in key steps of the translation process. The increase in levels of 80S ribosomes and the significant reduction in polyribosomal fractions in the BY47yS5-30 and BY47yS5-46 mutant strains (Figure 3) suggest a significant defect in the translation initiation step. It has previously been shown that altered expression of almost any single rp of the small subunit (including rpS5) in S. cerevisiae affects ribosome biogenesis and limits ribosome production (34). Interestingly, Woolford et al. (33) reported that the carboxy-terminal extension of rpS14 is required for maturation of 43S preribosomes in yeast and is important for the late (cytoplasmic) steps of 40S subunit assembly. RpS14 resides on the 40S subunit’s platform in close proximity to the E-site and rpS5 (6). Although the exact location of the rpS5 N-terminal region is not known (especially relative to rpS14’s carboxy-terminus), it could not be excluded that the N-terminal extension of rpS5 might play a role in 40S subunit maturation. However, the 40S:60S subunit and 18S:25S rRNA ratios were not significantly affected in the mutant strains (Figures 4 and 5), indicating that the increase in 80S ribosomes and reduction in the polyribosomal pool in these strains most likely reflect the inability of the mutant 40S subunits to function properly in translation, rather than the impairment of their biogenesis. This in turn suggests that the N-terminal region of rpS5 is essential for translational activity, rather than for ribosomal biogenesis. However, although we did not observe a major ribosome biogenesis defect in mutant strains, the slightly reduced levels of 40S ribosomes, particularly in the BY47yS5-30 and BY47yS5-46 strains (Figure 4), might nevertheless contribute to the translation initiation defect.
The results of polyribosome analysis were corroborated by our experiments directly assaying cap-dependent initiation (Figure 6A), which showed that this process was almost unaffected in the BY47yS5-13 and BY47yS5-24 strains, but was reduced in the BY47yS5-30 and BY47yS5-46 strains by ∼30% and more than 50%, respectively (Figure 6A). Interestingly, although translation driven by the URE2 IRES was also reduced by 50% in the BY47yS5-46 strain, in contrast to cap-mediated translation, it was enhanced by ∼50% in the mutant BY47yS5-13, BY47yS5-24 and BY47yS5-30 strains. However, it is difficult to comment on the basis for the observed differences between cap-dependent and IRES-mediated translation in these three mutant strains because the mechanism of initiation on the URE2 IRES is unknown. Reduction of both cap-dependent and IRES-mediated translation (by about 50%) in the BY47yS5-46 strain, on the other hand, suggests more general translation defects, which might be attributable to the affected association of the general translation initiation factors eIF2 and eIF3 with the mutant 40S subunits, which is discussed below.
The translational fidelity of the mutant strains was altered relative to the wt Y47yS5-0 strain, displaying moderately to severely increased levels of frameshifting (Figure 7A) as well as a mildly altered ability to recognize stop codons (Figure 7B). Interestingly, all effects were rather minor in the BY47yS5-13, BY47yS5-24 and BY47yS5-30 strains, whereas the BY47yS5-46 strain displayed a more than 10-fold increase in programmed −1 frameshifting and a ∼2-fold increase in the read-through of the UGA stop codon. However, the +1 frameshifting and recognition of UAG and UAA stop codons were less affected in all the mutant yeast strains compared to the wt strain. The observation that the translational fidelity of mutant ribosomes with a modified rpS5 (and hence a modified E-site) was generally altered is consistent with previous suggestions that the E-site is allosterically coupled to the A-site and that this coupling controls the ability of the ribosomes to discriminate between cognate and noncognate tRNAs at the A-site (41). It cannot be excluded that the altered structure of the E-site might also affect the structure of the adjacent P-site due to its proximity. Codon–anticodon mismatches in the P-site have been found to compromise tRNA selection fidelity (42).
However, the observation that initiation was significantly more affected by mutation of rpS5 than elongation or termination steps suggests that the N-terminal region of eukaryotic rpS5 that is absent in prokaryotes (in particular, residues 25–46) may have evolved specifically to control certain steps in eukaryotic initiation, possibly involving ribosomal recruitment of initiation factors. Consistently, association of eIF3 with mutant 40S subunits decreased, whereas association of eIF2 increased in the BY47yS5-30 and BY47yS5-46 strains (Figure 8). In this respect, it is significant that the five-lobed mammalian eIF3 was modeled onto a protein-rich area on the solvent side of the 40S subunit with its left ‘arm’ in close proximity to rpS5 (43). eIF3 is critical for ribosomal recruitment of mRNA, and simultaneous depletion of its eIF3a and eIF3b subunits in yeast led to a strong reduction in the formation of 48S initiation complexes and to accumulation of mRNA-free 80S ribosomes (44). The overall increase in mRNA-free 80S ribosomes observed in the rpS5 mutant stains is consistent with reduced levels in them of eIF3 associated with 40S subunits, and as a consequence, with a presumable defect in recruitment of mRNA to 40S ribosomal complexes. However, it was previously reported that although a 90% reduction in eIF3 binding to yeast 40S subunits had more a dramatic effect on recruitment of mRNA to 40S subunits (also a 90% reduction), it also resulted in a ∼50% reduction in ribosomal recruitment of eIF2–ternary complexes (44), whereas in our study, the level of association of eIF2 with 40S subunit in mutant stains actually increased. In mammalian ribosomal initiation complexes, the eIF2 α-subunit is situated in very close proximity to rpS5 because both, rpS5 and eIF2α, cross-linked to the same –3 position on mRNA (45). Thus, N-terminal truncation of rpS5 could potentially either directly affect recruitment of eIF2-ternary complexes to 40S subunits or influence ribosomal association of eIF2 by modulating eIF5-stimulated hydrolysis of eIF2-bound GTP or subsequent dissociation of eIF2-GDP. Further in vitro biochemical analysis is required to determine the exact steps in initiation that are affected by truncation of rpS5.
In conclusion, taken together, the data presented here indicate that the N-terminal region of eukaryotic rpS5 is important for efficient translation initiation, maintenance of the translation reading frame, stop codon recognition and likely also influences ribosomal association of initiation factors eIF3 and eIF2.
FUNDING
Funding for open access charge: The National American Heart Association (grant 0730120N to A.A.K.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Drs Jonathan Warner, Jonathan Dinman, Shuetsu Fukushi, Tom Dever and Alan Hinnebusch for their generous gifts of antibodies and plasmids used in this study. Drs Valentin Boerner and Ms Aekam Barot are gratefully acknowledged for their helpful advice regarding tetrad dissection analysis. We thank Drs Tom Dever, Vyacheslav Kolb and Alexander Spirin for helpful discussions. We are especially grateful to Drs Tatyana Pestova and Christopher Hellen for their critical reading of the manuscript and many valuable suggestions.
REFERENCES
- 1.Lecompte O, Ripp R, Thierry JC, Moras D, Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 2002;30:5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts E, Sethi A, Montoya J, Woese CR, Luthey-Schulten Z. Molecular signatures of ribosomal evolution. Proc. Natl Acad. Sci. USA. 2008;105:13953–13958. doi: 10.1073/pnas.0804861105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dresios J, Panopoulos P, Synetos D. Eukaryotic ribosomal proteins lacking a eubacterial counterpart: important players in ribosomal function. Mol. Microbiol. 2006;59:1651–1663. doi: 10.1111/j.1365-2958.2006.05054.x. [DOI] [PubMed] [Google Scholar]
- 4.Wool IG, Chan YL, Gluck A. Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol. 1995;73:933–947. doi: 10.1139/o95-101. [DOI] [PubMed] [Google Scholar]
- 5.Ignatovich O, Cooper M, Kulesza HM, Beggs JD. Cloning and characterisation of the gene encoding the ribosomal protein S5 (also known as rp14, S2, YS8) of Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:4616–4619. doi: 10.1093/nar/23.22.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galkin O, Bentley AA, Gupta S, Compton BA, Mazumder B, Kinzy TG, Merrick WC, Hatzoglou M, Pestova TV, Hellen CU, et al. Roles of the negatively charged N-terminal extension of Saccharomyces cerevisiae ribosomal protein S5 revealed by characterization of a yeast strain containing human ribosomal protein S5. RNA. 2007;13:2116–2128. doi: 10.1261/rna.688207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 8.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae–tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 9.Robert F, Brakier-Gingras L. A functional interaction between ribosomal proteins S7 and S11 within the bacterial ribosome. J. Biol. Chem. 2003;278:44913–44920. doi: 10.1074/jbc.M306534200. [DOI] [PubMed] [Google Scholar]
- 10.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 11.Rose MD, Winston F, Heiter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 12.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;53:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller PP, Harashima S, Hinnebusch AG. A segment of GCN4 mRNA containing the upstream AUG codons confers translational control upon a heterologous yeast transcript. Proc. Natl Acad. Sci. USA. 1987;84:2863–2867. doi: 10.1073/pnas.84.9.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komar AA, Lesnik T, Cullin C, Merrick WC, Trachsel H, Altmann M. Internal initiation drives the synthesis of Ure2 protein lacking the prion domain and affects [URE3] propagation in yeast cells. EMBO J. 2003;22:1199–1209. doi: 10.1093/emboj/cdg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komar AA, Gross SR, Barth-Baus D, Strachan R, Hensold JO, Goss Kinzy T, Merrick WC. Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. J. Biol. Chem. 2005;280:15601–15611. doi: 10.1074/jbc.M413728200. [DOI] [PubMed] [Google Scholar]
- 16.Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harger JW, Dinman JD. Evidence against a direct role for the Upf proteins in frameshifting or nonsense codon readthrough. RNA. 2004;10:1721–1729. doi: 10.1261/rna.7120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foiani M, Cigan AM, Paddon CJ, Harashima S, Hinnebusch AG. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva AM, Wang D, Komar AA, Castilho BA, Williams BR. Salicylates trigger protein synthesis inhibition in a protein kinase R-like endoplasmic reticulum kinase-dependent manner. J. Biol. Chem. 2007;282:10164–10171. doi: 10.1074/jbc.M609996200. [DOI] [PubMed] [Google Scholar]
- 20.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dever TE, Yang W, Aström S, Byström AS, Hinnebusch AG. Modulation of tRNA(iMet), eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2.GTP.Met-tRNA(iMet) ternary complexes. Mol. Cell. Biol. 1995;15:6351–6363. doi: 10.1128/mcb.15.11.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phan L, Zhang X, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell. Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alwine JC, Kemp DJ, Stark GR. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc. Natl Acad. Sci. USA. 1977;74:5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch FF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Jacobs JL, Dinman JD. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 2004;32:e160. doi: 10.1093/nar/gnh157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.elBaradi TT, van der Sande CA, Mager WH, Raue HA, Planta RJ. The cellular level of yeast ribosomal protein L25 is controlled principally by rapid degradation of excess protein. Curr. Genet. 1986;10:733–739. doi: 10.1007/BF00405095. [DOI] [PubMed] [Google Scholar]
- 28.Maicas E, Pluthero FG, Friesen JD. The accumulation of three yeast ribosomal proteins under conditions of excess mRNA is determined primarily by fast protein decay. Mol. Cell Biol. 1988;8:169–175. doi: 10.1128/mcb.8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsay YF, Thompson JR, Rotenberg MO, Larkin JC, Woolford J.L., Jr Ribosomal protein synthesis is not regulated at the translational level in Saccharomyces cerevisiae: balanced accumulation of ribosomal proteins L16 and rp59 is mediated by turnover of excess protein. Genes Dev. 1988;6:664–676. doi: 10.1101/gad.2.6.664. [DOI] [PubMed] [Google Scholar]
- 30.Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N. Revealing modular organization in the yeast transcriptional network. Nat. Genet. 2002;31:370–377. doi: 10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- 31.Vilardell J, Warner JR. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol. Cell. Biol. 1997;17:1959–1965. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mager WH, Planta RJ, Ballesta JG, Lee JC, Mizuta K, Suzuki K, Warner JR, Woolford J. A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:4872–4875. doi: 10.1093/nar/25.24.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakovljevic J, de Mayolo PA, Miles TD, Nguyen TM, Léger-Silvestre I, Gas N, Woolford J.L., Jr The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol. Cell. 2004;14:331–342. doi: 10.1016/s1097-2765(04)00215-1. [DOI] [PubMed] [Google Scholar]
- 34.Ferreira-Cerca S, Pöll G, Gleizes PE, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005;20:263–275. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Perreault A, Bellemer C, Bachand F. Nuclear export competence of pre-40S subunits in fission yeast requires the ribosomal protein Rps2. Nucleic Acids Res. 2008;36:6132–6142. doi: 10.1093/nar/gkn625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Hershey JWB, Merrick WC. The pathway and mechanism of eukaryotic protein synthesis. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 33–38. [Google Scholar]
- 38.Beringer M, Rodnina MV. The ribosomal peptidyl transferase. Mol. Cell. 2007;26:311–321. doi: 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 39.Moore PB. The ribosome returned. J. Biol. 2009;8:8. doi: 10.1186/jbiol103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodnina MV, Wintermeyer W. Recent mechanistic insights into eukaryotic ribosomes. Curr. Opin. Cell. Biol. 2009;21:435–443. doi: 10.1016/j.ceb.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 41.Nierhaus KH. Decoding errors and the involvement of the E-site. Biochimie. 2006;88:1013–1019. doi: 10.1016/j.biochi.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 42.Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 44.Jivotovskaya AV, Valásek L, Hinnebusch AG, Nielsen KH. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol. Cell. Biol. 2006;26:1355–1372. doi: 10.1128/MCB.26.4.1355-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. Specific functional interactions of nucleotides at key -3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]