Abstract
Using a comparative genomics approach to reconstruct the fate of genomic regulatory blocks (GRBs) and identify exonic remnants that have survived the disappearance of their host genes after whole-genome duplication (WGD) in teleosts, we discover a set of 38 candidate cis-regulatory coding exons (RCEs) with predicted target genes. These elements demonstrate evolutionary separation of overlapping protein-coding and regulatory information after WGD in teleosts. We present evidence that the corresponding mammalian exons are still under both coding and non-coding selection pressure, are more conserved than other protein coding exons in the host gene and several control sets, and share key characteristics with highly conserved non-coding elements in the same regions. Their dual function is corroborated by existing experimental data. Additionally, we show examples of human exon remnants stemming from the vertebrate 2R WGD. Our findings suggest that long-range cis-regulatory inputs for developmental genes are not limited to non-coding regions, but can also overlap the coding sequence of unrelated genes. Thus, exonic regulatory elements in GRBs might be functionally equivalent to those in non-coding regions, calling for a re-evaluation of the sequence space in which to look for long-range regulatory elements and experimentally test their activity.
INTRODUCTION
Long-range cis-regulation is of central importance in evolution, embryonic development, and human disease. The loci of many developmental transcription factor genes (1) are spanned by clusters of highly conserved non-coding elements (HCNEs) (2), which demarcate the region containing long-range enhancer elements that regulate the gene’s expression (3,4). The spanned regions can extend more than a megabase around the corresponding target gene and are often gene-poor, or contain gene deserts. A substantial number of these regions, however, contain other unrelated genes whose introns contain HCNEs but which apparently are not subject to their regulatory effects. We have termed these genes bystander genes to distinguish them from target genes which are under HCNE-mediated regulation (5). We refer to the entire arrangement of target genes, bystander genes or gene deserts spanned by HCNE arrays as genomic regulatory blocks [GRBs, (5)].
One unresolved question concerning the evolution of HCNEs is when and how the HCNEs appeared in their current locations. The observation that whole-genome duplication (WGD) can disentangle HCNEs from bystander genes points to the possibility that HCNEs appeared in the region within ‘striking distance’ to their target gene. Since the sequences of most HCNEs in genomes with no recent WGD are unique, they might have been conscripted from the original intronic sequence of either the target or the neighboring (bystander) gene, or the intergenic sequence between them. Moreover, they kept emerging over the course of vertebrate (and Metazoan) evolution: there is evidence that many of these elements might have appeared in the tetrapod lineage after its separation from fish (6). Since these elements cluster around target genes and cover the entire span of GRBs, in some cases the spatial arrangement of HCNEs might play a role (unknown as of yet) in their regulatory mechanism that leads to turnover and recruitment of new elements. In accord with this possibility, it has been shown recently that many old repetitive elements in GRBs are also under purifying selective pressure (7), and that there are cases of cis-regulatory elements recruited from transposable element sequences (8,9).
The above observations led us to speculate that some of these regulatory elements might have been employed from DNA that already served other functions. The ability to code for protein is one of the most suitable functions to test this hypothesis, due to the characteristic evolutionary signature of selection on protein coding sequence. Therefore, we set out to investigate whether one of the most obvious functional elements in GRBs—coding exons—might show evidence for additional non-coding evolutionary pressure and thus indicate that they double as parts of regulatory elements of the same type and origin as their HCNE neighbors. A number of cases of ‘enhancers in protein-coding sequence’ have been studied individually at different levels (see Discussion section). Two recent studies (10,11) identified a putative Hox-Pbx responsive cis-regulatory sequence, which resides in the coding sequence of Hoxa2 and is an important component of Hoxa2 regulation in rhombomere 4. The authors found that this Hox-Pbx exonic element is embedded in a large 205 bp long ultraconserved genomic element shared by all vertebrate genomes, which suggests superimposed functional and evolutionary constraints on both coding and non-coding function.
GRBs have properties that allow us to identify cases of evolutionary separation of overlapping functional elements: they are the regions with the longest conserved gene order across distant vertebrates, with bystander genes apparently ‘locked’ into the conserved syntenic arrangement by the requirement that HCNEs remain in cis to their target gene (5). The support for the latter is provided by analysis of the fate of GRBs after WGD followed by partial rediploidization, where a fraction of bystander genes becomes disentangled from the ancestral lock-in with HCNEs controlling the target gene, as described in (5). Analogous examples of physical separation of intercalated functional elements have been described for protein-coding genes encoding intronic small nucleolar RNAs (snoRNAs) (12–14). Similarly, our model of GRBs makes an interesting prediction about the fate of overlapping coding and non-coding functions in exons of bystander genes after WGD: since the non-coding (regulatory) information is likely to target a neighboring (GRB target) gene, rediploidization will lead to the separation of the two functions at the duplicated loci in a subset of cases. The non-coding function should remain active in cis to the target gene, while the coding information for the bystander gene can remain functional at the other locus (Figure 1A).We should therefore be able to detect such overlaps computationally on a genome-wide scale, and, importantly, pinpoint those for which WGD resulted in separation of non-coding and protein-coding function. The GRB model also predicts that exons of target genes might have acquired this function, as corroborated by the Hox-Pbx exonic element described above. To make the detection of these elements and their interpretation as unambiguous as possible, we focused on coding exons of bystander (apparently unaffected by long-range regulation) genes and followed their fate after WGD in teleost fish. Since many bystander genes are broadly expressed (‘housekeeping’) genes that are likely to rediploidize (i.e. lose one of the two copies) following WGD (5), we expect the overlapping regulatory function in the ‘decayed’ copy that is in cis with the neighboring target gene to remain conserved as an isolated exonic remnant that could be tested for enhancer activity. In this article, we use a genome-wide computational approach to present evidence in support of this hypothesis.
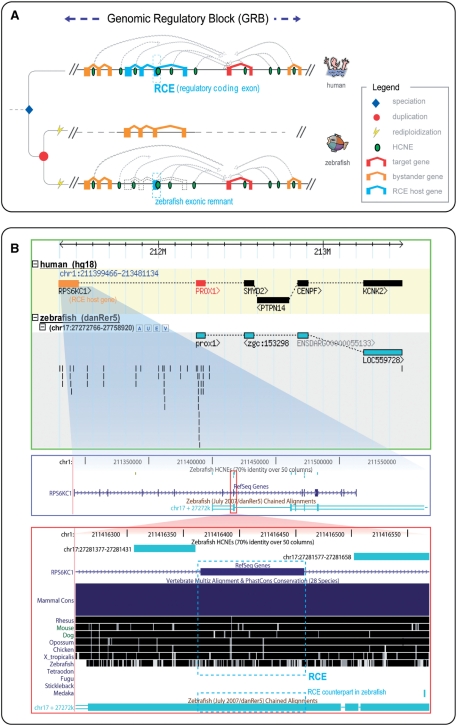
Figure 1.
The GRB model, the evolutionary scenario to define RCEs and an example of an RCE. (A) A GRB is defined as a genomic region where a target gene (red) receives long-range regulatory inputs from an array of HCNEs (green ovals) that span the entire GRB and often intertwine with exons of unrelated bystander genes (orange). The regulatory elements need to stay in cis to their target gene to function, leading to the conservation of synteny between the target and its long-range regulatory inputs. In the evolutionary scenario illustrated, teleost WGD (red circle) and subsequent rediploidization (yellow fork) resulted in each gene being retained in a single functional copy. However, one exon fragment (blue dashed frame) that overlaps a regulatory element was retained in duplicate, with one copy remaining in conserved synteny with the target gene just like the HCNEs, and the other remaining as part of a functional gene elsewhere in the genome. We named such zebrafish exonic remnants and their vertebrate orthologs RCEs, and named the genes they are or were part of ‘RCE host genes’ (blue). (B) The PROX1 - RPS6KC1 locus. The prospero homeobox protein PROX1, which is essential for early development of the central nervous system (CNS), is an example of a 1-to-1 GRB orthology scenario. PROX1 has a bystander gene RPS6KC1 in the synteny block defined by PROX1 and the HCNEs spanning the locus. RPS6KC1 encodes a ribosomal protein kinase, which has no evidence for involvement in CNS development or for being tightly regulated in general. In this case, RPS6KC1, as the bystander gene, was lost in the zebrafish synteny block, leaving several human–zebrafish HCNEs in the gene desert created by its disappearance. Interestingly, three out of 15 exons were also kept as highly conserved remnants in the zebrafish (referred as RCE 9, 10, 11 in Supplementary Table S1).
MATERIALS AND METHODS
Data extraction
We downloaded genomic sequences (hg18, mm9 and danRer5 for human, mouse and zebrafish respectively) and whole-genome alignments from the UCSC Genome Browser Database (15), and transcriptome mapping information from Ensembl (release 49) (16). Using the method described in Ancora (17), human–zebrafish HCNEs were extracted by scanning pairwise human–zebrafish UCSC net alignments for minimal regions >50 nt with at least 70% identity.
Defining synteny blocks and their gene content
Starting from 215 putative GRB target genes selected from zebrafish–human HCNE density peaks in Ancora (17) and their gene functions in development and/or as transcription factors, we retrieved the human-zebrafish synteny blocks overlapping with the target genes. Synteny blocks were calculated from UCSC Genome Browser human–zebrafish net alignments (15) joined in a graph-based procedure using a gap threshold of 450 k bp in the human genome and 150 k bp in the zebrafish genome, as previously described (5). This procedure allows for inversions and other local rearrangements such that syntenic blocks are separated by macro-rearrangements rather than smaller insertions and alignment gaps.
For each synteny block overlapping with the target gene, we retrieved all Ensembl protein-coding genes in the block [excluding the target gene(s)] as putative bystander genes. For each bystander gene, we checked its orthologous gene(s) in zebrafish, using a complete ortholog set composed by known Ensembl ortholog set plus an additional ortholog prediction set defined with reciprocal exon alignment coverage (18). Those bystander genes, which did not have an ortholog in the corresponding synteny block in zebrafish, were labeled as candidate RCE host genes. Many of those candidate genes were however not lost from the zebrafish genome, but had orthologs elsewhere, outside of the ancestral block of conserved synteny.
Detection of exon remnants of bystander genes
We identified putative exonic remnants of bystander genes by counting the conservation of each exon for each candidate RCE host gene using human–zebrafish orthologous UCSC chain alignments. For each exon, if <15% of base pairs were aligned, it was counted as completely lost; otherwise, the aligned part in the exon was extracted and defined the remnant part of the host gene regulatory coding exon (RCE). The RCE term we use is broadly related to the coding regions under non-coding selection (‘CRUNCS’) used in Chen et al. (19), but we use a different definition here. To make the subsequent reading frame consistency check easier, we cropped 1 or 2 nucleotides from the ends of the region if it did not begin and end with complete codon. To remove exons that might still be functional as part of expressed transcripts, we excluded all candidates that showed evidence of expression in zebrafish by genomic overlap (≥1 bp) with known zebrafish spliced ESTs and mRNAs (as of 22 May 2009) obtained from the UCSC Genome Browser (15).
Background sets
As a proxy for neutrally evolving sequence, we used the human–mouse–dog ancestral repeats (ARs) obtained from Hardison et al. (20). To obtain an estimate of the local neutral rate whose variance is matched to the substitution rate estimate for the RCE regions, we selected the nearest local AR and trimmed it to a total ungapped alignment length matching that of RCE region. In this process, the local ARs had to fulfill each of four criteria: (i) no overlap with its local RCE segment, (ii) length of at least 100 bp, (iii) is longer than the locally matched RCE segment and (iv) is as close as possible to its locally matched RCE segment.
For each RCE region, we extracted a random subsequence of the same length as the RCE from the remainder of the full-length coding sequence (CDS) of its host gene. The orthologous sequences of randomCDS in mouse were extracted using UCSC chained alignment data.
Nucleotide substitution rates and conservation score in RCE
We extracted the human RCE regions (cropped to start and end with complete codons according to the reading frame) and identified putatively orthologous genomic regions in mouse using the human–mouse BlastZ net alignment from UCSC Genome Browser Database (15). Regions without a human–mouse alignment were excluded from the analysis.
As an indicator of selection pressure on an amino acid, the non-synonymous substitution rates to synonymous substitution rates ratio (Ka/Ks) was calculated using codeml in pair-wise mode (runmode = −2, model = 0). To compare the protein-coding selection pressure on the genes containing RCE segments, we calculated the Ka/Ks ratio for the RCE host gene and took two genes from the same GRB as references; the target gene (we took the one closest to the RCE host gene if more than one putative target gene was found inside the defined GRB), and a randomly chosen bystander gene that had a mouse ortholog.
The nucleotide substitution rates between human and mouse RCE orthologous pairs were computed by using baseml with REV substitution model and enforced molecular clock (runmode = 0, model = 7, clock = 1, ndata = 1).
We calculated the conservation score for each RCE, randomCDS and local AR alignment by dividing the total number of aligned identical nucleotides with the total length of alignment.
Nucleotide distance of 4D sites
We extracted all 4-fold degenerate (4D) synonymous sites from RCE segments, the RCE host genes, and a background gene set consisting of 1000 randomly chosen genes from the whole set of human protein-coding genes that had Ensembl orthologs in mouse. For each dataset, the 4D sites (and their orthologous sites in mouse) were retrieved and concatenated together to make a new alignment.
The nucleotide distance between orthologous 4D sites was computed with JC69 model (21), using the formula
where D is the expected distance; x represents the number of different nucleotides, and n is the total number of nucleotides. Since the distance is directly determined by the difference rate (x/n) rather than the length of the sequence, correction for the sequence length (i.e. normalization) was not necessary.
Scanning of transcription factor binding sites and protein domains
To analyze the transcription factor binding site (TFBS) content in the RCEs, we scanned the human–mouse alignment of the regions using JASPAR Core TFBS position weight matrices (PWMs) and a 90% relative score threshold (22). Putative TFBSes matching the RCE segments were extracted using Perl with TFBS modules (23). For comparison to the TFBS content in the RCE, we took two background sets, (i) the random CDS (defined as above) and (ii) the nearest HCNEs. The nearest HCNEs fulfilled the following criteria: (a) They were part of the set of HCNEs between human-mouse (≥50 bp, ≥98%) (17), (b) they were as close as possible to its local RCE, and (c) trimmed to the same length as its local RCE segment.
To analyze the over-represented TFBS familial profiles in the RCE, we scanned the three sets (RCE, randomCDS and nearest HCNE) with JASPAR_FAM TFBS matrix profiles representing generalized core motifs for 11 structural classes of transcription factors (22), and computed z-score as a measure of over-representation (24). We used the Perl module Statistics::Distributions from CPAN to calculate the P-value.
We also checked if any putative protein domains overlapped the RCE regions. We first scanned the RCE host gene with Pfam [release 23.0 (25)] with default parameters (global & local merged strategy, E-value: 1.0), keeping only those domains that overlapped with the RCE regions (by at least one amino acid). To obtain significant hits, we further filtered the results by limiting the E-value to ≤0.001.
Testing DNA elements for enhancer activity by transgenesis in zebrafish
Structure of the Tol2-based enhancer test vector
The basic enhancer test DNA vector contains the Gateway® C1 cassette, the zebrafish gata2 promoter (26) coupled to the GFP gene and the polyA signal, flanked by Tol2 terminal repeats, into which the test sequence was placed by LR recombination (27).
Microinjections and zebrafish handling
The mixture of DNA construct and Tol2 transposase mRNA was injected at a concentration of 25 ng/µl each into one-cell stage wild-type fertilized zebrafish eggs using glass capillaries. Injected fish were observed at 1 dpf and 2 dpf for GFP expression, raised to sexual maturity and screened to isolate transgenic lines. Detailed description of all procedures can be found in Navratilova et al. (27). Sequences were tested in at least four independent transgenic lines.
RESULTS
A candidate set of exonic remnants of ancestral bystander genes
We hypothesized that a subset of the sequences that contain coding exons in a GRB, in either target or bystander genes, has been recruited over time into regulatory elements functionally equivalent to those detected in HCNEs. After WGD, some of the bystander genes rediploidized such that one copy was inactivated, releasing the coding pressure on the embedded regulatory element and other exons in cis to it. Over time, the coding sequence deteriorated and left behind a remnant of the exon with a regulatory role only (targeting the GRB target gene). The approach is illustrated in Figure 1A. We investigated 215 curated GRB target genes [‘Materials and Methods’ section and (5,28)] spanned by arrays of human–zebrafish HCNEs for evidence of such a scenario. We defined the corresponding zebrafish-human synteny blocks around each target gene, and identified zebrafish orthologs and paralogs for every human gene inside the span of these synteny blocks (‘Materials and methods’ section). We identified a total of 38 zebrafish exonic remnants (Supplementary Table S1) that were retained in the zebrafish synteny block, but were no longer part of a functional coding transcript (Figure 1A and ‘Materials and Methods’ section). As evident from the human annotation, which we take to represent the ancestral (pre-3R WGD) gene state, the remnants were derived from 19 host genes (‘Host gene’ in Supplementary Table S1). In most cases, 1 or 2 individual exons of each host gene remained (Supplementary Figure S1). In many of them, the conservation extends into one or both flanking introns, in agreement with the idea that the sequence might have been recruited into its non-coding function independently of the exon’s coding role. We named the 38 originating exons RCEs (see full definition in ‘Materials and Methods’ section); again, their orthologous exonic remnants detected in zebrafish do not code for protein any more. We further characterized these regions using a series of computational approaches, to establish (i) if the zebrafish remnants are under non-coding selection only, (ii) if their mammalian orthologs still show evidence of dual coding + noncoding selection and (iii) if they have sequence properties equivalent to those of regulatory HCNEs in the region.
The GRB target genes can be divided into two main groups based on whether they have either one (1-to-1, singletons) or two zebrafish orthologs (1-to-2, co-orthologs), where both copies of the gene have been retained after teleost WGD; in other words, a 1-to-2 orthology means that one human (tetrapod) gene has two orthologs in a zebrafish (teleost) that are paralogous to each other: the two zebrafish paralogs are more closely related to each other than to their (common) tetrapod ortholog. These groups form a basis for interpretation of origin of RCEs from the bystander genes that were lost from the GRB in fish [see ref. (5) for examples]. Examples of GRB target genes with 1-to-1 orthology scenarios are PROX1 (Figure 1B), OTX2 and TSHZ1, while examples of the 1-to-2 scenario are PAX6 (Supplementary Figure S2), ZIC2, LHX1, SP3, etc.
Additional information about the 38 remnants of coding regions and the corresponding potential RCEs in human, including genomic coordinates in hg18 and danRer5, sequences, alignments, the number of HCNEs within the orthologous human host gene, and the primers for PCR amplification for possible experiments are given in the Supplementary Data file.
Assessment of coding potential of exon remnants
The annotation of the zebrafish genome is presently incomplete, making unambiguous ortholog gene assignment difficult, especially in the case of single-exon genes. To assure that the detected zebrafish exonic remnants do not code for protein any longer, we used three evaluation criteria. Specifically, using the human:zebrafish alignment for each retained zebrafish exon and the open reading frame (ORF) of the corresponding human gene, we searched the retained zebrafish sequence for: (i) splice site conservation: nearly all eukaryotic nuclear introns begin with the nucleotide sequence GU and end with AG (the GU–AG rule); conservation of this splice site identification signal indicates that the adjacent exons might still be transcribed/spliced; (ii) reading frame conservation: any insertions/deletions (indels) resulting in a disrupted ORF, which indicate that the exon may no longer be coding; (iii) presence of point mutations resulting in a stop codon.
Based on these three tests, we labeled the strength of evidence that coding potential of a zebrafish exon remnant was abolished. This was set as ‘class I’ for 28 of 38 remnant exons, indicating that a splice site was mutated, the ORF disrupted by indels, or that an internal stop-codon was found. Otherwise we assigned a lower confidence ‘class II’ level of evidence (the remaining 10 of 38 regions in Table 1). For the bystander gene(s) containing ‘class II’ RCEs, we investigated whether (i) they had an ortholog in the corresponding branch in at least two other teleost genomes (medaka, fugu, stickleback and tetraodon), (ii) the zebrafish ortholog (if any) of the originating host gene was outside the synteny block of the corresponding target gene. According to the GRB model (Figure 1A), if the gene is still functional elsewhere in the genome (e.g. outside the GRB), it is more likely to have disappeared from its original syntenic location, leaving behind regulatory elements (originally intertwined with its exons) in cis to the target gene. Besides the lack of known evidence of transcription (ESTs, mRNA) for the exonic remnants (see the Supplementary Data for details), these additional criteria suggest that all 38 RCEs, including those in class II, have lost their protein-coding potential in zebrafish.
Table 1.
The candidate set of RCE host genes
| ID | Target gene (Tg) | RCE host gene (Bg) | Number of total exons | Number of remnant exons | Number of class I(II) remnants | Number of HCNEs within host gene |
|---|---|---|---|---|---|---|
| 1 | PAX6 | ELP4 | 12 | 2 | 2 (0) | 36 |
| 2 | WT1 | EIF3M | 11 | 2 | 2 (0) | 2 |
| 3 | ZIC2 | PCCA | 25 | 3 | 1 (2) | 33 |
| 4 | EYA1 | KCNB2 | 2 | 1 | 1 (0) | 0 |
| 5 | PROX1 | RPS6KC1 | 15 | 3 | 3 (0) | 9 |
| 6 | TWIST1 | HDAC9 | 25 | 2a | 1 (1) | 2 |
| 3b | 3 (0) | 7 | ||||
| 7 | FOXP2 | PPP1R3A | 4 | 1 | 1 (0) | 0 |
| 8 | LHX1 | ACACA | 56 | 2 | 1 (1) | 13 |
| 9 | NR2F1 | O94914_HUMAN | 9 | 3 | 1 (2) | 1 |
| 10 | IRX3 | RPGRIP1L | 26 | 1 | 1 (0) | 8 |
| 11 | GSX2 | CHIC2 | 6 | 2c | 0 (2) | 3 |
| 12 | TSHZ1 | ZNF407 | 7 | 1 | 1 (0) | 36 |
| 13 | EVX2 | MTX2 | 10 | 2 | 2 (0) | 4 |
| 14 | ZNF536 | C19orf2 | 11 | 1 | 1 (0) | 1 |
| 15 | FIGN | KCNH7 | 17 | 1 | 1 (0) | 0 |
| 16 | LBXCOR1 | MAP2K5 | 22 | 3 | 1 (2) | 15 |
| 17 | SMAD3 | IQCH | 21 | 1 | 1 (0) | 2 |
| 18 | SP3 | OLA1 | 10 | 2 | 2 (0) | 12 |
| 19 | DLX2 | SLC25A12 | 18 | 2 | 2 (0) | 0 |
Summary table for 19 RCE host genes (Bg) and the corresponding target genes (Tg). For each RCE host gene, the number of total exons and retained exons are given. Each retained exon remnant was assigned to class I or II, according to the strength of evidence for loss of their coding potential. The number of intragenic HCNEs from each host bystander gene is given as an indicator of potential regulatory content of the host gene’s introns.
aBoth HDAC9 orthologs were lost from the synteny blocks of GRB target gene TWIST1 in zebrafish, which left two exonic remnants on zebrafish chromosome 19.
bThree on chromosome 16.
cIn the CHIC2 gene, there are two remnants on chr20:20493 k branch and they are identical in sequence and closely located in the chr20:23213 k branch in the Zv7 assembly, but both on the same branch are mapped to the same position in Zv8 assembly, which we considered as a Zv7 assembly error, and reported only one from each branch.
RCEs in mammals have low nucleotide substitution rates
Our next task was to investigate whether there is evidence for overlapping coding and non-coding selection pressure in mammalian orthologs of these exon remnants, i.e. potential RCEs that should correspond to their bifunctional ancestral (pre-WGD) state. We first investigated if the RCE regions were under purifying selection pressure by comparing the estimated rate of nucleotide substitution between human:mouse orthologs. The human:mouse comparison is suitable for several reasons; (i) zebrafish is a phylogenetic out-group relative to human and rodents, (ii) the rodent-specific rate of loss of HCNEs conserved in human and zebrafish is very low (29), and (iii) human and mouse are at an evolutionary distance that was shown to satisfactorily discriminate conserved regulatory elements from non-conserved flanking regions (30). We compared the nucleotide substitution rate for each RCE sequence (dRCE) to that of neutrally evolving ancient repeats (20,31) (dAR) from the genomic neighborhood, and to that of randomly sampled CDS from the corresponding host gene (drandomCDS) (‘Materials and Methods’ section). Ancient repeats (ARs) are non-coding and assumed to be non-functional, and most should reveal the baseline neutral nucleotide substitution rate for the examined genomic regions. Assuming that RCEs are uniformly distributed along the genes’ coding sequences, or that there is no positional bias for purifying selection pressure relative to bystander gene start, the drandomCDS is the expected substitution rate for coding sequence in the same genomic neighborhood. Thus, the ratios dRCE/dAR and dRCE/drandomCDS are expected to be close to 1 if selection has not distinguished between substitutions within RCE and substitutions within nearby ARs or random CDS. If any of these ratios were significantly lower than 1, this would be an indication that purifying selection on substitutions has been more prevalent in RCEs, or that underlying mutation rates are lower in RCEs.
Since RCEs are coding sequences in human and mouse genomes, their nucleotide substitution rates should be much lower (confirmed by median dRCE/dAR = 0.145, P < 7 × 10−12; paired Wilcoxon test, Figure 2A), and their sequence conservation in human:mouse alignments much higher (confirmed by P < 7 × 10−12; paired Wilcoxon test, Figure 2B) than that in the local (positionally matched) ancient repeats. Comparison of the transversion and transition rates of RCE regions with that of corresponding background sets also show significant difference (Supplementary Figure S3).
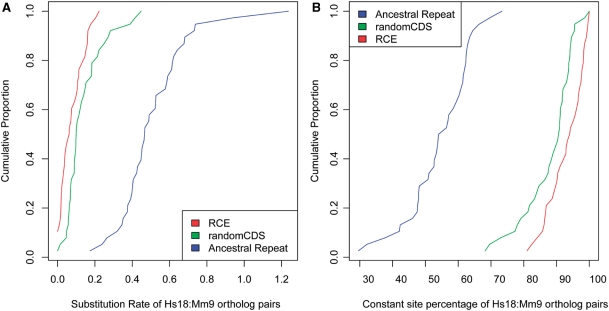
Figure 2.
Comparison of sequence conservation of RCE versus ancestral repeats and randomCDS. (A) Cumulative distribution of nucleotide substitution rates for 38 pairs of RCE region and local ancient repeats, and 38 randomly selected CDS regions from the same host genes. (B) Cumulative distribution of conservation scores for 38 pairs of RCE region and local ancient repeats, and randomly selected CDS regions from the same host genes.
We used an additional independent test with essentially the same result. Instead of using ancient repeats, we took all 4D synonymous sites from the ENCODE project (32) as a neutrally evolving reference (33), and computed the human:mouse conservation P-value [using phyloP (34)] for all RCEs and randomCDS regions. Similarly, we found that most RCEs and randomCDS regions are significantly more conserved than the ENCODE 4D sites, while the ancient repeats used in the previous test had similar P-values as 4D sites. Out of 34 RCE regions with significant conservation P-value (≤0.05), 26 had a smaller P-value than the paired randomCDS reference (Supplementary Figure S4).
The substitution rates in the human:mouse orthologs of the total set of 38 identified exon remnants were also significantly lower than that in paired random CDS regions (median dRCE/drandomCDS = 0.424, P = 6 × 10−4; paired Wilcoxon test) (Table 2, Figure 2A). The conservation scores were also significantly higher (P = 4 × 10−4; paired Wilcoxon test, see Table 2, Figure 2B). The difference was not substantially larger if we considered only the set of 28 RCEs with unambiguous loss of coding capacity (marked ‘class I’ in Supplementary Table S1). The median value of substitution rates ratio was 0.135 for dRCE/dAR (P < 7 × 10−9; paired Wilcoxon test, Table 2), and 0.424 for dRCE/drandomCDS (P = 0.002; paired Wilcoxon test). Both ratios were close to the corresponding ratio for the full set of RCEs, indicating that the two categories (‘class I’ and ‘class II’) of RCEs have similar constraint properties. We also compared class I to class II directly, which showed that there is no significant difference between them (P = 0.486, Wilcoxon test), adding confidence that ‘class II’ elements are indeed exonic remnants with non-coding function only.
Table 2.
RCE regions, random CDS and ancient repeats
| RCE region | Random CDS | P-value | Ancient repeat | P-value | |
|---|---|---|---|---|---|
| Median substitution rate | |||||
| All | 0.062 | 0.120 | 0.0006 | 0.467 | <7e-12 |
| Only high | 0.066 | 0.108 | 0.002 | 0.467 | <7e-09 |
| Median conservation score (%) | |||||
| All | 94.1 | 89.3 | 0.0004 | 53.7 | <7e-12 |
| Only high | 93.8 | 90.1 | 0.007 | 53.5 | <1e-10 |
Human:mouse substitution rate and conservation scores for RCE regions compared to random coding regions from their host genes. Remnants of bystander genes have significantly lower substitution rates and higher conservation than referenced CDS regions and ancient repeats (paired Wilcoxon tests).
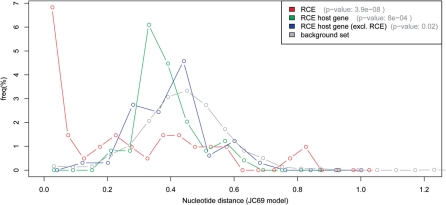
To account for the possibility that selection pressure could be heterogeneously distributed across protein-coding sequence of the considered genes, we investigated the selection pressure on their 4D sites, which should not be influenced by protein-coding selection pressure. We extracted all 4D sites within the RCE segments, the entire RCE host genes, and a background set of 1000 randomly sampled human genes. We retrieved the alignment of each human sequence and its corresponding mouse ortholog, and calculated the nucleotide distance for the 4D sites in each alignment (‘Materials and methods’ section). The nucleotide distances distribution 4D sites shows a clear peak towards zero for most human–mouse RCE pairs (median = 0.216), which was significantly lower than for RCE host genes (median = 0.398, P = 0.002) and the random background (median = 0.440, P < 4 × 10−8) (Figure 3). Importantly, the distribution of nucleotide distances for RCE host genes was also significantly different from the background (P = 8 × 10−4), underlining the characteristic properties of RCE regions. To exclude contribution of detected RCEs to its host gene, we also repeated the analysis excluding the 4D sites of RCEs from that of the RCE host gene set, showing that the peak of distance distribution for the remainder of the host gene shifted towards the center of the background set (median = 0.414, P = 0.02, see Figure 3, Supplementary Table S2). Thus, the 4D synonymous sites within RCE regions are under significantly stronger purifying selection pressure than both other regions in the same gene and the genome average.
Figure 3.
Nucleotide distance of 4D sites. Histogram of nucleotide distances of RCE 4D sites (red line), RCE host gene 4D sites (green line), RCE host gene excluding the RCE (blue line), and the 4D sites from 1000 randomly selected human:mouse orthologous gene pairs (grey line). The P-value in the legend represents the significant difference level between the corresponding set and the random background set.
TFBS evidence and protein domain content on RCEs
The underlying reason for the observed additional purifying selection pressure acting on RCE regions remains to be explained (Introduction section). To examine the possibility that a high density of putative TFBS could partially account for non-coding selection pressure, we compared the TFBS composition within RCEs to random CDS regions from the genes hosting the RCEs, and the nearest HCNEs using JASPAR_FAM familial TFBS profiles (35) (‘Materials and Methods’ section). We performed relative over-representation analysis on them by using the JASPAR_FAM database (22). We found that three out of 11 TFBS familial profiles (ETS, REL and MADS) show significant difference (P < 0.05) between the RCE set and the randomCDS set. Among them, the ETS class also showed a significant difference between RCE and HCNE sets (see Supplementary Table S3). Due to relatively small number of RCEs, however, the observed differences are not conclusive.
Protein domain content of RCEs
Since it can be envisaged that the amount and distribution of the regulatory sequence that can be accommodated in an overlap with protein-coding information will depend on evolutionary constraints on the underlying protein sequence, we investigated whether any of the known protein domains or types of domains were prevalent in overlap with RCEs. In total, half (19 out of 38) RCEs were found to overlap partially or completely with one or two out of 21 protein domains from the Pfam database with significant E-value (E < 0.001) (‘Materials and methods’ section and Supplementary Table S4). Based on this limited sample, there does not seem to be a preference for a particular protein domain to host overlapping regulatory regions.
Effect of RCEs on the Ka/Ks ratio of the underlying protein-coding sequence
The ratio of the rate of non-synonymous substitutions (Ka) to the rate of synonymous substitutions (Ks) is frequently used as an indicator of selection pressure acting on protein-coding genes. To investigate to which extent the RCE regions have influenced the fate of its host bystander gene, we compared the Ka/Ks ratios (also denoted ω or dN/dS) for each RCE host gene to the target gene and a randomly chosen bystander gene from the corresponding GRB (‘Materials and Methods’ section). For any given pair, a Ka/Ks < 1 is indicative of purifying selection and a Ka/Ks > 1 is consistent with positive selection (36,37).
Since target genes of long-range cis-regulation most often encode transcription factors and other development-related genes with clear orthologs of related function present across Metazoa (38), one might expect them to be more constrained and to have stronger selection pressure on them than the other genes in the corresponding GRB. For a gene with an exonic region doubling as a regulatory element (e.g. RCE host gene), one expects that the additional constraint would give rise to a stronger purifying selection, which is eventually reflected in a lower Ka/Ks ratio. Indeed, we found that the Ka/Ks values were 3.7-fold lower for target genes when compared to randomly chosen bystander genes (P = 3.4 × 10−3, Wilcoxon test), but only 1.6-fold lower than for bystander genes containing RCEs (P = 0.1, Wilcoxon test) (Table 3, Supplementary Figure S5A). We also compared the distributions of conservation scores, and again observed differences (P = 6.0 × 10−3) for random bystanders versus target genes, but not for RCE host genes versus target genes (P = 0.3, Wilcoxon test) (Table 3, Supplementary Figure S5B).
Table 3.
RCE host genes and random bystander genes
| Target gene | RCE host gene | P-value | Random bystander gene | P-value | |
|---|---|---|---|---|---|
| Median Ka/Ks ratio | 0.048 | 0.062 | 0.1 | 0.153 | 0.003 |
| Median conservation score (%) | 90.2 | 88.5 | 0.3 | 85.2 | 0.060 |
Human:mouse Ka/Ks and conservation scores for RCE host genes and random bystander genes compared to target genes from the corresponding GRBs. P-values for the comparisons were computed by a paired Wilcoxon test.
We did not observe any overall significant difference in either in Ka/Ks ratio or in conservation score between the 19 RCE host genes and other, randomly sampled, bystander genes. However, there was a trend for enrichment of low Ka/Ks ratios (for example, Ka/Ks < 0.17, see the dotted gray line in Supplementary Figure S5A) for RCE host genes, compared with randomly chosen bystander genes. As the Ka/Ks ratio drops (stronger purifying selection pressure), the RCE host genes, unlike other bystander genes, show a composition of constraints similar to that observed for target genes. This biphasic property of the bystander genes indicates that they probably represent a mix of constrained (regulatory element overlapping) and non-constrained cases.
Chromatin signature evidence for RCE function
From the evolutionary behavior of RCE regions, it appears likely that they do have a cis-regulatory role, even if they are not independent enhancers. To probe further into data supporting that the RCEs are regulatory, we looked for epigenetic marks that are hallmarks for enhancers: p300 (39), H3K4me1 (in absence of H3K4me3) (40), and also for CTCF binding sites (41). Several recent studies have used ChIP-Seq technology to generate high-throughput data for these markers; currently, data are available only for a handful of cell lines/tissues, but helpful enough for a preliminary analysis.
We found support for 13 of the RCEs to have epigenetic signatures of enhancers, with no conflicting epigenetic marks that we could find (Supplementary Table S5). Among them, eleven RCEs were found to overlap with enhancers predicted by the presence of H3K4me1 in the absence of H3K4me3 (see Supplementary Figure S6A for example cases in gene RPS6KC1).
To make sure that this observation is indicative of enhancer activity of the RCEs, we verified to which extent the overlapping of tissue- or cell-type-specific chromatin enhancer marker(s) is a general feature of developmental enhancers. We checked the p300 sites overlapping with known developmental enhancers from the VISTA Enhancer database (42), which contains human non-coding conserved fragments whose enhancer activity was tested experimentally in 11.5 day mouse embryos. Out of 496 positive enhancers in the database (as of 25 September 2009), 202 (40.7%) were found to overlap with p300 sites in at least one of the three embryonic tissues (limb, midbrain, and hindbrain) in which p300 binding was determined by ChIP-seq (39). This indicates that the tissue-specific p300 enhancer data can be used for a general enhancer verification purpose on large enough collections of elements. We also found that 33.7% (167 out of 496) VISTA enhancers overlapped with regions marked by H3K4me1 (in the absence of H3K4me3), a pattern argued to denote enhancers active in a specific cell type [see ref. (40)]. This indicates that, on the whole, the patterns appear to be different between cell types, but not necessarily that each mark is present exclusively in one cell type.
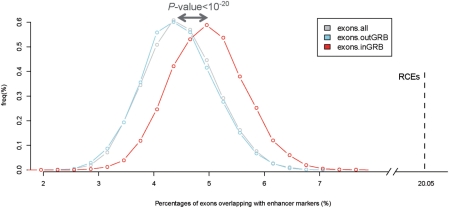
To investigate whether the overlap of RCEs with p300 and H3K4me1/-H3K4me3 marks was greater than expected by chance, we compared the RCEs with several background sets using a random sampling approach. We extracted all human exons (from Ensembl protein-coding genes, ‘exons.all’) and divided them into two groups (‘exons.inGRB’ and ‘exons.outGRB’), depending on overlap with any of the GRB loci used in this study (‘Materials and methods’ section). We randomly sampled 1000 exons from each of the sets (exons.inGRB, exons.outGRB and exons.all) and computed how many overlapped with at least one of the enhancer markers [p300 from Visel et al. (39), H3K4me1 in the absence of H3K4me3 from Heintzman et al. (40)]. We repeated the random sampling 10 000 times and compared the distributions of overlapping percentages for each set. A significantly higher fraction of exons in GRBs overlap with those regions, compared to the other two sets (Figure 4, sampling P < 10–20 in both comparisons, Wilcoxon tests). This is consistent with enhancers being enriched in GRBs, compared to the rest of the genome. But, in addition, it suggests that epigenetic marks indicative of enhancer function also overlap with coding sequence, and more often so in GRB regions. This overlap is even more pronounced if we consider only the RCE subset of exons, where our evolutionary analysis indicated overlapping coding and non-coding function. A total of 8 out of 38 (21%) of RCEs overlap with enhancer marks (Figure 4). This is much higher than the maximum value (∼8%) observed for the ‘exons.inGRB’ set, and reveals that the RCE set has a significant over-representation of characteristics that indicate enhancer function, compared to other exonic regions.
Figure 4.
Fraction of exons overlapping with enhancer markers. Histogram of percentages of exons in GRBs (red), exons outside of GRBs (light blue) and all exons (grey) that overlap with enhancer marks (p300 and/or H3K4me1). The percentages were calculated based on 10 000 sets of 1000 randomly sampled exons for each category. The percentage of RCEs overlapping with enhancer marks is indicated by a vertical dotted line.
To further demonstrate the over-representation of enhancer marks in RCEs, we compared the fraction of RCEs overlapping with H3K4me1 (but not H3K4me3) to that of HCNEs. Many HCNEs were found to act as enhancers (about 50% based on conservation only and practically all where the conservation was accompanied by p300 binding (39). We took 12 HCNE datasets from Ancora (17) with different sequence identity thresholds between human and other four species (mouse, dog, chicken and zebrafish, Supplementary Figure S7). For each set, we computed the percentage of HCNEs in GRB regions overlapping with H3K4me1. The percentages ranged between 8.0% and 10.4%, with a mean of 9.7%. The RCE set had an even higher percentage of H3K4me1 marks (15.8%) than any of the HCNE sets. This adds further evidence in favor of the hypothesis that many RCEs act as enhancers in a manner equivalent to the neighboring HCNEs in GRBs.
Transgenic evidence for the enhancer activity of ELP4 RCE1 and its zebrafish exonic remnant
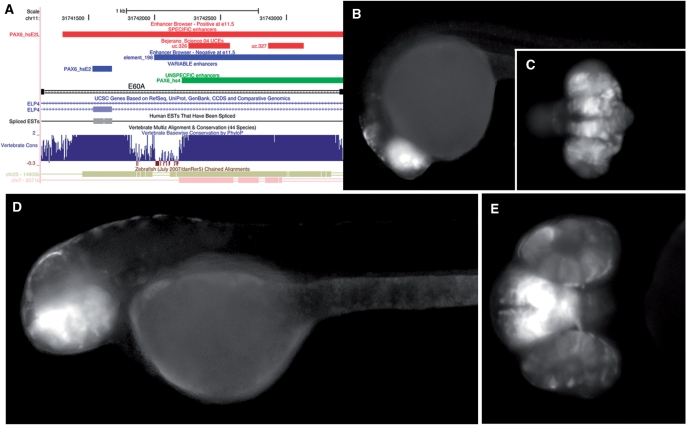
While systematic tests for RCE activity are unavailable, a larger 700 bp zebrafish region containing the exonic remnant of one of our RCE elements (RCE1) was recently tested for enhancer activity by Kleinjan et al. (43). In their study of subfunctionalization of duplicated zebrafish pax6 genes (pax6a and pax6b) by cis-regulatory divergence, they tested an HCNE-containing region (labeled E60A) next to pax6a without noting that it contains an exonic remnant of the bystander gene elp4. E60A drove expression in optic cup and forebrain, which are both parts of the expression pattern of the target gene PAX6 [Figure 5 in ref. (43)]. The result is interesting, but not conclusive regarding the role of the RCE sequence: the E60A fragment contains an HCNE, an exonic remnant, and another conserved region (Figure 5A), making it difficult to say which one is the core regulator to drive the expression pattern, or if they perform the regulatory function cooperatively. To examine the role of the RCE, the region was tested at a higher resolution: several independent transgenic lines for each sequence tested using the reporter method proven to be efficient and reliable for identifying vertebrate enhancer activity (27). While the exact RCE sequence (PAX6_hsE2) resulted in strong, but inconsistent expression patterns, the sequence extended to cover the flanking intronic HCNE (PAX6_hs4), labeling as PAX6_hsE2L in Figure 5A, drove reporter expression with high specificity and reproducibility in the retina and telencephalon, domains of the PAX6 gene endogenous expression (Figure 5B–E). The flanking intronic part of the overall conserved sequence alone did not show any enhancer activity in zebrafish. The result demonstrates that the RCE is an integral part of the regulatory element in question that is necessary, but not sufficient, to drive part of the PAX6 expression.
Figure 5.
Transgenic experimental evidence for one RCE element. (A) Screenshot from the UCSC browser (hg18) showing sequences tested, and results from the zebrafish enhancer assay (PAX6_hsE2L—specific, PAX6_hsE2—variable, PAX6_hs4—unspecific). Other tracks visualize UCRs (51), enhancer test results from the VISTA Enhancer browser (4) and an in silico PCR mapping of the sequence E60A tested by Kleinjan et al. (43). (B–E) Zebrafish transgenic lines expressing EGFP driven by PAX6_hsE2L. (B) Lateral view, 1dpf; (C) ventral, 1dpf; (D) lateral, 2dpf; (E) ventral, 2dpf.
The enhancer function of RCE1 is additionally supported by a large p300 binding site [forebrain/midbrain but not limb, mouse data from Visel et al. (39)] that coincides with the neighboring HCNE (PAX6_hs4, the region that did not drive expression on its own), but does not extend to cover the HCNEs immediately adjacent to the elp4 exon or the exon itself. (For the available cell lines, there are no signals for any of the chromatin state markers we examined in this region.).
RCEs inferred from the early vertebrate (2R) WGD
It has long been hypothesized that the increased complexity and genome size of vertebrates has resulted from (now firmly established) two rounds (1R and 2R) of WGD occurring in early chordate/vertebrate evolution, providing the requisite raw materials for the developmental regulatory networks of higher complexity (44). By plotting the genomic map positions of only the subset of paralogous genes that were duplicated prior to the fish–tetrapod split, Dehal et al. (45) showed that their global physical organization provides unmistakable evidence of two distinct genome duplication events early in vertebrate evolution indicated by clear patterns of four-way paralogous regions covering a large part of the human genome.
By analogy with the 3R (teleost WGD), exonic remnants revealing RCEs could have arisen in earlier WGDs as well, although most would be expected to have diverged beyond recognition by present day. However, for a number of large GRBs with the highest density of HCNEs [e.g. MEIS1/MEIS2 IRXa/IRXb clusters, (1)] there are still paralogous HCNEs with detectable similarity at the sequence level. We analyzed 2R paralogous loci for exonic remnants equivalent to those in zebrafish; in the 2R case, however, the RCEs should be present in all jawed vertebrates, including human (Supplementary Figure S7A). Using the UCSC selfChain data in human genome (hg18) (15), we investigated all possible paralogous GRBs and extracted all alignable regions that are exonic in one locus, but non-exonic in the other paralogous locus. We defined them as ancient RCEs, which are candidates for regulatory coding elements originating before the 2R WGD. We estimated the minimal GRB region by union of all human:teleost synteny blocks, which is expected to be smaller than the minimal synteny block size between human and pre-2R chordates (e.g. lamprey, a jawless vertebrate with a pre-2R WGD common ancestor with human).
We found three ancient RCEs in bystander genes by checking the alternative loss of coding property for the selfChain regions in the GRB regions for each paralogous target gene pair. Each of them overlaps an exonic region of a bystander gene, but its paralogous region in the human genome is not coding any longer. For example, the synteny block of SP3-CDCA7 is paralogous to that of SP4-CDCA7L (Supplementary Figure S7B), also supported by Ensembl phylogenetic tree for both SP3 and CDCA7 protein family (Supplementary Figure S8). Their chain alignment to lamprey shows that they both align to the same region of lamprey (Supplementary Figure S7B), which also reveals that the paralogous relationship is the result of the 2R WGD. DNAH11, a bystander gene located between SP3 and CDCA7, does not have a paralog in the intergenic region of SP4-CDCA7L block; however one of its exonic regions (chr7: 21569357-21570551 in Table 4) is found to align well to a non-coding region (chr2:174454487-174455587) between SP4 and CDCA7L. We predicted this non-coding region to be an exonic remnant left from rediploidization after 2R WGD. The other two cases of the exonic remnants are also found in bystander genes of the MEIS1/MEIS2 (gene C15orf41), and BARHL1/BARHL2 (gene C1orf146) GRBs (Table 4). Even though the function of the latter two bystander genes is unknown, their protein sequence is conserved across all vertebrates.
Table 4.
RCEs predicted in early vertebrate WGD
| Target gene 1 | RCE 1 coordinates | Host gene 1 | Target gene 2 | RCE 2 coordinates | Host gene 2 |
|---|---|---|---|---|---|
| SP4 | chr7:21569357-21570551 | DNAH11 | SP3 | chr2:174454487-174455587 | N/A |
| MEIS2 | chr15:34723710-34724239 | C15orf41 | MEIS1 | chr2:67348036-67348590 | N/A |
| BARHL2 | chr1:92478271-92479605 | C1orf146 | BARHL1 | chr9:134560677-134561963 | N/A |
RCEs predicted to have originated from early vertebrate (2R) WGD. The coordinates are based on UCSC human genome hg18.
We also found that the paralogous counterpart for one of these RCEs overlaps with the enhancer epigenetic marks mentioned above. The bystander gene C15orf41 in the GRB of MEIS2 has lost its paralogous gene in the ‘sister’ MEIS1 GRB, but one of its exons is still retained and conserved along most vertebrates (Supplementary Figure S6B). A strong signature of H3K4me1 binding (in the absence of H3K4me3) suggests it functions as part of an enhancer. Prediction data of regulatory potential (46) also suggests this is a regulatory element (the light blue track in Supplementary Figure S6B).
DISCUSSION
Using a hypothesis-driven comparative genomics approach, we detected a number of exonic remnants which, prior to the WGD in the teleost lineage, were likely bifunctional—coding exons doubling as regulatory elements or parts thereof. We corroborated this observation by showing evidence that the corresponding exons in mammals are still under both coding and non-coding selection pressure. The non-coding pressure was indicated by their significantly decreased nucleotide substitution rates and nucleotide distances of synonymous sites, when compared to neutrally evolving and protein-coding regions in the same genomic regions.
The idea that some coding exons might be under a combination of coding and noncoding selection pressure has recently received some attention. Xing and Lee (47,48) demonstrated that non-coding selection pressure can distort Ka/Ks values, making the metric unsuitable for annotating some exons in the genome or estimating the functional significance of amino acid residues encoded by them. More recently, several different probabilistic models were suggested for exons under different modes of selection pressure (4,19,49).
In particular, many facultative (occasionally skipped) exons were shown to have a high conservation of synonymous sites (50,51), presumably because the coding information is overlapped by regulatory inputs governing inclusion or skipping of these exons during splicing. However, under our model, this explanation for the noncoding conservation component is implausible since we explicitly detected exon remnants that lack evidence for being transcribed in zebrafish according to the UCSC genome browser ‘known zebrafish spliced ESTs’ and mRNA annotation (accessed 22 May 2009, ‘Materials and Methods’ section).
These observations imply that additional (non-coding) purifying selection pressure acts on RCE regions. This does not necessarily mean that all RCEs in our set have been subject to evolutionary constraint throughout the ∼500 Myr separating humans and zebrafish from their last common ancestor. While it is possible that some exonic remnants are indeed wholly or partly unannotated non-coding RNA, and others may have more recently lost their protein-coding ability, the available sequence evidence—including the absence of most of the other exons of the ancestral gene, frequent disruption of ancestral splice sites, and lack of EST support—indicate that this is a highly unlikely explanation for the majority of detected cases.
If the RCE regions have been subject to extra purifying selection from non-coding functional components, what is their function? Like the HCNEs that function as long-range regulatory sequences for their target gene(s) (2,5), the RCE regions appear to be part of the same array of conserved elements around a target gene responsive to long-range developmental regulation. Many of those elements have been shown to possess enhancer activity [from 50% in mouse (4,42) to close to 80% in zebrafish reporter assays (27,52)]. The conservation of detected RCEs often extends significantly into one or both of the flanking introns in tetrapod genomes, which indicates that the whole region must have been recruited into its non-coding function at some point. It was apparently not an obstacle that (part of) it coded for a functional part of a protein (Supplementary Table S4). This does not necessarily suggest that the entire lengths of exons that gave rise to RCEs, or that their—still exonic—orthologs in tetrapod are regulatory—the most we can claim without additional evidence is that the part of the ancestral exon that has been retained as an exonic remnant in zebrafish most likely has regulatory function.
Overlap between coding and regulatory sequence has been observed in genomes of bacteria (53) and viruses (54–57), and was explained as a way to minimize genome size. For vertebrates, where protein-coding regions make up only a small percentage of the genome, coding + regulatory overlap is not likely to be a space-saving strategy. Even so, the number of reported individual cases of such arrangements is growing. An early study revealed that interaction of transcription factor B-Myb with HSS8 (a hypersensitive site mapped to exon 2 of the Bcl-2 gene) may enhance Bcl-2 gene expression by cooperating with its promoter (58). Barthel and Liu (59) computationally identified a regulatory region associated with the gene ADAMTS5 that encompasses the entirety of the essential coding exon 2. The APOE gene was also found to contain an enhancer in its coding region for the E4 allele, which is associated with Alzheimer’s disease (60).
In this work, we did not attempt to find the RCEs overlapping the exons of the GRB target genes, since they cannot be detected as exonic remnants under non-coding selection. However, the high density of HCNEs in introns of target genes, as well as low rate of synonymous substitution at many of their exons indicates that exons of GRB targets might often overlap their own regulatory elements. The recently reported ultraconserved element in Hoxa2 (10) is one example of this. On the other hand, even though exons can be targets of RNA-mediated posttranscriptional regulation (10,61), this type of regulation requires the RCE to be transcribed, which cannot explain the selection pressure on isolated and apparently un-transcribed exonic remnants studied in this article.
Our results add support to the idea that HCNEs were recruited from existing sequences within regulatory reach of their target genes. A recent study demonstrated that a large number of repeat elements in regions that we now know as GRBs are also undergoing purifying selection (7). These findings should provide an incentive to test experimentally the detected exon remnants in zebrafish and their orthologs in human for the presence of enhancer activity. Suitable test systems exist in zebrafish (62), medaka (63) and mouse (4). If proven able to drive expression in a spatiotemporal pattern that recapitulates a subset of expression patterns of the neighboring gene, this would mean that we have to modify our view of both how protein sequences evolve and where to look for regulatory elements in vertebrate genomes. For protein sequences, it would mean that the non-coding component might mask the effect on selection at the protein level to an extent where it might be difficult to draw conclusions about functional importance of a part of a protein sequence based on its evolutionary conservation. For regulatory information, this will demonstrate that these exons are an integral part of the arrays of HCNEs, and that the non-coding component of the selection pressure that acts on them is equivalent to the pressure that kept HCNEs highly conserved for hundreds of millions of years. It would also suggest that the bystander genes were in place (i.e. in synteny to the neighboring HCNE target) before the HCNEs themselves appeared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Norwegian Research Council (170508); Bergen Research Foundation (BFS) (to D.F., Ø.D., B.L.); Young Future Research Leaders (YFF) program of the Norwegian Research Council (NFR); Sars Centre and the University of Bergen (to B.L., P.N., T.S.B.). Funding for open access charge: YFF grant 180435 from Norweign Research Council (NFR) awarded (to B.L.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Pär Engström for preliminary analyses related to this research, as well as Altuna Akalin, Jan Christian Bryne and other members of the Lenhard group for valuable advice and discussion. We also thank the ENCODE consortium, Broad Institute and Bradley E. Bernstein for making publicly available ChIP-Seq data used for checking epigenetic signatures of individual RCEs in this study.
REFERENCES
- 1.Sandelin A, Bailey P, Bruce S, Engstrom PG, Klos JM, Wasserman WW, Ericson J, Lenhard B. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics. 2004;5:99. doi: 10.1186/1471-2164-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimura-Yoshida C, Kitajima K, Oda-Ishii I, Tian E, Suzuki M, Yamamoto M, Suzuki T, Kobayashi M, Aizawa S, Matsuo I. Characterization of the pufferfish Otx2 cis-regulators reveals evolutionarily conserved genetic mechanisms for vertebrate head specification. Development. 2004;131:57–71. doi: 10.1242/dev.00877. [DOI] [PubMed] [Google Scholar]
- 4.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 5.Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engstrom PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephen S, Pheasant M, Makunin IV, Mattick JS. Large-scale appearance of ultraconserved elements in tetrapod genomes and slowdown of the molecular clock. Mol. Biol. Evol. 2008;25:402–408. doi: 10.1093/molbev/msm268. [DOI] [PubMed] [Google Scholar]
- 7.Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc. Natl Acad. Sci. USA. 2007;104:8005–8010. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishihara H, Smit AF, Okada N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006;16:864–874. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie X, Kamal M, Lander ES. A family of conserved noncoding elements derived from an ancient transposable element. Proc. Natl Acad. Sci. USA. 2006;103:11659–11664. doi: 10.1073/pnas.0604768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lampe X, Samad OA, Guiguen A, Matis C, Remacle S, Picard JJ, Rijli FM, Rezsohazy R. An ultraconserved Hox-Pbx responsive element resides in the coding sequence of Hoxa2 and is active in rhombomere 4. Nucleic Acids Res. 2008;36:3214–3225. doi: 10.1093/nar/gkn148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tumpel S, Cambronero F, Sims C, Krumlauf R, Wiedemann LM. A regulatory module embedded in the coding region of Hoxa2 controls expression in rhombomere 2. Proc. Natl Acad. Sci. USA. 2008;105:20077–20082. doi: 10.1073/pnas.0806360105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown JW, Clark GP, Leader DJ, Simpson CG, Lowe T. Multiple snoRNA gene clusters from Arabidopsis. RNA. 2001;7:1817–1832. [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CL, Chen CJ, Vallon O, Huang ZP, Zhou H, Qu LH. Genomewide analysis of box C/D and box H/ACA snoRNAs in Chlamydomonas reinhardtii reveals an extensive organization into intronic gene clusters. Genetics. 2008;179:21–30. doi: 10.1534/genetics.107.086025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tycowski KT, Shu MD, Steitz JA. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 15.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engstrom PG, Fredman D, Lenhard B. Ancora: a web resource for exploring highly conserved noncoding elements and their association with developmental regulatory genes. Genome Biol. 2008;9:R34. doi: 10.1186/gb-2008-9-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong X, Fredman D, Lenhard B. Synorth: exploring the evolution of synteny and long-range regulatory interactions in vertebrate genomes. Genome Biol. 2009;10:R86. doi: 10.1186/gb-2009-10-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Blanchette M. Detecting non-coding selective pressure in coding regions. BMC Evol. Biol. 2007;7(Suppl 1):S9. doi: 10.1186/1471-2148-7-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardison RC, Roskin KM, Yang S, Diekhans M, Kent WJ, Weber R, Elnitski L, Li J, O'C;onnor M, Kolbe D, et al. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 2003;13:13–26. doi: 10.1101/gr.844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z. Computational Molecular Evolution. Oxford: Oxford University Press; 2006. [Google Scholar]
- 22.Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenhard B, Wasserman WW. TFBS: Computational framework for transcription factor binding site analysis. Bioinformatics. 2002;18:1135–1136. doi: 10.1093/bioinformatics/18.8.1135. [DOI] [PubMed] [Google Scholar]
- 24.Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng A, Tang H, Ong BA, Farrell MJ, Lin S. Promoter analysis in living zebrafish embryos identifies a cis-acting motif required for neuronal expression of GATA-2. Proc. Natl Acad. Sci. USA. 1997;94:6267–6272. doi: 10.1073/pnas.94.12.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navratilova P, Fredman D, Hawkins TA, Turner K, Lenhard B, Becker TS. Systematic human/zebrafish comparative identification of cis-regulatory activity around vertebrate developmental transcription factor genes. Dev. Biol. 2009;327:526–540. doi: 10.1016/j.ydbio.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 28.Fredman D, Engstrom PG, Lenhard B. Web-based tools and approaches to study long-range gene regulation in Metazoa. Brief Funct. Genomic Proteomic. 2009;8:231–42. doi: 10.1093/bfgp/elp023. [DOI] [PubMed] [Google Scholar]
- 29.McLean C, Bejerano G. Dispensability of mammalian DNA. Genome Res. 2008;18:1743–1751. doi: 10.1101/gr.080184.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenhard B, Sandelin A, Mendoza L, Engstrom P, Jareborg N, Wasserman WW. Identification of conserved regulatory elements by comparative genome analysis. J. Biol. 2003;2:13. doi: 10.1186/1475-4924-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 32.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong WS, Nielsen R. Detecting selection in noncoding regions of nucleotide sequences. Genetics. 2004;167:949–958. doi: 10.1534/genetics.102.010959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siepel A, Pollard K, Haussler D. New methods for detecting lineage-specific selection. Res. Comput. Mol. Biol. 2006;3909:190–205. [Google Scholar]
- 35.Sandelin A, Wasserman WW. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J. Mol. Biol. 2004;338:207–215. doi: 10.1016/j.jmb.2004.02.048. [DOI] [PubMed] [Google Scholar]
- 36.Li WH. Molecular Evolution. Sunderland, Mass: Sinauer Associates; 1997. [Google Scholar]
- 37.Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18:486. doi: 10.1016/s0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- 38.Vavouri T, Walter K, Gilks WR, Lehner B, Elgar G. Parallel evolution of conserved non-coding elements that target a common set of developmental regulatory genes from worms to humans. Genome Biol. 2007;8:R15. doi: 10.1186/gb-2007-8-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleinjan DA, Bancewicz RM, Gautier P, Dahm R, Schonthaler HB, Damante G, Seawright A, Hever AM, Yeyati PL, van Heyningen V, et al. Subfunctionalization of duplicated zebrafish pax6 genes by cis-regulatory divergence. PLoS Genet. 2008;4:e29. doi: 10.1371/journal.pgen.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohno S. Evolution by Gene Duplication. Berlin, New York: Springer; 1970. [Google Scholar]
- 45.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xing Y, Lee C. Can RNA selection pressure distort the measurement of Ka/Ks? Gene. 2006;370:1–5. doi: 10.1016/j.gene.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 48.Xing Y, Lee C. Alternative splicing and RNA selection pressure—evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- 49.Blanchette M, Bataille AR, Chen X, Poitras C, Laganiere J, Lefebvre C, Deblois G, Giguere V, Ferretti V, Bergeron D, et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xing Y, Lee C. Assessing the application of Ka/Ks ratio test to alternatively spliced exons. Bioinformatics. 2005;21:3701–3703. doi: 10.1093/bioinformatics/bti613. [DOI] [PubMed] [Google Scholar]
- 51.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 52.Antonellis A, Huynh JL, Lee-Lin SQ, Vinton RM, Renaud G, Loftus SK, Elliot G, Wolfsberg TG, Green ED, McCallion AS, et al. Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet. 2008;4:e1000174. doi: 10.1371/journal.pgen.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagase T, Nishio S, Itoh T. Essential elements in the coding region of mRNA for translation of ColE2 Rep protein. Plasmid. 2008;59:36–44. doi: 10.1016/j.plasmid.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 54.Verdin E, Becker N, Bex F, Droogmans L, Burny A. Identification and characterization of an enhancer in the coding region of the genome of human immunodeficiency virus type 1. Proc. Natl Acad. Sci. USA. 1990;87:4874–4878. doi: 10.1073/pnas.87.12.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sokolowski M, Tan W, Jellne M, Schwartz S. mRNA instability elements in the human papillomavirus type 16 L2 coding region. J. Virol. 1998;72:1504–1515. doi: 10.1128/jvi.72.2.1504-1515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oberg D, Collier B, Zhao X, Schwartz S. Mutational inactivation of two distinct negative RNA elements in the human papillomavirus type 16 L2 coding region induces production of high levels of L2 in human cells. J. Virol. 2003;77:11674–11684. doi: 10.1128/JVI.77.21.11674-11684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Man M, Epel BL. Characterization of regulatory elements within the coat protein (CP) coding region of Tobacco mosaic virus affecting subgenomic transcription and green fluorescent protein expression from the CP subgenomic RNA promoter. J. Gen. Virol. 2004;85:1727–1738. doi: 10.1099/vir.0.79838-0. [DOI] [PubMed] [Google Scholar]
- 58.Lang G, Gombert WM, Gould HJ. A transcriptional regulatory element in the coding sequence of the human Bcl-2 gene. Immunology. 2005;114:25–36. doi: 10.1111/j.1365-2567.2004.02073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barthel KK, Liu X. A transcriptional enhancer from the coding region of ADAMTS5. PLoS ONE. 2008;3:e2184. doi: 10.1371/journal.pone.0002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen HP, Lin A, Bloom JS, Khan AH, Park CC, Smith DJ. Screening reveals conserved and nonconserved transcriptional regulatory elements including an E3/E4 allele-dependent APOE coding region enhancer. Genomics. 2008;92:292–300. doi: 10.1016/j.ygeno.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 61.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl Acad. Sci. USA. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de la Calle-Mustienes E, Feijoo CG, Manzanares M, Tena JJ, Rodriguez-Seguel E, Letizia A, Allende ML, Gomez-Skarmeta JL. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061–1072. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Conte I, Bovolenta P. Comprehensive characterization of the cis-regulatory code responsible for the spatio-temporal expression of olSix3.2 in the developing medaka forebrain. Genome Biol. 2007;8:R137. doi: 10.1186/gb-2007-8-7-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.