Abstract
Common fragile sites (CFS) are chromosomal regions that exhibit instability during DNA replication stress. Although the mechanism of CFS expression has not been fully elucidated, one known feature is a severely delayed S-phase. We used an in vitro primer extension assay to examine the progression of DNA synthesis through various sequences within FRA16D by the replicative human DNA polymerases δ and α, and with human cell-free extracts. We found that specific cis-acting sequence elements perturb DNA elongation, causing inconsistent DNA synthesis rates between regions on the same strand and complementary strands. Pol δ was significantly inhibited in regions containing hairpins and microsatellites, [AT/TA]24 and [A/T]19–28, compared with a control region with minimal secondary structure. Pol δ processivity was enhanced by full length Werner Syndrome protein (WRN) and by WRN fragments containing either the helicase domain or DNA-binding C-terminal domain. In cell-free extracts, stalling was eliminated at smaller hairpins, but persisted in larger hairpins and microsatellites. Our data support a model whereby CFS expression during cellular stress is due to a combination of factors—density of specific DNA secondary-structures within a genomic region and asymmetric rates of strand synthesis.
INTRODUCTION
Genetic instability is a hallmark of tumor initiation and progression. Multiple genomic changes, both at the nucleotide level as well as the chromosomal level, must occur for a cell to become cancerous. These cells, once transformed into a malignant state, are often very aneuploid and contain translocations, large deletions, amplifications, and other structural changes (1). Chromosomal fragile sites are loci that are prone to gaps or breaks within metaphase chromosomes under specific cell culture conditions, and may represent a particular mechanism for initiating genomic instability (2). Common fragile sites (CFS) are found in all individuals and are often deleted or altered within a broad range of cancers (3). Many CFS are located near or within tumor suppressor genes. Cellular treatment with aphidicolin (APH), an inhibitor of replicative polymerases α, δ and ε, induces breaks at CFS regions. FRA16D, among the most highly expressed CFS, is located within the WWOX tumor suppressor gene (4,5). This fragile site contains at least three distinct regions of homozygous deletion in the AGS stomach carcinoma derived cell line, and is near an miRNA gene (6). Non-small cell lung cancer cell lines frequently show absence of FHIT staining and loss of heterozygosity at 3p14.2, the location corresponding to FRA3B (7). This region is a location of deletion and translocation breakpoints in cancer cell lines (8). CFS also serve as preferred sites of sister chromatid exchange (9) and naturally occurring human papilloma virus 16 integration (10).
Numerous studies have set out to define common DNA sequence elements within CFS that are responsible for the observed instability. Rare fragile sites result from the genetic expansion of mini- and microsatellite sequences (11). CFS, in contrast, have not been shown to contain expanded repeat elements. Since the locations of these sites were determined cytogenetically, they often span megabases in length. Stable integration of FRA3B at ectopic sites in the genome resulted in increased DNA breakage and chromosomal rearrangements at the integration site, suggesting that intrinsic DNA features of CFS are critical for the observed chromosomal instability (12). Cell hybrids with chromosome 3 homologues containing various deletions in FRA3B have been studied in order to define the minimally essential region of fragility. These studies have shown mixed results whereby in most cases, fragility was reduced but not eliminated when a portion of FRA3B was lost (2,13). Complete deletion of FRAXB, however, prevented chromosomal breaks at this location (14). Computational studies to identify sequence features have also been mostly inconclusive. A large percentage of FRA3B contains LINE1 elements but these sequences were poorly represented in FRA16D and other sites (4). Conversely Alu repeats dominate in FRA16D (4). The Twistflex program, which calculates the twist angle between dinucleotide base pairs, was developed to measure flexibility within fragile sites (15). Seven CFS have been analyzed with this program and all have shown a high number of fragility peaks compared to surrounding regions (2). These peaks are enriched in A + T content, since the AT base pair has the greatest twist angle (16), and many contain AT/TA dinucleotide repetitive elements.
The rate of replication fork progression is not uniform through the eukaryotic genome (17). CFS have been found in late-replicating regions of the genome and may still be unreplicated by the end of G2 phase (18). CFS expression in vitro is induced primarily by cellular treatment with APH which may slow replicative DNA polymerases. In cells, clusters of flexibility peaks can act as sinks for the superhelical density generated ahead of the replication fork as the DNA unwinds (19), which may hinder efficient topoisomerase activity and decrease the processivity of the polymerase complex (20). Thus, inhibition or delay of DNA replication fork progression within CFS may be the underlying mechanistic basis of chromosomal instability, as stalled forks can lead to strand breaks that initiate chromosomal rearrangements (21).
Various proteins involved in DNA replication and cellular stress responses are critical for maintaining CFS stability. ATR and BRCA1 proteins play a role in the S- and G2/M-phase checkpoints which act to stabilize stalled replication forks, and knockdown of either protein increases fragile site expression in cells (22,23). The WRN helicase is also essential in CFS maintenance (24). WRN can interact with the p50 subunit of human pol δ (25). WRN can also support Saccharomyces cerevisiae pol δ during replication of tetraplex and hairpin structures formed by d(CGG)n trinucleotide repeats (26). The phenotype of WRN deficiency in cells includes replication defects and genomic instability, similar to the expression of fragile sites (27). Furthermore, loss of the WRN protein results in the cancer prone premature aging disorder Werner syndrome.
Here, we have directly tested the ability of pol δ and components of the replication machinery to replicate regions of FRA16D. Although replication stalling by way of polymerase stalling is a core requirement of hypotheses concerning CFS expression, to the best of our knowledge, no study has shown the biochemistry of polymerase progression through fragile site sequences at the nucleotide level. This study clarifies the molecular basis of CFS expression by elucidating the interactions of replicative enzymes and cis-acting elements during FRA16D replication.
MATERIALS AND METHODS
Proteins and reagents
Recombinant 4 subunit, human pol δ and PCNA were purified as previously described (28). Recombinant histidine-tagged WRN protein and the exonuclease-dead E84A mutant (X-WRN) were purified using a baculovirus/insect cell expression system and an AKTA Explorer FLPC (GE Healthcare, Piscataway, NJ), as previously described (29,30). The concentration of active protein was determined by Bradford Assay (BioRad, Hercules, CA) and standard helicase reactions with a 16-bp forked duplex. Purity was determined by SDS–PAGE and Coomassie staining. RFC was a kind gift from Dr Gregory D. Bowman. WRN fragments were expressed in Escherichia coli and purified as described previously (31,32). Human pol α-primase was purchased from Chimerx (Milwaukee, WI). Replication competent HeLa cytoplasmic extracts were purchased from Chimerx (SV40 DNA Replication Assay Kit).
Preparation of replication competent U2OS cytoplasmic extract
U2OS cells were cultured in T150 flasks at 37°C in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 5% fetal bovine serum. Pre-confluent cells were harvested using trypsin (Gibco) and cytoplasmic extraction was performed using the established protocol (33).
Vector constructs
Oligonucleotides (IDT, Coralville, IA) corresponding to sequences within FRA16D, (Genebank Accession Number AF217490), were as follows (FRA16D sequences, 5′–3′, contained in the final construct are indicated in CAPS):
Region1:AATCGTAAGTCATATAAGTACCTTTATATAAA ATATATATATATATATATGTGTATATTTATTTGAGTATAAATACTAA GGGTAATTCGTTAATAACTGATAGTTGAATACAATATCTTCTAGTGACGTACGGCGTTGCGC;
Region2:ggatccCCCCAAAACGGTGCAACCGGTCCGAC CAAAGTTTAAGGGCTGGAGGTCACTAGGAGGGTGGAGTC GGAGGGTTTCACGACCTTAATGTCTGTACTCGGTGGGGTGGACCGGATTGTGAA TTTACTTCTCAACTCCTTTGAAACGGGTCggatccgtgccaccg
Region3:ggatccAGAGTACAGAGTCGGAGGGTT CATCGACCCTGATGTTCCCGTCCGGGGGTACGTACCGATG TATATATATATATATATATATATATATATATATATATATAAAT ATATATGAAAAAAAAAAAAAAAAAAAAAAAAAAAATCATCTTTAggatccgtgccaccg
Region4:ggatccCTATATGTGGACCTCACACACGGGAAAA GATGAACGTAATGTGAAAAAAAAAAAAAAAAAAACTCTACCGCA GAACGAGACAACGGGTCCGACCTCACGTCACCGCTAGAGTCGA ATGACGTTGGAGGCTGAGGGTCCAAGTTCGCTAggatccgtgccaccg
Region5:AAAACTTCTGGTTTAACAAGGTAGACCTTTTTAAA TTCGTTGTTTTATTTGTTTCATCATATCCTGAATATTGTATTTCGTAT TTTATTTACAGGTACTCACATGGCACTATCTTTATTTACTAATTTATTTATT TACGTACGGCGTTGCGC.
T7 polymerase was used to synthesize the complementary strand using the primers, 5′-GCGCAACGCCGTACG-3′ for R1 and R5, and 5′-CGGTGGCACGGATCC-3′ for R2, R3 and R4. Fragile site sequences were inserted into the HincII or BamHI restriction site within the multiple cloning site of the pGEM3zf(−) vector (Promega Corporation). The presence and validity of the insert in pGEM3zf(−) was confirmed by restriction enzyme analysis followed by DNA sequence analyses. Constructs in either orientation were obtained for subsequent analyses of both complementary DNA strands. Log-phase cultures of plasmid-bearing E. coli strain DH5αIQ were infected with R408 helper phage for 3 h for the production of ssDNA, with the exception of R3 which was infected for 16 h due to difficulties in ssDNA production. ssDNA sequence integrity was reconfirmed by dideoxy sequence analysis.
Polymerase stalling assay
The G40 primer, 5′-GCATGCCTGCAGGTCG-3′, which initiates DNA synthesis at position 40 of pGEM3zf(−), was 5′ end-labeled with [32P] and hybridized to each ssDNA template. This positioning allowed for a minimum of a 14 bp sequence, designated as the running start, before the start of the fragile site sequence. Unless indicated otherwise, polymerase δ was added at a 20:1 molar ratio of enzyme:template, along with 1 µg of PCNA per 100 fmol of template DNA. We performed preliminary experiments with the control vector using different time points and concentrations of pol δ to optimize the reaction conditions necessary to obtain> 90% of extended primer products that have completely replicated the target region. Our in vitro experiments require high concentrations of pol δ to determine the intrinsic ability of the polymerase to cope with microsatellite sequences. High pol δ concentrations have been used in other studies, e.g. during extension of primed oligonucleotide or M13 DNA templates (28). The standard reaction conditions were 40 mM Tris pH 7.8, 1 mM DTT, 0.2 mg/ml BSA, 10 mM MgCl2, 0.5 mM ATP, 5 mM NaCl and 250 µM dNTPs. Reactions proceeded at 37°C and were terminated using stop dye. Reaction products were separated using an 8% denaturing polyacrylamide gel, and quantitated using a Molecular Dynamics Phosphorimager (Sunnyvale, CA). To analyze polymerase transit through specific regions, DNA reaction products were grouped into three classes for quantitation: 3′ to the CFS sequence (running start), within the CFS insert, and 5′ to the CFS insert [within the pGEM3zf(−) vector]. The number of DNA molecules within each class was determined by ImageQuant software quantitation, and corrected for background pixels. We defined the percentage transit through the CFS as the number of molecules 5′ to the CFS insert divided by the total number of molecules within the CFS insert + 5′ to the insert [×100]. Reactions with pol δ/PCNA were performed a minimum of three times. Error bars represent the standard deviation. Statistical significance was determined using a two-sample t-test with a P < 0.05.
Reactions with WRN helicase, exonuclease-deficient WRN (X-WRN), or WRN fragments were performed with a 1:1 molar ratio of enzyme to DNA. Reactions that contained WRN, X-WRN, or WRN fragments, as well as corresponding controls, contained 40 mM Tris pH 7.8, 1 mM DTT, 0.2 mg/ml BSA, 5 mM MgCl2, 5 mM NaCl, 2 mM ATP, 250 µM dNTPs, 1 µg PCNA, 0.05 mg/ml yeast tRNA at 37°C. Reactions using HeLa cell-free extracts contained 30 mM HEPES, pH 7.5, 7 mM MgCl2, 0.5 mM DTT, 4 mM ATP, 100 µM dNTPs, 40 mM phosphocreatine, 0.625 units creatine phosphokinase, 100 fmol template DNA and 12 µl extract (140 μg) in 25 µl total volume at 37°C.
Western analysis
Nuclear fractions were isolated from U2OS cells using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, IL) with addition of a protease inhibitor, according to the manufacturer’s protocol. Western analysis was performed on HeLa and U2OS replication competent cytoplasmic extracts and U2OS nuclear extract using 20 µg of total protein per sample as described previously (34). The membrane was incubated overnight at 4°C with a rabbit polyclonal antibody to WRN (Santa Cruz Biotechnology, Santa Cruz, CA), or mouse monoclonal antibodies to pol δ, pol ε, PCNA (BD Biosciences, San Jose, CA); pol α, RPA (Abcam, Cambridge, MA); RFC (Santa Cruz Biotechnology). Immunoreactive proteins were visualized using ECL+ (Amersham Biosciences, Piscataway, NJ) and scanned on a Molecular Dynamics Storm phosphorimager.
RESULTS
Development of the CFS replication assay
FRA16D is contained within a 270 kb region of chromosome 16q23.2 and has been extensively characterized (4,35–37). Sequences within a region between 170 and 195 kb of FRA16D, each ∼150 bp in length, were chosen to study DNA replication at the nucleotide level (Figure 1A), hereafter referred to as fragile regions. During molecular characterization of this CFS, the authors noted the presence of several AT-rich sequences associated with increased DNA flexibility, a physical feature that may contribute to DNA breakage. Our substrates include sequences identified as flexibility peaks in the original study (4). In addition, sequences were chosen based on their A + T content, ability to form secondary structure, microsatellite composition, and association with deletion endpoints in cancer cell lines (Table 1). All regions were predicted to form secondary structure by Mfold to some extent, although this potential varied greatly between sequences.
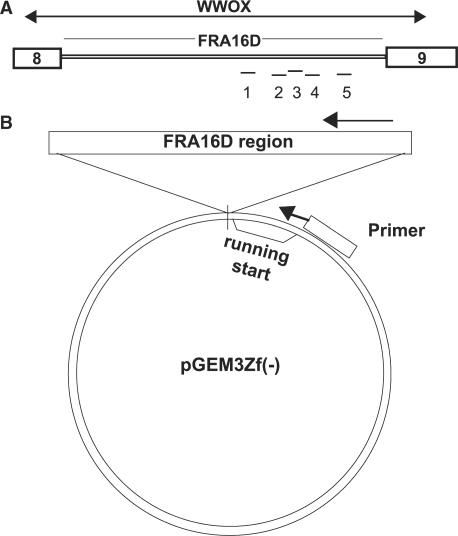
Figure 1.
Schematic of experimental design. (A) FRA16D is located within intron 8 of the WWOX gene. Approximate location of regions 1–5 (each ∼150 bp in length) within FRA16D are noted. Regions 2, 3 and 4 are continuous segments. (B) Oligonucleotides corresponding to each region were used to create double-stranded DNA fragements which were inserted into the multiple cloning site of pGEM3zf(−) vector, in both orientations. The final vector constructs contain common sequences both proximal and distal to the FRA16D region. DNA synthesis reactions are initiated from a common primer sequence (running start) within each vector.
Table 1.
Properties of FRA16D experimental sequences
| Region | Locationa | Flexibility Valueb | A + T (%) | Microsatellitesc | Predicted stem-loopsd |
|---|---|---|---|---|---|
| 1 | 170246 | 13.889 | 79 | [AT/TA]9 | 5 (3–7 bp) |
| 2 | 191417 | 8.163 | 47 | None | 6 (3–6 bp) |
| 3 | 191565 | 12.245 | 74 | [A/T]28, [AT/TA]24i | 4 (2–21 bp) |
| 4 | 191713 | 8.163 | 53 | [A/T]19 | 4 (2–12 bp) |
| 5 | 199256 | 10.526 | 75 | None | 3 (3–4 bp) |
aStart of region from WWOX sequence between exons 8 and 9, Genebank Accession No. AF217490.
bFlexibility values were determined using the Twistflex program. http://margalit.huji.ac.il/TwistFlex/index.html
cRepeative DNA sequence with≥ 4 units are catagorized as a microsatellite.
dTotal number of stem-loop structures capable of forming under the most thermodynamically favorable conditions as predicted by Mfold. Numbers in parentheses indicate the range of stem sizes in basepairs.
Region 1 contains an area of high flexibility, identified as Twistflex Peak 3 in Reid et al. (4), as well as an [AT/TA]9 microsatellite. Region 3 also contains a Twistflex peak (peak 5) and contains a [A/T]28 mononucleotide followed by an interrupted [AT/TA]24 dinucleotide microsatellite. Deletion breakpoints have been mapped in the AGS tumor cell line that are in the vicinity of this region (4,36). Regions 2 and 4 are sequences that immediately flank region 3, such that regions 2–4 represent 444 bp of contiguous sequence within FRA16D. Region 2 does not contain microsatellites but does have many pairs of small inverted repeats (IRs) that are <5 base pairs. Region 4 contains an [A/T]19 repeat and significant hairpin forming capability. Region 5 contains none of the characteristic properties of the other regions except that it is A + T rich, and serves as an internal control for DNA synthesis progression (Table 1). All regions were inserted within the pGEM3zf(−) vector which contains a primer site located 14–20 bases 5′ to the beginning of the fragile site sequence insert (Figure 1B). Regions were designated as the ‘A’ or ‘B’ orientation corresponding to a pair of complementary sequences. Our strategy was to synthesize oligonucleotides containing variable internal sequences that correspond to FRA16D, plus common flanking sequences (Figure 1B). In this way, we can utilize the common DNA sequences to prime and initiate synthesis, and normalize the extent of DNA synthesis that proceeds through the CFS sequence into a common flanking sequence.
Our initial observations showed that T7 polymerase required greater time to fully replicate regions 2, 3 and 4, with regions 3 and 4 showing the greatest amount of incomplete synthesis (Data not shown). ssDNA production was also less efficient for region 3, requiring a greater time of viral infection to obtain workable amounts of DNA. These observations indicate that regions 3 and 4 are inherently difficult to replicate.
Regions 2, 3 and 4 are inhibitory to polymerase replication
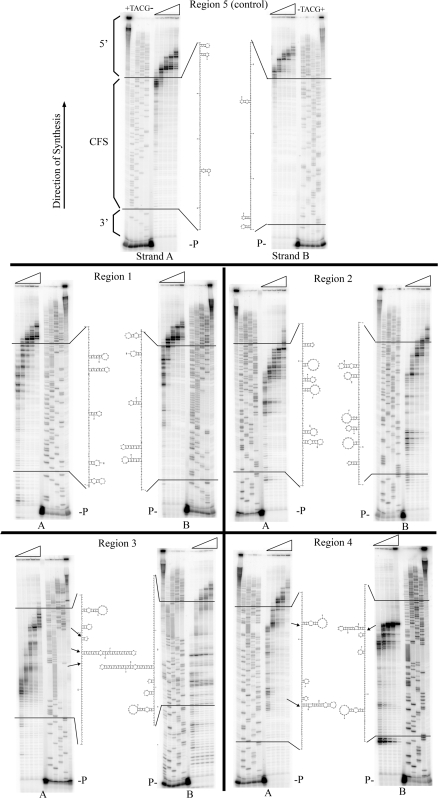
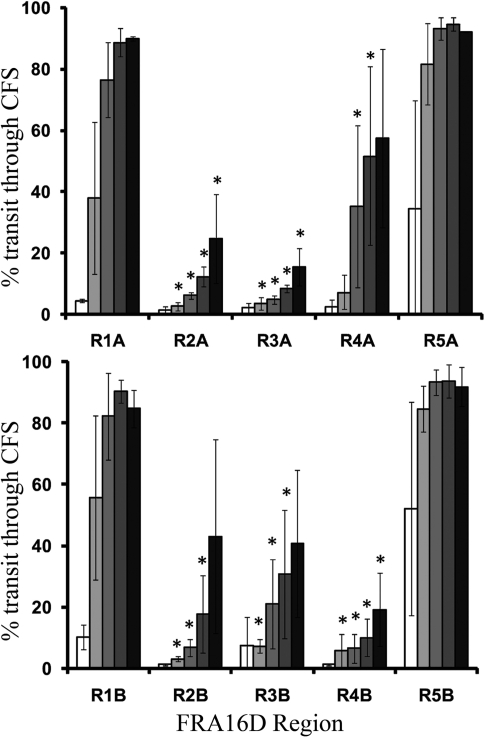
We investigated the progression of recombinant 4 subunit human DNA polymerase δ (pol δ) in the presence of PCNA through regions 1–5 of FRA16D under multiple hit conditions. An excess enzyme over template DNA was used to study polymerase synthesis progression, since it is known that the polymerase disassociates when stalled at repetitive elements (38). Under excess polymerase conditions, the polymerase will rebind to terminated nascent DNA and continue synthesis when the majority of the primer has been extended. Thus, the accumulation of DNA extension products of similar size should represent template sequences of slowed polymerase elongation or slowed translocation to the next template site, rather than sites of enzyme termination/dissociation. We refer to such product accumulation as polymerase stalling throughout this study. For our control region 5, located within FRA16D, we observed minimal stalling within the fragile region sequences at all time points and in either orientation, indicating that the polymerase was able to replicate the region quickly and without significant impediment (Figure 2, top panel). Quantitation, using the mean of at least three experiments, showed ∼80–90% of products represented completed transit through this control region within 5 min (Figure 3). Replication through region 1 showed stalling just before encountering the hairpin formed by the [AT/TA]9 repeat in orientation A, although by 10 min most of the stalling was alleviated (see Supplementary Figure S1 for details of secondary structures). At 15 min, 90% of the molecules had completed transit through this fragile region, similar to the control region 5. Somewhat surprising was that in orientation B, very little stalling was seen near the [AT/TA]9 run, but instead TTGXXXXXCAA and other small IRs were hotspots, albeit only at the lowest time points. However, no statistically significant differences were observed at any time points comparing region 1 to 5 (P > 0.05, two sample t-test).
Figure 2.
DNA polymerase δ stalling analysis within regions 1–5 of FRA16D. Reactions were performed using 100 fmol of single-stranded DNA primed with a 32P end-labled 15-mer which binds 14–22 bases before the inserted FRA16D region. DNA synthesis reactions contained 2 pmol pol δ, 1 µg PCNA and 250 µM dNTPS with standard reaction conditions. Representative polyacrylamide gels show polymerase δ synthesis progression through the indicated regions of FRA16D, in both the ‘A’ and ‘B’ complementary orientations, at time intervals of 2, 5, 10, 15 and 30 min (indicated by triangles). A sequencing ladder (TACG) is shown for each template along with no polymerase (−) and excess polymerase (+) controls. Total template-primer extension was 85–100% for all reactions. Sequences corresponding to FRA16D are enclosed between the black bars. Locations of the primer (P) and common sequences located in the vector that are proximal (3′) and distal (5′) to the CFS are indicated. The most stable secondary-structures predicted by Mfold at the standard reaction conditions are included (see Supplementary Figure S1 for higher magnification of sequences and secondary structures). Arrows refer to areas of stalling with predicted structural elements within the ssDNA template.
Figure 3.
Quantitation of pol δ transit through FRA16D regions. Reaction products such as shown in Figure 2 were quantitated for total radioactivity by Phosphorimager analyses, and grouped into three classes: proximal (3′) to the CFS insert, within the CFS, and distal (5′) to the CFS insert. The percentage transit through the CFS was calculated as the number of DNA molecules [distal to CFS/(within CFS + distal to CFS)] × 100. Analysis is shown for the ‘A’ (top panel) and ‘B’ (bottom panel) strand orientations. Increasing reaction times are indicated by increasing shades of gray from white (2 min) to black (30 min). Each bar represents the average of at least three independent experiments along with the standard deviation. Asterisks indicate statistical significance (P < 0.05, two-sample t-test), compared to region 5.
Regions 2, 3 and 4 are contiguous sequences within FRA16D, and increased pol δ stalling was observed within all three regions. Region 2 showed multiple, distinct areas of polymerase stalling (Figure 2). Due to the high number of small hairpins present throughout this region (Supplementary Figure S1), as well as the possibility of alternate secondary structures, it was impossible to pinpoint specific motifs responsible for stalling. At 15 min, only 12–18% of products had completed transit through region 2 (Figure 3). For region 3, we observed a significantly different rate of polymerase progression. Stalling occurred throughout the sequence in both ‘A’ and ‘B’ strands, but was clustered at IRs; notably at CCCXXXXXGGG and [AT/TA]24. Significant stalling was also observed at the [A/T]28 repeat. The stalling pattern at [AT/TA]24 revealed a periodic nature of progression, possibly arising as a consequence of partial unwinding of the hairpin stem ahead of the polymerase. The pattern was similar for the [A/T]28 sequence; since hairpin formation is not likely, this may be occurring as the polymerase bypasses other types of DNA secondary structure in short bursts. Overall, only 8% (strand A) or 30% (strand B) of the products had completed transit through the CFS sequence after 15 min. Region 4 had two major stall sites: within and towards the end of the [A/T]19 in the ‘A’ direction and at the base of the GGAGGTTGCAGTAAGCXXXXXXXGCCACTGCACTCC IR in the ‘B’ direction. Only a slight effect of this IR on pol δ progression was observed in the ‘A’ direction. Quantitatively, a strand bias was observed where 51% of product had completed transit through the CFS sequence in ‘A’ orientation at 15 min, but only 10% for the ‘B’ orientation; both results were significantly different from the control region 5 (P = 0.030 and 0.0001, respectively). Altogether, a total of 9–11 sequence motifs with statistically significant inhibition of pol δ synthesis were observed within a 444 bp region of FRA16D (Table 2). Although DNA synthesis inhibition was not absolute within any one region, the overall effect is expected to be additive on replication dynamics through this chromosomal location. The observed replication slow zone of ∼400–500 base pairs within FRA16D (regions 2–4) would result in progressively more inhibition as synthesis proceeded through the adjacent regions, although this prediction needs to be directly tested in future experiments.
Table 2.
Summary of inhibitory sequence motifs identified in FRA16D
| Inhibition observedb |
||||
|---|---|---|---|---|
| Region | Sequence | pol α | pol δ | Cell Extracts |
| 2 | GTCG(X)10CGAC | −/+ | −/+ | −/− |
| TTGXGAA(X)5TTCXCAA | −/+ | −/+ | −/− | |
| CGGT(X)3ACCG | +/+ | +/+ | −/− | |
| CACT(X)5GGTG | +/+ | +/+ | −/− | |
| 3 | [A]28 (peak c)a | +/+ | +/+ | +/− |
| [TA]23i (peak b) | +/+ | +/+ | +/+ | |
| hairpin cluster (peak a) | +/+ | +/+ | +/− | |
| 4 | [A]19 (peak d) | +/+ | +/+ | −/+ |
| Long IR (peak e) | +/+ | −/+ | −/+ | |
| GC(X)5GC | −/+ | −/+ | −/− | |
aPeaks refer to motifs indicated in Supplementary Figure S1.
b−/+ refer to inbition in complementary strands A/B.
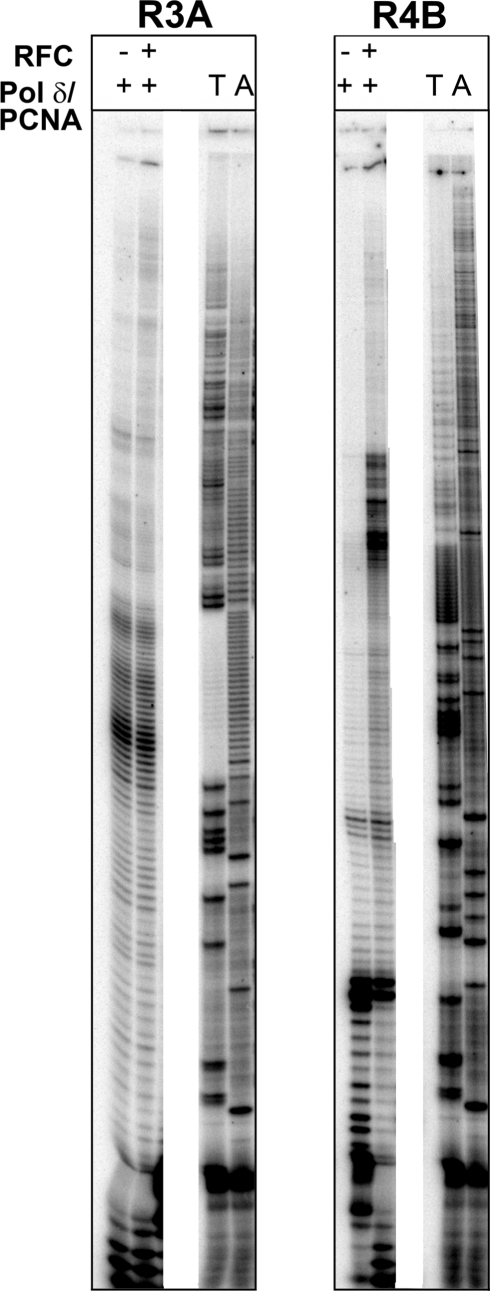
To assess whether other replication components could relieve pol δ stalling in difficult templates such as R3A and R4B, we tested extension with pol δ/PCNA along with the PCNA clamp loader RFC. For R3A where stalling occurred within the [A/T]28 and just prior to [AT/TA]24i, addition of RFC had no effect (Figure 4). In R4B where stalling by pol δ was near the beginning of the target sequence, RFC allowed further extension up to the base of the long IR near the end. Addition of SSB inhibited synthesis for both templates, possibly due to the lack of a mechanism to displace SSB.
Figure 4.
Replication by the pol δ holoenzyme. Effects of the clamp loader RFC on pol δ/PCNA processivity were observed. Reactions proceeded for 30 mins and contained 100 fmol template DNA, 200 fmol pol δ, 250 nM PCNA, 0.003 mg/ml RFC in 20 µl volume as indicated.
Synthesis through FRA16D regions 1–5 were performed with pol α-primase to determine if the stalling seen using pol δ was strictly sequence related, or due to both structure and polymerase biochemistry (Supplementary Figure S2). Pol α-primase showed minimal stalling within region 5, similar to pol δ reactions. Region 1A had stall sites near the short, double AT hairpins whereas R1B lacked any apparent inhibition. Stalling was present throughout region 2; again, due to the large number of hairpins and different conformation variances that are predicted, we cannot determine specific inhibitory structures in this region. Region 3 displayed strong pausing at the (A)28 in R3A. Stalling near the beginning of the (AT)24i hairpin was much less, compared to pol δ, and the periodic nature within the tract was not present. Higher amounts of primer extension in R3B allowed for the detection of various other, more minor, stall sites compared to R3A. Region 4 showed a prominent stall site at the large hairpin (arrow, Supplementary Figure S2) which was present in both ‘A’ and ‘B’ orientations; this is very different from the strand bias seen with pol δ, in which stalling is only seen near this hairpin in the ‘B’ strand. Very intense stalling was also observed at the [A/T]19 repeat, as seen with pol δ. These data show that the pol α-primase stalling profile is very similar to that of pol δ, aside from the mentioned differences. This suggests that replication rates are highly dependent upon DNA sequence, although each polymerase may also impart subtle differences.
Werner syndrome protein relieves stalling of polymerase δ
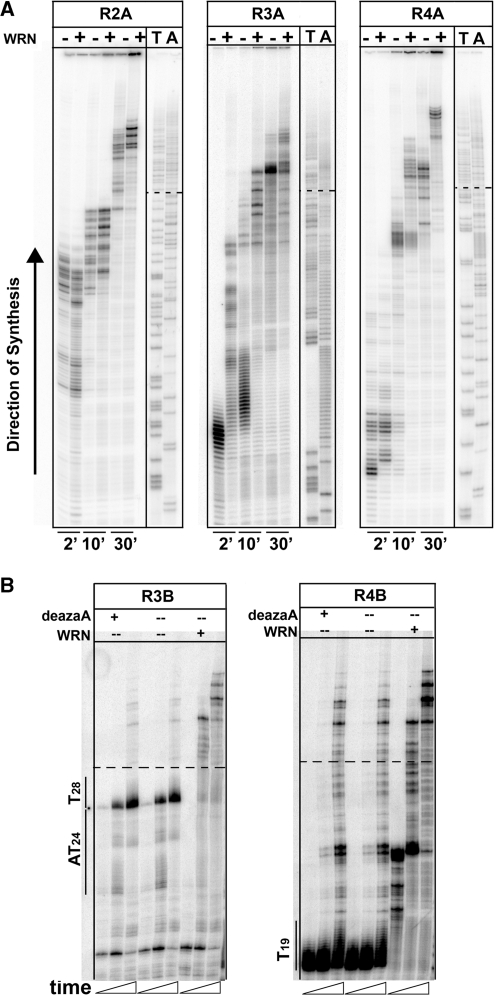
During the course of replication, cellular factors exist that specialize in aiding progression through complex sequences. Normally, fragile sites are well maintained in the absence of exogenous or endogenous stress. A mechanism in healthy cells to maintain fragile sites must involve bypass of inhibitory DNA elements. WRN is required for fragile site stability in vivo and is known to interact with pol δ. We studied the importance of WRN protein for allowing bypass of structures that were detrimental to pol δ progression. We found that WRN can alleviate stalling in FRA16D regions that are inhibitory to pol δ. This result is seen most dramatically in regions 3 and 4 at the [A/T] and [AT/TA] microsatellites (Figure 5A) where products of longer length are observed in the presence of WRN with pol δ/PCNA compared to pol δ/PCNA alone. Since the poly(dA/dT) tract cannot form hairpin structures, we sought to explain the pausing and subsequent alleviation by WRN. One possibility is the formation of triplex DNA whereby on the ‘A’ strand the template (A)n can fold back onto the nascent T·A primer-template duplex through Hoogsteen base pairing, creating an A–A·T triplet. On the ‘B’ strand, a similar situation can occur where the template (T)n strand can fold around the nascent strand and Hoogsteen base pair with the nascent DNA, forming a T–A·T triplet. To test for this on the ‘B’ strands, we substituted dATP with deaza-dATP which disrupts Hoogsteen pairing but does not alter Watson–Crick pairing. As seen in Figure 5B, disruption of Hoogsteen base pairing did not alleviate pol δ stalling within this sequence, suggesting that triplex DNA is not responsible for pausing. Again, however, pausing was alleviated by the presence of WRN at the poly(dA/dT) tracts.
Figure 5.
WRN stimulates pol δ progression through specific FRA16D sequence motifs. (A) Regions 2A, 3A and 4A were synthesized using pol δ + PCNA alone (−) or in combination with WRN (+). All reactions contained 100 fmol template DNA, 2 pmol pol δ and 1 µg PCNA. WRN (100 fmol) was supplemented as indicated. Reactions proceeded for 2, 10 and 30 min. Dashed lines: boundary of CFS insert. (B) Experimental testing for the presence of triplex DNA forming within the poly(dT) tract in regions 3B and 4B. Reactions as described in part A were repeated, except that deaza-dATP was substituted for dATP. Location of microsatelite DNA is indicated.
Next we examined whether other replicative polymerases could be aided by WRN during replication. We used pol α to replicate template 4B and observed a stalling profile similar to that with pol δ, with the two major sites of stalling at the end of [T]19, extending a few nucleotides beyond it, and at the base of the large IR described above (Supplementary Figure S3). Reactions were complemented with WRN to determine any effect through a protein complex. Although synthesis did not extend any further compared to pol α alone, stalling was reduced at the two major sites with the addition of WRN. WRN also reduced polymerization time through region 4; 50% transit occurred in 10 min with pol α/WRN compared to 30 min with pol α alone. This indicates that WRN can stimulate pol α polymerization, although the mechanism remains to be determined.
WRN increases DNA polymerization independent of its helicase or exonuclease activities
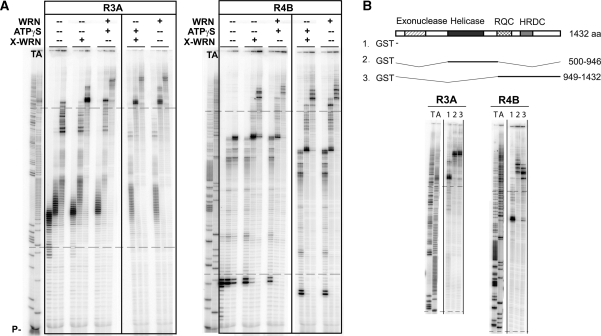
We sought to discriminate the particular function of WRN that contributes to the increased progression of pol δ through inhibitory sequences. One possibility is that the helicase domain unwinds secondary structure ahead of the polymerase. Alternatively, the exonuclease function of WRN may degrade the nascent DNA and allow the polymerase to restart synthesis. To test these possibilities, regions 3A and 4B were synthesized using pol δ plus wild-type WRN, while substituting ATPγS for ATP to inhibit helicase activity. Surprisingly, reactions with ATPγS were slightly more processive than reactions with ATP, especially for region 3A (Figure 6A). To confirm that ATPγS is inhibitory to helicase activity under the polymerase reaction conditions, an unwinding assay was performed using the exonuclease-dead variant (X-WRN) with a forked DNA substrate. With ATP in the reaction, 37–88% of substrate was unwound, compared to 1.6–7.2% with ATPγS (Supplementary Figure S4). We also substituted wild-type WRN with exonuclease-dead X-WRN, and again, the stalling profile was similar to wild-type WRN (Figure 6A). Finally, we used X-WRN and ATPγS to eliminate both the exonuclease and helicase functions. The stalling profile was once again unchanged, relative to the wild-type WRN. Overall, these results indicate that neither the helicase nor the exonuclease activities are necessary for WRN stimulation of pol δ mediated progression through these FRA16D sequences.
Figure 6.
WRN stimulates pol δ independently of its helicase or exonuclease activities. (A) Reactions contained pol δ/PCNA alone or with, wild-type WRN, or exonuclease-dead WRN (X-WRN) as indicated and ATPγS was included, as shown, to inhibit helicase function. Reactions proceeded for 2, 10 and 30 mins and contained 100 fmol template DNA, 2 pmol pol δ and 100 fmol WRN or X-WRN, as indicated. (B) Replication extension by WRN fragments. 1, GST control, 2. WRN500–946, or 3. WRN949–1432 were added (100 fmol) to pol δ/PCNA reactions. Gels in ‘B’ are cropped prior to the start of the fragile site sequence. Dashed lines: boundary of CFS insert.
To determine the mechanism by which WRN aids pol δ progression, we observed replication using GST fusion WRN fragments, WRN500–946 and WRN949–1432. WRN949–1432 contains an HRDC and RQC domain that are involved in protein–DNA and protein–protein interactions (39). A pol δ interaction domain was previously identified within the C-terminus of WRN, using a yeast two-hybrid approach (25). This C-terminal domain fragment was added to pol δ reactions on templates R3A and R4B (Figure 6B) to observe processivity in the absence of helicase activity. We observed products of higher length with WRN949–1432 compared to GST only. WRN500–946 contains the helicase domain. To test the ability of WRN to stimulate synthesis by unwinding secondary-structure, we tested stimulation of pol δ with this fragment and compared the extension profile with only GST added to the polymerase reaction. WRN500–946 was also able to stimulate pol δ progression. In R3A, the extension profiles were quite similar regardless of the WRN fragment. In R4B, higher length products were observed with the helicase containing WRN500–946, compared to WRN949–1432. This difference could reflect the necessity of particular enzymatic activities on certain templates. Regardless, both WRN fragments increased pol δ processivity, indicating that either fragment can act to traverse fragile site impediments (Figure 6B).
Specificity of WRN mediated pol δ stimulation during replication of inhibitory sequences
We wished to examine any stimulatory effect of pol δ by WRN in R5A, a template that is highly A+T rich, but contains no microsatellites, has minimal secondary-structure potential, and was not very inhibitory to pol δ replication progression. The addition of WRN did not alter the replication stalling profile of R5A, in which nascent strand termination was observed primarily 5′ to the CFS region (Supplementary Figure S5A). We further sought to determine if the stimulatory effect of WRN was confined to this specific protein, or if other helicases of the same family shared this function. The addition of E. coli RecQ to pol δ/PCNA reactions replicating R3A resulted in a pol δ stalling profile that is very similar compared to that of pol δ/WRN through this region (Supplementary Figure S5B). The stimulation by RecQ was not dependent upon its helicase activity, as demonstrated by the substitution of ATPγS for ATP. This result is consistent with our observation that the helicase is not essential for the stimulatory effect of WRN.
Replication progression using human cell-free extracts
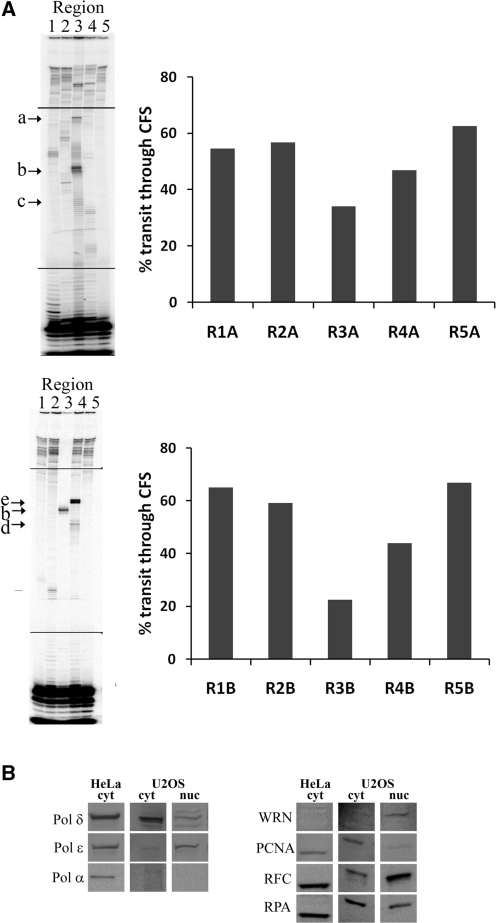
Multiple polymerases and protein complexes are involved in DNA replication, aside from pol δ and pol α-primase. To determine whether the sequence-specific stall sites observed for replicative polymerases in vitro also exist in the context of cellular replication proteins, we examined primer-extension reactions in replication competent HeLa cell-free extracts (Figure 7A). We confirmed the presence of replication proteins pol δ, pol ε, pol α, PCNA, RF-C and RP-A (Figure 7B). It was evident that some of the stall sites observed in reactions with pol δ alone were absent in the extract reactions (for example, region 2). This suggests that the less intense sites observed with pol δ alone may be overcome by other proteins present in the holoenzyme or cell extract. Importantly, however, stalling at the most inhibitory sites observed with pol δ (within regions 3 and 4) persisted in the extracts. In region 3A, stalling was at the very end of the [A]28, just prior to the start of the [AT]24 (peak b). In the complementary strand where the polymerase encounters the dinucleotide repeat before the mononucleotide repeat, stalling was prior to the [TA]24. In region 4A, only very minor stalling was noted, while in the complementary strand 4B, stalling was observed near the base of the hairpin GGAGGTTGCAGTAAGCXXXXXXXGCCACTGCACTCC (peak e) and, to a lesser degree, at the end of the [T]19 (peak d). Since WRN was undetectable in the extracts, we supplemented these reactions with a WRN:template ratio ranging from 0.02:1 to 10:1 (data not shown). No additional primer extension was observed with any concentration of WRN. Addition of APH to the extracts reduced the overall extent of DNA synthesis but did not alter the sites of stalling (data not shown). Finally, we observed a similar pausing profile using extracts from the U2OS cell line (Supplementary Figure S6). Overall, sites that are most inhibitory remain inherently difficult to replicate even in the presence of the holoenzyme including DNA pol δ, PCNA, RFC and RPA.
Figure 7.
CFS replication in HeLa cell extracts. (A) Replication-competent cell-free extracts were used for synthesis of regions 1–5 in both ‘A’ and ‘B’ orientations. Extract (140 µg protein) was supplemented with 4 mM ATP, 40 mM phosphocreatine and 0.625 units creatine phosphokinase. DNA synthesis reactions were stopped 1 min after the addition of 100 fmol primer-template DNA and 100 µM dNTPs. The fragile regions are enclosed in black bars. Arrows indicate sites of replication stalling at specific motifs: (a), near group of small inverted repeats; (b), base of [AT/TA]24i stem; (c), within poly(dA); (d), end of poly(dT); (e), base of long inverted repeat (see Supplmentary Figure S1 for sequences). Histogram shows percent of product that has completely replicated through the indicated FRA16D region, quantitated as described above. (B) Western analyses of HeLa replication-competent (RC) extract (20 µg), compared to U2OS cytoplamic RC extract and nuclear fractions (for full western blots, Supplementary Figure S7). Blots were probed with the indicated antibodies.
DISCUSSION
We directly tested the involvement of sequence composition upon DNA replication progression within a CFS, FRA16D. In our approach, we quantitated DNA synthesis through five distinct regions of FRA16D (Table 1). We have shown at the molecular level that regions containing poly(dA/dT) tracts or putative hairpin structures are inhibitory to replication of FRA16D, relative to regions devoid of such sequences (Figures 2, 3 and 7). Increased DNA flexibility and accompanying high A+T content was not sufficient to inhibit DNA synthesis within a DNA region. A total of 9–11 sequence motifs causing significant inhibition of human pol α-primase and pol δ synthesis were observed within a 444 bp region of FRA16D (Table 2). DNA synthesis inhibition due to any one sequence motif was not absolute. However, we propose that the overall effect is additive on replication dynamics through this chromosomal location, and the observed replication slow zone of ∼400–500 base pairs within FRA16D (regions 2–4) would result in progressively more inhibition as synthesis proceeds through the adjacent regions. Furthermore, we demonstrate that pol δ stalling is alleviated in the presence of the WRN protein (Figures 5 and 6), directly confirming its involvement in CFS maintenance at the nucleotide level.
Current models for the mechanistic basis of fragility have focused on secondary-structure formation, namely hairpins and triplex DNA (40). A particular sequence motif of high inhibition identified in this study was long IR sequences. Such motifs included the [AT/TA]24i microsatellite in region 3 (peak b, Figure 7A) and the imperfect IR in region 4 (peak e, Figure 7A; see Supplementary Figure S1 for sequences and structures). These sequences are inherently difficult to replicate, as significant stalling was observed in the presence of PCNA and RF-C (Figure 4) and in cell-free extracts containing pol δ, PCNA, RF-C and the single-strand binding protein RPA (Figure 7). We observed pol δ stalling at smaller IRs with fewer than 5 bp in the stem during DNA synthesis (Figure 2), but this stalling was mostly absent in cell extracts (Figure 7A). Previous studies have also shown a correlation of E. coli pol III and Drosophila melanogaster polymerase α pause sites with regions of base pair homology (41,42). Stable hairpin formation in vivo requires an IR size of at least 7 bp (43). IRs of this size are very prevalent throughout the genome, suggesting that under normal conditions, mechanisms are present which can largely bypass small hairpins. We have focused on polymerases proposed to be responsible for synthesis of the lagging DNA strand during replication: pol α-primase and pol δ. Secondary-structure formation is expected to occur more frequently on the lagging strand, since the DNA is often single-stranded. Furthermore, the size of our FRA16D regions is very comparable to eukaryotic Okazaki fragment lengths of 100–200 bp. Nonetheless, future studies are needed to observe replication progression by the replicative pol ε through CFSs.
Expanded [AT/TA] microsatellite sequences can form hairpin or cruciform structures, which may impede the progressing polymerase and delay replication (44,45). We observed strong pol δ stalling within the [AT/TA]24i microsatellite of region 3. However, only transient stalling was observed at the [AT/TA]9 microsatellite in region 1 (Figure 2). Mfold predictions place the observed stalling at the base of hairpin structures, suggesting that stems with lower thermodynamic stability are less inhibitory to pol δ. A length dependant relationship of [AT/TA] sequences and replication stalling in FRA16D was seen in yeast using 2D gel analysis (46). A study examining the progression of S. cerevisiae pol δ also found strong pause sites at the beginning of hairpin and tetraplex structures (26).
We observed prominent polymerase stalling within the [A/T]19 and [A/T]28 microsatellite sequences present in regions 3 and 4. While these sequences do not form any apparent secondary structures at the reaction conditions and sequence length described in this study, poly(dA/dT) tracts are capable of forming bent DNA. Structural analysis of bent DNA reveals a progressive narrowing of the minor groove from the 5′ end to the 3′ end of the tract (47). DNA bending has been shown to be toward the minor groove at the center of A6–8-tracts (47–49). We propose that for longer tracts, bending may occur in waves where the minor groove is most compressed every 6–8 bases. It has been previously suggested that structural changes to the duplex primer-template stem caused by DNA bending may disrupt polymerase–DNA interactions (50). The periodic nature of polymerase stalling at the long [A/T] microsatellites suggests that it may be due to narrowing and widening of the minor groove within poly(dA) tracts. These mononucleotide tracts cause termination by other polymerases as well, including pol α and pol κ (50). Difficulty during PCR synthesis has also been noticed for various segments of FRA16D containing poly(dA) and poly(dT) homopolymers (4). Together, these studies suggest a common cis-acting mechanism for stalling, rather than a specific polymerase related phenomenon.
We demonstrate here that the WRN protein may have redundant activities that aid pol δ during replication of FRA16D sequences, especially poly(dA/dT), observed with the WRN500–946 and WRN949–1432 fragments (Figure 6B). Increased replication rates by WRN500–946, which contains the helicase domain, may result from unwinding of secondary structure ahead of the fork, although direct polymerase stimulation is also possible through unidentified pol δ interaction sites. In the absence of the helicase, binding of WRN with pol δ may increase the rate of polymerization by modifying replisome conformation. Our study with full length WRN protein and ATPγS to inhibit helicase activity confirms that the helicase is not required for polymerase stimulation through FRA16D (Figure 6A). The effects on pol δ may be a general property of RecQ helicases, as the E. coli RecQ protein also was able to stimulate polymerization, both with and without ATPγS (Supplementary Figure S5B). A previous study demonstrated that WRN was able to increase the efficiency of primer extension by S. cerevisiae pol δ without the addition of ATP/dATP needed for helicase activity (51). Moreover, both the WRN and E. coli RecQ proteins have been previously shown to support yeast pol δ during replication of tetraplex and hairpin structures formed by d(CGG)n trinucleotide repeats (26), consistent with our observations. Stimulation by WRN may be most efficient at these difficult sequences, as replication through the secondary-structure barren sequence, R5A, was not enhanced (Supplementary Figure S5A). Fine mapping of pol δ interaction sites on the WRN protein and subsequent abolishment of these sites is required to validate the importance of direct WRN/polymerase binding for replication stimulation.
WRN is versatile, in that it has been shown to stimulate DNA synthesis by various polymerases. Similar to our findings here for pol δ, synthesis by pol η is enhanced with WRN without requiring its helicase or exonuclease activities (52). We observed a slight increase in pol α polymerization during synthesis of region 4B in the presence of WRN. A previous study failed to detect a WRN dependent affect on primer extension with either pol α or pol ε (51). The discrepancy between our study and this previous study for pol α may be due to differences in the nature of the experimental template or in the reaction conditions. A greater polymerase:template ratio used in the current study may be driving the reaction towards synthesis instead of degradation by the exonuclease.
The WRN helicase has been shown to be essential for CFS maintenance in cells (24), consistent with our in vitro experiments. Intriguingly, we did not observe changes to the DNA synthesis stalling profile when HeLa extracts were supplemented with the exogenous addition of WRN. We propose two alternative explanations for these results. First, we have not directly identified the DNA polymerase responsible for the DNA synthesis observed in the cell-free extracts. Because we are using a primed ssDNA template, it is likely that pol δ is the primary polymerase, due to its high affinity for such templates. However, we cannot rule out a contribution of pol ε at this time, and WRN has been reported to have no effect on pol ε extension synthesis (51). A second possibility is that WRN can only complex with the complete cellular pol δ replisome under circumstances of cellular stress. Checkpoint responses mediated by ATR are activated during fragile site expression (53). Activation of ATR results in rapid degradation of the p12 subunit of pol δ (54), which may remodel the cellular replisome structure to allow WRN entry and binding. It would be interesting to observe replication stimulation and fragile site maintenance by other RECQ helicases, as BLM also interacts with pol δ (55) and is able to unwind certain motifs with DNA secondary structure. Other RECQ proteins, RECQ1, RECQ4 and RECQ5 may have helicase activity in vivo although it has not been extensively demonstrated thus far [reviewed in ref. (56)].
We have measured replication progression through complementary strands of FRA16D, by both, pol α-primase and pol δ/PCNA, and in cell-free extracts. In both systems, we observed a consistent trend indicating strand asymmetry during synthesis of specific structures in region 3 and especially region 4 (Figures 2 and 7A). In region 3A, stalling is observed in multiple locations (peaks a–c, Figure 7A) while only one stall site is seen in region 3B (peak b). In region 4B, stalling is most intense at the longest IR and to a lesser degree at the (T)19 sequence (peaks d and e, Figure 7A). In the complementary region 4A, pol δ bypasses this hairpin with relative ease while stalling within the (A)19 (Figure 2). In the HeLa cell extracts, no significant stalling is observed in region 4A. Strand asymmetry may be due to the presence of smaller IRs in region 4B directly preceding the large inhibitory hairpin, causing the polymerase synthesis rate to slow prior to encountering the large hairpin, making it more detrimental. During cellular DNA replication, transient uncoupling of leading and lagging strand synthesis may contribute to the inhibition of fork progression through FRA16D.
Based on the results of this study, we propose that late replication within FRA16D represents the cumulative biochemical effect of several cis-acting elements acting sequentially over several hundred base pairs to slow/inhibit replicative polymerases. Future studies are needed to test this model, and should observe replicative progression in a continuous sequence composed of larger DNA regions to overcome limitations of the current study (which only tested relatively small target sequences). Nevertheless, our direct experimental evidence is consistent with the previous suggestion that the broad composition of FRA16D gives rise to chromosomal fragility, and that localized DNA sequence elements play a determining role in breakage (4). We have not observed a correlation between DNA pol δ synthesis inhibition and sequences of high flexibility. Region 1 contains Twistflex Peak 3 (4), but pol δ transit through this region is not different from that of a control region (Figure 2). We acknowledge that the inhibitory elements we have identified (Table 2) are also present in other, non-fragile areas of the genome. We have observed that inhibition in any single sequence motif is not absolute within the regions tested. Therefore, we propose that specific sequence motifs are detrimental only when present in a high density along a given chromosomal region and in the presence of replicative stress. Our model is supported by previous studies where fragility is reduced, but not eliminated, when portions of FRA3B are deleted (2,13). Complete deletion of FRAXB, however, prevented chromosomal breaks at this location (14) indicating that CFS contain multiple areas that are inhibitory to replication. Bioinformatics studies are currently being conducted to compare the presence and density of inhibitory motifs in known CFS relative to other genomic regions in order to test our model. Validation of potential CFS regions containing these motifs will be performed experimentally by polymerase stalling analysis.
In summary, our study has identified a region within FRA16D (191 417–191 860 bp) that contains multiple IRs and microsatellite sequences clustered in ∼450 bp. Moreover, we observed consistent DNA strand biases for DNA synthesis progression through this region. We propose the following biochemical model to describe DNA synthesis progression through CFS. The additive effect of polymerase stalling at specific DNA sequence motifs, together with asymmetric rates of leading- and lagging-strand synthesis, results in a significantly slowed rate of replication fork progression through FRA16D during S-phase, relative to surrounding chromosomal regions. RFC increases pol δ efficiency until it encounters DNA that is deviated from the standard B-form. During replication stress, the ATR checkpoint response may be activated, allowing WRN entry into the replisome. WRN is then able to alleviate some of this stalling by direct DNA polymerase interaction and stimulating polymerization through poly(dA/dT) and other difficult to replicate sequences. When such mechanisms that normally act to bypass inhibitory structures fail, the replication fork arrests, which has been proposed to precede replication fork collapse (46). Through homologous recombination, a vast array of chromosomal rearrangements can occur, contributing to the phenotype seen during CFS expression. Understanding DNA replication dynamics through additional CFS will help to elucidate the mechanisms and endogenous factors responsible for inducing DNA breakage at these critical chromosomal sites.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [grant numbers R01 CA100060 to K.A.E., GM31973 to M.Y.W.T.L., ES0515052 to P.L.O.]; and the Jake Gittlen Cancer Research Foundation [K.A.E.]. Funding for open access charges: National Institutes of Health [grant no. CA100060].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Gregory D. Bowman for kindly providing us with RFC, and Dr Jim Keck for providing us with purified RecQ protein.
REFERENCES
- 1.Venkitaraman AR. Chromosomal instability in cancer: causality and interdependence. Cell Cycle. 2007;6:2341–2343. doi: 10.4161/cc.6.19.4753. [DOI] [PubMed] [Google Scholar]
- 2.Durkin SG, Glover TW. Chromosome fragile sites. Annu. Rev. Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 3.Smith DI, McAvoy S, Zhu Y, Perez DS. Large common fragile site genes and cancer. Semin. Cancer Biol. 2007;17:31–41. doi: 10.1016/j.semcancer.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S, Nancarrow JK, Woollatt E, Kremmidiotis G, Gardner A, Venter D, et al. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum. Mol. Genet. 2000;9:1651–1663. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- 5.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60:2140–2145. [PubMed] [Google Scholar]
- 6.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl Acad. Sci. USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geradts J, Fong KM, Zimmerman PV, Minna JD. Loss of Fhit expression in non-small-cell lung cancer: correlation with molecular genetic abnormalities and clinicopathological features. Br. J. Cancer. 2000;82:1191–1197. doi: 10.1054/bjoc.1999.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mimori K, Druck T, Inoue H, Alder H, Berk L, Mori M, Huebner K, Croce CM. Cancer-specific chromosome alterations in the constitutive fragile region FRA3B. Proc. Natl Acad. Sci. USA. 1999;96:7456–7461. doi: 10.1073/pnas.96.13.7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glover TW, Stein CK. Induction of sister chromatid exchanges at common fragile sites. Am. J. Hum. Genet. 1987;41:882–890. [PMC free article] [PubMed] [Google Scholar]
- 10.Dall KL, Scarpini CG, Roberts I, Winder DM, Stanley MA, Muralidhar B, Herdman MT, Pett MR, Coleman N. Characterization of naturally occurring HPV16 integration sites isolated from cervical keratinocytes under noncompetitive conditions. Cancer Res. 2008;68:8249–8259. doi: 10.1158/0008-5472.CAN-08-1741. [DOI] [PubMed] [Google Scholar]
- 11.Lukusa T, Fryns JP. Human chromosome fragility. Biochim. Biophys. Acta. 2008;1779:3–16. doi: 10.1016/j.bbagrm.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Ragland RL, Glynn MW, Arlt MF, Glover TW. Stably transfected common fragile site sequences exhibit instability at ectopic sites. Genes Chromosomes Cancer. 2008;47:860–872. doi: 10.1002/gcc.20591. [DOI] [PubMed] [Google Scholar]
- 13.Corbin S, Neilly ME, Espinosa R., 3rd, Davis EM, McKeithan TW, Le Beau MM. Identification of unstable sequences within the common fragile site at 3p14.2: implications for the mechanism of deletions within fragile histidine triad gene/common fragile site at 3p14.2 in tumors. Cancer Res. 2002;62:3477–3484. [PubMed] [Google Scholar]
- 14.Arlt MF, Miller DE, Beer DG, Glover TW. Molecular characterization of FRAXB and comparative common fragile site instability in cancer cells. Genes Chromosomes Cancer. 2002;33:82–92. doi: 10.1002/gcc.10000. [DOI] [PubMed] [Google Scholar]
- 15.Mishmar D, Rahat A, Scherer SW, Nyakatura G, Hinzmann B, Kohwi Y, Mandel-Gutfroind Y, Lee JR, Drescher B, Sas DE, et al. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc. Natl Acad. Sci. USA. 1998;95:8141–8146. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarai A, Mazur J, Nussinov R, Jernigan RL. Sequence dependence of DNA conformational flexibility. Biochemistry. 1989;28:7842–7849. doi: 10.1021/bi00445a046. [DOI] [PubMed] [Google Scholar]
- 17.Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- 18.Le Beau MM, Rassool FV, Neilly ME, Espinosa R., 3rd, Glover TW, Smith DI, McKeithan TW. Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: implications for the mechanism of fragile site induction. Hum. Mol. Genet. 1998;7:755–761. doi: 10.1093/hmg/7.4.755. [DOI] [PubMed] [Google Scholar]
- 19.Zlotorynski E, Rahat A, Skaug J, Ben-Porat N, Ozeri E, Hershberg R, Levi A, Scherer SW, Margalit H, Kerem B. Molecular basis for expression of common and rare fragile sites. Mol. Cell Biol. 2003;23:7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bacolla A, Gellibolian R, Shimizu M, Amirhaeri S, Kang S, Ohshima K, Larson JE, Harvey SC, Stollar BD, Wells RD. Flexible DNA: genetically unstable CTG.CAG and CGG.CCG from human hereditary neuromuscular disease genes. J. Biol. Chem. 1997;272:16783–16792. doi: 10.1074/jbc.272.27.16783. [DOI] [PubMed] [Google Scholar]
- 21.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 23.Arlt MF, Xu B, Durkin SG, Casper AM, Kastan MB, Glover TW. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol. Cell Biol. 2004;24:6701–6709. doi: 10.1128/MCB.24.15.6701-6709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pirzio LM, Pichierri P, Bignami M, Franchitto A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J. Cell Biol. 2008;180:305–314. doi: 10.1083/jcb.200705126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szekely AM, Chen YH, Zhang C, Oshima J, Weissman SM. Werner protein recruits DNA polymerase delta to the nucleolus. Proc. Natl Acad. Sci. USA. 2000;97:11365–11370. doi: 10.1073/pnas.97.21.11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamath-Loeb AS, Loeb LA, Johansson E, Burgers PM, Fry M. Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem. 2001;276:16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- 27.Dhillon KK, Sidorova J, Saintigny Y, Poot M, Gollahon K, Rabinovitch PS, Monnat RJ., Jr Functional role of the Werner syndrome RecQ helicase in human fibroblasts. Aging Cell. 2007;6:53–61. doi: 10.1111/j.1474-9726.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- 28.Xie B, Mazloum N, Liu L, Rahmeh A, Li H, Lee MY. Reconstitution and characterization of the human DNA polymerase delta four-subunit holoenzyme. Biochemistry. 2002;41:13133–13142. doi: 10.1021/bi0262707. [DOI] [PubMed] [Google Scholar]
- 29.Sowd G, Lei M, Opresko PL. Mechanism and substrate specificity of telomeric protein POT1 stimulation of the Werner syndrome helicase. Nucleic Acids Res. 2008; 36:4242–4256. doi: 10.1093/nar/gkn385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Opresko PL, Sowd G, Wang H. The Werner syndrome helicase/exonuclease processes mobile D-loops through branch migration and degradation. PLoS ONE. 2009;4:e4825. doi: 10.1371/journal.pone.0004825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brosh RM, Jr, von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. Embo J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Kobbe C, Karmakar P, Dawut L, Opresko P, Zeng X, Brosh R.M., Jr, Hickson ID, Bohr VA. Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J. Biol. Chem. 2002;277:22035–22044. doi: 10.1074/jbc.M200914200. [DOI] [PubMed] [Google Scholar]
- 33.Brush GS, Kelly TJ, Stillman B. Identification of eukaryotic DNA replication proteins using simian virus 40 in vitro replication system. Methods Enzymol. 1995;262:522–548. doi: 10.1016/0076-6879(95)62043-5. [DOI] [PubMed] [Google Scholar]
- 34.Shah SN, Eckert KA. Human postmeiotic segregation 2 exhibits biased repair at tetranucleotide microsatellite sequences. Cancer Res. 2009;69:1143–1149. doi: 10.1158/0008-5472.CAN-08-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finnis M, Dayan S, Hobson L, Chenevix-Trench G, Friend K, Ried K, Venter D, Woollatt E, Baker E, Richards RI. Common chromosomal fragile site FRA16D mutation in cancer cells. Hum. Mol. Genet. 2005;14:1341–1349. doi: 10.1093/hmg/ddi144. [DOI] [PubMed] [Google Scholar]
- 36.Paige AJ, Taylor KJ, Stewart A, Sgouros JG, Gabra H, Sellar GC, Smyth JF, Porteous DJ, Watson JE. A 700-kb physical map of a region of 16q23.2 homozygously deleted in multiple cancers and spanning the common fragile site FRA16D. Cancer Res. 2000;60:1690–1697. [PubMed] [Google Scholar]
- 37.Palakodeti A, Han Y, Jiang Y, Le Beau MM. The role of late/slow replication of the FRA16D in common fragile site induction. Genes Chromosomes Cancer. 2004;39:71–76. doi: 10.1002/gcc.10290. [DOI] [PubMed] [Google Scholar]
- 38.Viguera E, Canceill D, Ehrlich SD. Replication slippage involves DNA polymerase pausing and dissociation. Embo J. 2001;20:2587–2595. doi: 10.1093/emboj/20.10.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Kobbe C, Thoma NH, Czyzewski BK, Pavletich NP, Bohr VA. Werner syndrome protein contains three structure-specific DNA binding domains. J. Biol. Chem. 2003;278:52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- 40.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 41.LaDuca RJ, Fay PJ, Chuang C, McHenry CS, Bambara RA. Site-specific pausing of deoxyribonucleic acid synthesis catalyzed by four forms of Escherichia coli DNA polymerase III. Biochemistry. 1983;22:5177–5188. doi: 10.1021/bi00291a018. [DOI] [PubMed] [Google Scholar]
- 42.Kaguni LS, Clayton DA. Template-directed pausing in in vitro DNA synthesis by DNA polymerase a from Drosophila melanogaster embryos. Proc. Natl Acad. Sci. USA. 1982;79:983–987. doi: 10.1073/pnas.79.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nag DK, Petes TD. Seven-base-pair inverted repeats in DNA form stable hairpins in vivo in Saccharomyces cerevisiae. Genetics. 1991;129:669–673. doi: 10.1093/genetics/129.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Handt O, Baker E, Dayan S, Gartler SM, Woollatt E, Richards RI, Hansen RS. Analysis of replication timing at the FRA10B and FRA16B fragile site loci. Chromosome Res. 2000;8:677–688. doi: 10.1023/a:1026737203447. [DOI] [PubMed] [Google Scholar]
- 45.Hewett DR, Handt O, Hobson L, Mangelsdorf M, Eyre HJ, Baker E, Sutherland GR, Schuffenhauer S, Mao JI, Richards RI. FRA10B structure reveals common elements in repeat expansion and chromosomal fragile site genesis. Mol. Cell. 1998;1:773–781. doi: 10.1016/s1097-2765(00)80077-5. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Freudenreich CH. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol. Cell. 2007;27:367–379. doi: 10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strahs D, Schlick T. A-Tract bending: insights into experimental structures by computational models. J. Mol. Biol. 2000;301:643–663. doi: 10.1006/jmbi.2000.3863. [DOI] [PubMed] [Google Scholar]
- 48.Koo HS, Wu HM, Crothers DM. DNA bending at adenine - thymine tracts. Nature. 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 49.Zinkel SS, Crothers DM. DNA bend direction by phase sensitive detection. Nature. 1987;328:178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]
- 50.Hile SE, Eckert KA. DNA polymerase kappa produces interrupted mutations and displays polar pausing within mononucleotide microsatellite sequences. Nucleic Acids Res. 2008;36:688–696. doi: 10.1093/nar/gkm1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamath-Loeb AS, Johansson E, Burgers PM, Loeb LA. Functional interaction between the Werner syndrome protein and DNA polymerase delta. Proc. Natl Acad. Sci. USA. 2000;97:4603–4608. doi: 10.1073/pnas.97.9.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamath-Loeb AS, Lan L, Nakajima S, Yasui A, Loeb LA. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc. Natl Acad. Sci. USA. 2007;104:10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 54.Zhang S, Zhou Y, Trusa S, Meng X, Lee EYC, Lee M.YWT. A novel DNA damage response: rapid degradation of the p12 subunit of DNA polymerase {delta} J. Biol. Chem. 2007;282:15330–15340. doi: 10.1074/jbc.M610356200. [DOI] [PubMed] [Google Scholar]
- 55.Selak N, Bachrati CZ, Shevelev I, Dietschy T, van Loon B, Jacob A, Hubscher U, Hoheisel JD, Hickson ID, Stagljar I. The Bloom's; syndrome helicase (BLM) interacts physically and functionally with p12, the smallest subunit of human DNA polymerase {delta} Nucleic Acids Res. 2008;36:5166–5179. doi: 10.1093/nar/gkn498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.