Abstract
Filamentous phage display has been extensively used to select proteins with binding properties of specific interest. Although many different display platforms using filamentous phage have been described, no comprehensive comparison of their abilities to display similar proteins has been conducted. This is particularly important for the display of cytoplasmic proteins, which are often poorly displayed with standard filamentous phage vectors. In this article, we have analyzed the ability of filamentous phage to display a stable form of green fluorescent protein and modified variants in nine different display vectors, a number of which have been previously proposed as being suitable for cytoplasmic protein display. Correct folding and display were assessed by phagemid particle fluorescence, and with anti-GFP antibodies. The poor correlation between phagemid particle fluorescence and recognition of GFP by antibodies, indicates that proteins may fold correctly without being accessible for display. The best vector used a twin arginine transporter leader to transport the displayed protein to the periplasm, and a coil-coil arrangement to link the displayed protein to g3p. This vector was able to display less robust forms of GFP, including ones with inserted epitopes, as well as fluorescent proteins of the Azami green series. It was also functional in mock selection experiments.
INTRODUCTION
Phage display using filamentous phage vectors is a widely used method for the selection of specific proteins and peptides from large libraries (1–4). Unlike lytic phage, filamentous phage is released by secretion. This imposes a constraint on displayed proteins, since they have to cross the inner membrane as part of phage assembly. While this does not appear to cause problems for antibody fragments [e.g. scFvs (5–7) or Fabs (8–11)] or other usually secreted proteins, such as the fibronectin domain (12), proteins which are normally expressed and fold within the cytoplasm can have more difficulty.
Filamentous phage have five coat proteins (p3, p6, p7, p8 and p9), each of which has been used for display in one form or another (Table 1), with display at the N terminus of p3 (13,14) being most commonly used. In most N terminal display systems [p8 (15), p7 (16), p9 (17) and p7 + p9 together (18)], the displayed protein is transported across the inner membrane to the periplasmic space using a Sec based leader, such as PelB. This also includes display with p7 and p9 (16–18), in which a Sec based leader is appended to the N terminus, even though the wild-type proteins lack such leaders. As a result, each of these display vectors is likely to suffer from the same constraints as the standard p3 display vectors. Display vectors relying on the C terminal end of p3 (19) or p6 (20–22) have been proposed as being more suitable for the display of cytoplasmic proteins, since the C terminus of these proteins is found in the bacterial cytoplasm prior to phage assembly (23), and folding of the displayed protein is therefore thought to occur within the cytoplasm. The C terminus of p8 also resides in the cytoplasm and has been used for display of both proteins and peptides (24, 25), however, its use for protein display appears to require tailored optimization for the specific protein being displayed. Of the remaining two minor coat proteins, for which C terminal display has not been attempted, the C terminus of p7 is extremely hydrophobic and thought to be buried within the membrane (26), while that of p9 is relatively hydrophilic, and may be found partly within the cytoplasm (26), suggesting that it too may be effective as a C terminal display protein.
Table 1.
Display proteins used in filamentous phage display
| Display protein | Natural leader | Leader used | Display format | Proteins displayed | References | |
|---|---|---|---|---|---|---|
| g3p | Sec | Sec | N terminus | Widely used: antibody fragments, peptides, other proteins | (6,13,102,103) | |
| Sec/Sec | N terminus, jun-fos linkage between displayed protein and g3p | cDNA libraries | (80,81) | |||
| TAT/Sec | cpGFP (but not fluorescent) | (43) | ||||
| Sec | C terminus | Tyrosine kinase | (19) | |||
| g6p | None | None | C terminus | cDNA libraries | (20,21) | |
| g8p | Sec | Sec | N terminus | Peptides, Antibody fragments | (15, 104–106) | |
| Sec | C terminus | Peptides | (24) | |||
| g7p | None | Sec | N terminus | VH | (18) | |
| g9p | None | Sec | N terminus | VL, scFv | (17,18) |
An alternative strategy to the use of different display proteins, is the use of different translocation systems to target the displayed protein to the periplasmic space. Most proteins that cross the inner bacterial membrane do so using the type II secretory system. This comprises three different pathways: Sec, SRP (Signal Recognition Particle) and TAT (Twin Arginine Transport). The Sec pathway translocates proteins post-translationally (27–29), the SRP pathway translocates co-translationally (30,31), and both pathways converge at the Sec translocon, which transports proteins in an unfolded state across the inner membrane. This is in contrast to the TAT pathway which only translocates proteins that have already folded in the cytoplasm across the inner membrane (32–35). This has been widely used to transport folded and fluorescent GFP from the cytoplasm to the periplasm (36–40), as well as in a genetic selection system for correctly folded proteins (41). Recently, in an attempt to display otherwise undisplayable proteins, SRP (42) and TAT (43) leaders were used in phage display constructs. The SRP leader was used to display ankyrin based binding proteins, while the TAT leader was used to display a circular GFP permutant. In both cases, the previous use of standard phage display vectors with the Sec leader was unsuccessful. Specific ankyrin based binders could be selected from phage libraries using SRP, but not from similar Sec based libraries, while in the case of the TAT based display vector, a peptide linker fused to a circular permutant of GFP could be specifically recognized on phage. However, the display of fluorescent GFP was not demonstrated, suggesting that correctly folded and functional GFP may not have been displayed in this latter study.
Although this represents an apparently wide choice of potential display systems, no systematic comparison has ever been made between them, and it is not at all clear which system, if any, is generally most appropriate for the display of proteins, which normally fold in the cytoplasm.
In addition to the filamentous phage platform, a number of lytic cytoplasmic phages have also been used for display purposes. T7, an extensively studied double-stranded DNA phage assembled in the cytoplasm and released by cell lysis (44), is the cytoplasmic phage most widely used for display purposes (45–53), in large part because it is available commercially. A comparison between T7 and filamentous phage g3p (with a Sec leader) for peptide display, showed significantly less bias with T7 (54). A similar comparison between the D protein of lambda and g3p or g8p of filamentous phage (using Sec leaders) for the display of a hepatitis C cDNA library (55), also showed the superiority of the lytic phage for display of the predominantly cytoplasmic proteins, and we have recently shown display of GFP libraries in T7 (56). The problem with lytic phage is the large size of their genomes, and the relative difficulty of preparing effective in vitro packaging mixtures (especially T7). Although this has been overcome by the availability of commercial kits, they tend to be expensive, and the use of this phage is to be contrasted with the relative ease, with which filamentous phage libraries can be prepared, propagated, selected and analyzed using phagemid vectors. Consequently, a filamentous phage system effective for cytoplasmic proteins would be extremely useful.
GFP is a widely used fluorescent protein marker, naturally expressed and folded in the cytoplasm, that has been used to track the distribution of many different predominantly cytoplasmic fusion partners, in many different organisms (57–60). In bacteria, functional fluorescent GFP can be easily expressed in the cytoplasm, but not the periplasm (61). As GFP only becomes fluorescent when correctly folded (62), it should be usable as a general surrogate to assess the effectiveness of different display systems to display cytoplasmic proteins. Although GFP, or variants of it, have been displayed on the surface of a number of organisms, including Escherichia coli (63,64), yeast (65,66), hepatitis B (67), baculovirus (68–70), T7 (56) and adenovirus (71) it is striking that no publication has described and analyzed the display of GFP on filamentous phage, suggesting that in its present formats, it is poorly suited for cytoplasmic protein display.
In order to determine whether it is possible to display cytoplasmic proteins using filamentous phage, we have created a set of vectors, based on our traditional p3 based display system (5). These nine vectors exploit different display proteins and different leaders, and in a first set of experiments were used to display superfolder GFP (sfGFP) (72), a particularly stable form of GFP, and derivatives of it. The best vectors were then also tested on other fluorescent proteins.
MATERIALS AND METHODS
Bacterial strains
DH5αF′: F′/endA1 hsdR17(rK−mK+) supE44 thi-1 recA1 gyrA (Na1r) relA1 D(lacZYAargF) U169(m80lacZDM15)
Omnimax: F′ {proAB lacIq lacZM15 Tn10(TetR) (ccdAB)} mcrA (mrr hsdRMS-mcrBC) 80(lacZ)M15 (lacZYA-argF)U169 endA1 recA1 supE44 thi-1 gyrA96 relA1 tonA panD
Construction of vectors and fluorescent protein based inserts
The sequences of all named oligos are provided in Table 2. The Np3-Sec vector used is pDAN5, our standard phage display vector for scFv antibody display. Np3-SRP was created by assembling the SRP leader from the DsbAss gene with two phosphorylated oligonucleotides, DsbAss S and DsbAss AS. When annealed these oligos create overhanging ends which can be ligated directly into pDAN5-D1.3 cut with HindIII and NheI, replacing the pelB leader with the DsbAss SRP leader. Np3-TAT was created by assembling three oligonucleotides (TorA1HindIII, TorA2BssHII and TorA3) that together encoded the TorA leader. This contained HindIII, 26 bp of the TorA 5′ untranslated region, including the Shine-Dalgarno sequence, the full TorA leader, the first four codons of the mature TorA protein and an in frame BssHII site. The first four codons of the mature TorA protein were included to facilitate translocation, as a result of which, the gly-ala-his-ala extension described above is expected to be retained, rather than removed, during translacation. The gene was cloned into pDAN5 using HindIII and BssHII. Np9-TAT-GFP was created from Np3-TAT by amplifying Np3-TAT-GFP with Np3TATNotI5′ and Np3TATBamHI3′, and amplifying M13mp19 with gene9 5′ and gene9 3′. The amplified g9 was cloned into the amplified plasmid using NotI and BamHI. This replaced the g3 with g9, and added a glycine/serine linker to the 5′-end of g9. It also eliminated the His6 tag in Np3-TAT. To create Cp3, gene 3 was amplified from pDAN5 using gene3Mlu1-5′ and gene3EcoRI-3′. The PCR product was cut with MluI and EcoRI and cloned into pDAN5 D1.3 cut with BssHII and EcoRI (MluI and BssHII are compatible) to create pMich1. This replaced scFv-gene 3 with gene 3 alone. sfGFP (obtained from Geoff Waldo) was amplified from pDAN5-GFP using pDPH5′ and His6-EcoRI-3′, cut with BssHII and EcoRI and cloned into pMich1 cut with the same enzymes. This placed sfGFP downstream of gene 3, flanked by our standard cloning sites BssHII and NheI. Cp6 was created by amplifying gene 6 from M13mp19 with gene 6 5′ and gene 6 3′, cut with HindIII and BssHII and cloned into pCp3 cut with the same enzymes. This resulted in the replacement of p3 with p6. In addition, an NdeI is put at the 5′ and a BamHI is put at the 3′ of p6, allowing subsequent cloning of other potential display proteins if required. Cp9 was created by amplifying gene 9 from M13mp19 with gene 9 5′ and gene 9 3′, cut with HindIII and BssHII and cloned into pCp3 cut with the same enzymes. Np3-TATcoil was created in a two step procedure by cleaving Np3-TAT with HindIII and NheI and cloning the insert into pDAN5 D1.3 E coil (73) cut with the same enzymes. This appended an E coil to the C terminus of sfGFP upstream of the SV5 tag. Np3-TATcoil was created by combining the PelB leader with the K coil by PCR assembly. The PelB leader was amplified from pDAN5 using PelB 5′ and PelB 3′, the K coil was amplified from pDAN5 D1.3 K coil (73) using Kcoil 5′ and Kcoil 3′, and the two were combined by PCR assembly using PelB 5′ and Kcoil 3′. The assembled fragment was cleaved with EagI, and cloned into Np3-TAT cleaved with NotI and dephosphorylated. Np3-TATcys was created by amplifying the PelB leader from pDAN5 D1.3 with PelBcys 5′ and PelBcys 3′, and cloning into Np3-TAT using NheI and NotI. The Azami Green series (74), superfolder GFP (72), and the sfGFP derivatives clys- GFP and myc-GFP (56) were cloned into the various display vectors using BssHII and NheI, from our pre-existing pET vectors (75). To clone wild type GFP and cycle 3 GFP into pDan5 and pHugh3, the GFP coding regions were amplified from the vector series pHisflexGFP with the primers Bad-Hugh3 5′ and Bad-Hugh3 3′. The amplicons were digested with BssHII and NheI, gel purified and cloned into the appropriately digested plasmids. DsRed T1 (76) was cloned similarly after amplification from DsRed Express (Clontech) using 5′ DsRedDanII and 3′ DsRedDanI, and cleavage with BssHII and NheI.
Table 2.
Oligonucleotides used
| Oligo name | Oligo sequence |
|---|---|
| DsbAss S | P-AGC TTG CCA AAT TCT ATT TCA AGG AGA CAG TCA TAA TGA AAA AAA TCT GGC TGG CGC TGG CAG GCC TGG TGC TGG CGT TTA G |
| DsbAss AS | P-CGC GCA AAC GCC AGC ACC AGG CCT GCC AGC GCC AGC CAG ATT TTT TTC ATT ATG ACT GTC TCC TTG AAA TAG AAT TTG GCA |
| TorA1HindIII | GAC GTA AAG CTT CAC GGC GAT AAG AAG GAA GAA AAA TAA TGA ACA ATA ACG ATC TCT TTC AGG CAT CAC GTC GG |
| TorA2BssHII | TTT CAG GCA TCA CGT CGG CGT TTT CTG GCA CAA CTC GGC GGC TTA ACC GTC GCC GGG ATG CTG GGG CCC TCA |
| TorA3 | TCC GGC ATG CGC GCC CGC CGC TTG CGC CGC AGT CGC ACG TCG CGG CGT TAA CAA TGA GGG CCC CAG CAT CCC |
| gene3Mlu1-5′ | CAT CAC CAC GCG TTG GCC GCC ACT GTT GAA AGT TGT TTA |
| gene3EcoRI-3′ | GAG AGA GAA TTC GGC ATG CGC GCC ACG ATC GGT TTC CGC GCT AGA ATA AGA CTC CTT ATT ACG CAG TAT G |
| pDpH 5′ | GCA GCC GCT GGA TTG TTA TTA |
| pDpH 3′ | TTG TCG TCT TTC CAG ACG TTA |
| His6-EcoRI-3′ | AGT AGC GAA TTC TTA ATG GTG ATG GTG ATG GTG AGT |
| gene 6 5′ | TTC TTA AGC TTG CCA AAT TCT ATT TCA AGG AGA CAG TAC ATA TGC CAG TTC TTT TGG GTA |
| gene 6 3′ | GGC ATG CGC GCC GCT ACC ACC ACC GCT ACC ACC ACC GGA TCC TTT ATC CCA ATC CAA ATA AGA |
| gene 9 Hind3 5′ | TTC TTA AGC TTG CCA AAT TCT ATT TCA AGG AGA CAG TAC ATA TGA GTG TTT TAG TGT ATT |
| gene 9 BssH2 3′ | GGC ATG CGC GCC GCT ACC ACC ACC GCT ACC ACC ACC GGA TCC TGA GGA AGT TTC CAT TA |
| gene 9 5′ | GGC GGT GGA TCC ATG AGT GTT TTA GTG TAT T |
| gene 9 3′ | CTC CAA GCG GCC GCA TAT GAC CGG TTT ATG AGG AAG TTT CCA TTA |
| Np3TATNotI5′ | TAA ACC GGT CAT ATG CGG CCG CTT GGA GAA TTC ACT GGC CGT CGT TTT AC |
| Np3TATBamH13′ | CTC ATG GAT CCA CCG CCA CCG CTA CCG CCA CCA CCA GTA CTA TCC AGG CCC AGC AGT G |
| PelB 5′ | TTC TTC TCG CGG CCG GGT GAT GCC AAA TTC TAT TTC AAG G |
| PelB 3′ | GGC GTG CGC ACC GCT TGC TGC |
| Kcoil 5′ | GCA GCA AGC GGT GCG CAC GCC AAA GTA AGC GCT CTC AAG GAA |
| Kcoil 3′ | AGA GAT CGG TAA GAA AAG CGG CCG CAG AAC CGC CAG AGC CAC CAC CCT CTT TCA GGG CGC TCA CAA A |
| PelBcys 5′ | TAC AAA GCT AGC AGC GGC AAA CCA ATC CCA AAC CCA CTG CTG GGC CTG GAT AGT ACT CAC CAT CAC CAT CAC CAT TGC TGA GCC AAA TTC TAT TTC AAG G |
| PelBcys 3′ | TTC AAC AGT AGC GGC CGC GCA GTA GTC GGC GTG CGC ACC GCT TGC TGC GAG TAA TA |
| Bad-Hugh3 5′ | GCA AGC GGC GCG CAT GCC GCA CTC GAG GGA GAA GAA CTT TTC ACT GGA GTT G |
| Bad-Hugh3 3′ | GCC GCT AGC TTT GTA GAG CTC ATC CAT GCC ATG TGT AAT CCC AGC AGC AGT TAC |
| 5′ DSRedDanII | GGT TAG CGC GCA AGC CTC CTC CGA GGA CGT CAT CAA GGA G |
| 3′ DSRedDanII | CGT AAG CTA GCC AGG AAC AGG TGG TGG C |
All constructs were transformed into DH5α F′ and sequenced prior to use.
ELISA with folded and denatured GFP
Non-denatured GFP contained 1 μg of purified sfGFP in 80 μl PBS. The SDS treated samples additionally contained 2% SDS. The heat-treated sample was heated to 100°C for 10 min. After these treatments, samples were added to Nunc Maxisorp plates and incubated for 1 h at 37°C for protein adsorption. Antigen bound plate was washed with PBS and blocked with 200 μl of wonder block (WB - 0.3% BSA, 0.3% milk, 0.3% fish gelatin) for 1 h at room temperature. The blocked plate was washed and incubated with primary antibodies, anti GFP 3E6 (0.5 μg/ml, Q-biogene Inc.) or anti GFP polyclonal (1 μg/ml, Rockland Inc.). The secondary antibody for 3E6 was horseradish peroxidase (HRP) labeled anti-mouse (0.5 μg/ml, Dako Inc.) and the secondary antibody for the anti-GFP polyclonal was HRP labeled anti-goat (0.5 μg/ml, Abcam Inc.). The plates were washed four times with PBST (Phosphate buffered saline containing 0.1% tween 20) and four times with PBS. 100 μl TMB (Sigma Inc.) was used as the HRP substrate. The reaction was quenched using 50 μl 1 M sulfuric acid and absorbance read at 450 nm.
Bacterial growth, phage production, phage precipitation and quantification
Bacteria containing the different constructs were grown overnight on amp/glu (50 μg/ml carbenicillin, 3% glucose) plates at 30°C. A small streak of bacteria was added to 10 ml amp/glu (50 μg/ml carbenicillin, 3% glucose) 2×TY and grown for 2 h at 37°C before infection with M13K07 helper phage at a multiplicity of infection of 10:1. After 30 min at 37°C, the bacteria were pelleted by centrifugation and re-suspended in 50 ml of amp/kan (50 μg/ml carbenecillin, 25 μg/ml kanamycin) 2×TY prior to overnight growth at 30°C for phage production. The overnight bacterial culture was centrifuged at 2500 rcf and the supernatant PEG precipitated twice by adding 20% volume of 20% PEG 8000, 2.5 M NaCl. Phage were resuspended in a final volume of 1 ml PBS. In order to calculate the number of phage particles in each solution, absorbance readings at 269 nm and 320 nm were taken. The titre was calculated using the following formula (Abs. 269 − Abs. 320) × 6 × 1016/plasmid size (77). We had independently verified that the number of phages calculated using this formula matched the number of phages on a titration plates (amp phage + kan phage) (data not shown).
Phage ELISA
MaxiSorp® plates (Nunc Inc.) were used for all ELISAs. In the antibody ELISA experiments biotinylated antibodies (100 μl of 10 μg/ml) were first bound to neutravidin adsorbed on Nunc. Maxisorp plates. 70 μl of phage (at a concentration of 1012/ml) and 25 μl of wonder block were added and incubated for 1 h at room temperature. Plates were washed with PBST (three times) and PBS (three times). Bound phage was revealed using an anti-M13 HRP (0.2 μg/ml Amersham Pharmacia Inc.). HRP activity was quenched using 1 M sulfuric acid. ELISA values are given as absorbance at 450 nm.
Measuring phage fluorescence
To measure phage fluorescence, bacterial growth, and phage quantification were carried out as described above. 1012 phage were used to measure phage fluorescence, at a gain of 98 for all measurements. For the mAG series and sfGFP and its variants, excitation was carried out at 488 nm, and emission was measured at 535 nm. For cycle 3 GFP, excitation was set at 400 nm and the emission value at 509 nm calculated from a 500–530 nm emission scan. The fluorescence of wild-type GFP was too low to measure. For DsRed phage, excitation was at 540 nm and emission was measured at 595 nm.
Phage titration
Ten microliter of phage solution was added to 90 μl of 2×TY media in a microtite well. A 10-fold dilution series was carried out in the microtitre plate. 100 μl of DH5α grown to 0.5 OD600 at 37°C was added to each well and phage infection was allowed to occur for 30 min at 37°C. 5 μl of the infected cells from each well were spotted on amp/glu plates and incubated at 30°C overnight. The total amount of phage was calculated from the dilution with the highest number of countable colonies.
Competitive selection
Phage were prepared in 10 ml volumes as described previously. After centrifugation, the culture supernatant was filtered using a 0.22 μM syringe filter. 100 μl of each of the phage were diluted into 700 μl of media and 0.1% BSA for 30 min. 200 μl of blocked phage were used as the input for each selection. 1 μg of biotinylated antigen was added to the phage mix and incubated for 1 h. The binding to streptavidin magnetic beads and subsequent washes (3× PBST, 3× PBS) were carried out using the KingFisher automated magnetic bead handler (ThermoFisher Inc.). Phage elution was carried out in 150 μl 0.1 N HCl and neutralized with 50 μl 1.5 M Tris pH 8.8. 50 μl of eluted phage were infected into 1 ml DH5α at 0.5 OD600 at 37°C for 30 min. 100 μl of infected cells were plated on amp/glu plates in order to pick single colonies for PCR based vector identification. The Np3-pelB and Np3-TATcoil vectors were recognized using pDpH 5′ and pDpH 3′ primers, while the Np3-SRP vector was identified using the DsbAss S primer as forward and pDpH 3′ as reverse primer.
Mock selection
Fifty milliliters of overnight phage cultures were prepared and purified as described above. 1013/ml phage solutions for GFP and mAG displaying phages were prepared and blocked with wonder block. sfGFP and mAG phage were mixed in the ratios indicated in Table 3. One hundred microliters of each phage mix was added to well A of the KingFisher instrument (ThermoFisher) microtiter plate. 1 μg of biotinylated mAb 3E6 was also added to the well as antigen and incubated for 1 h. Binding to streptavidin beads and selection were carried out as detailed in the previous section. Ten microliters of eluted phage were infected into 1 ml DH5α at 0.5 OD600 at 37°C for 30 min. About 1000 infected cells were plated on amp/glu plates on a filter and the filter was induced by transfer to amp/IPTG plates and incubated at 30°C for 1 h for protein induction. The GFP folds and fluoresces in 1 h while mAG takes about 4–5 h to fluoresce, allowing straightforward identification and counting of selected phage. The numbers were further confirmed by PCR using GFP specific primers. The total number of colonies was used to calculate eluted phage titers, and numbers from 1 h protein induction and PCR were used to calculate the ratios. Input and eluted phage titers were determined to calculate the sfGFP/mAG phage ratio (Table 3).
Table 3.
Mock selection
| Antigen | Input GFP phage | Input mAG phage | Ratio phage GFP:mAG | Output GFP phage | Output mAG phage | Ratio phage GFP:mAG |
|---|---|---|---|---|---|---|
| Bio-3E6 + strep beads | 1.8 × 109 | 0.6 × 109 | 3:1 | 1.8 × 104 | 0 | 1:0 |
| Bio-3E6 + strep beads | 1.8 × 108 | 1.2 × 1010 | 1:67 | 6 × 104 | 3 × 104 | 2:1 |
| Bio-3E6 + strep beads | 0.6 × 107 | 3 × 1010 | 1:5000 | 1.8 × 104 | 3.2 × 104 | 1:1.8 |
| Strep beads only | 1.8 × 109 | 0.6 × 109 | 3:1 | 0 | 450 | 0:1 |
| Strep beads only | 1.8 × 108 | 1.2 × 1010 | 1:67 | 4.5 × 103 | 9 × 104 | 1:20 |
| Strep beads only | 0.6 × 107 | 3 × 1010 | 1:5000 | 0 | 9 × 104 | 0:1 |
Western blot analysis of phage display
Bacterial growth and phage production was carried out as described above. Thirty microliters of PEG precipitated phage were electrophoresed using the Novex bis-tris gel system (Invitrogen Inc.). After electrophoresis, the proteins were transferred to nitrocellulose membrane using a semidry blot apparatus. The membrane was blocked with 3% milk PBST for 1 h at RT. This was then incubated in 1 μg/ml of anti-SV5 antibody in 2% milk PBST overnight at 4°C. The blot was washed 2× PBST and 1× PBS, 5 min each. Anti-mouse HRP (Santa Cruz biotech) at 1:2000 dilution was used as the secondary antibody, 1 h RT. After washing, the HRP activity was detected using SuperSignal West Dura (Pierce Inc.) and the Versa doc imager (Biorad Inc.).
RESULTS
Plasmid constructs
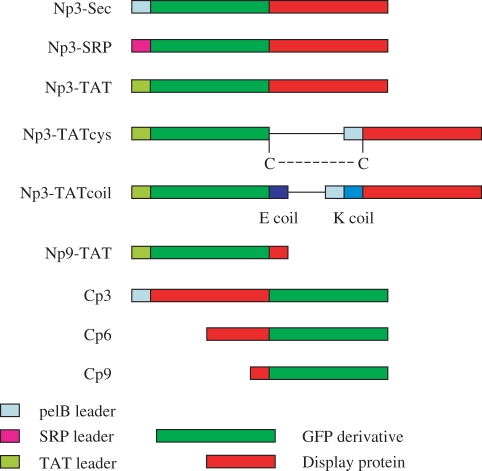
The constructs shown in Figure 1, each cloned between the HindIII/EcoRI sites of pUC119, were created by PCR of appropriate gene fragments from M13mp19 (78). In each case, the cloning sites used for the fluorescent protein were BssHII and NheI, the same as those used in pDAN5 (5). The BssHII site in pDAN5 is found within the 3′-end of the sequence encoding the leader, with additional nucleotides encoding gly-ala-his-ala found downstream. The whole of this sequence was used for all constructs. This creates a small N terminal extension for each of the GFP molecules. We would expect this to be removed in the case of the SRP and Sec leaders, during translocation to the periplasm, and retained in the case of the TAT leader (see materials and methods). In addition, all constructs contained the easily used SV5 peptide tag (79) at the C terminus of the displayed GFP. In the case of the p3C vector, the linker between p3 and the displayed protein was the same as that used by Fuh et al. (YSSAETDR) (19). For the p6C and p9C vectors, an identical linker (GSGGGSGGGS) was used between the C terminus and the GAHA sequence containing the BssHII site. For the p3 TAT based display vectors, an arrangement similar to that described by Paschke and Hohne (43) was used, with the torA leader driving the export of the fluorescent protein, and a Sec leader (that used in pDAN5), for translocation of p3. However, unlike Paschke and Hohne, who used a cys-coil-cys arrangement, based on jun/fos, we used two alternative coupling methods to link the exported GFP to p3. This is based on experiments (data not shown) indicating that the cys-coil-cys arrangement, also used in the pJUFO series of vectors (80–82), can lead to the formation of long cysteine bonded concatamers. In the first method (Np3-TATcys), a cysteine was placed at the C terminus of the fluorescent protein and at the N terminus of the mature p3 (i.e. after cleavage of the pelB leader). This is based on a successful p3 based disulfide bonded display system, and the sequence around the p3 cysteine was identical to that previously described (83). In the second method, E and K coiled coils [that have an affinity of 60 pM for one another (84)] were used, with the E coil fused to displayed protein and the K coil to the N terminus of p3. Although previous work (85) has shown that direct TAT-p3 fusions are unable to display, we also created a construct of this type, as well as a TAT-p9 fusion. All constructs were sequenced for verification.
Figure 1.
Display constructs created. A graphical representation of the different constructs created, showing the different leaders and display proteins used.
Display of sfGFP
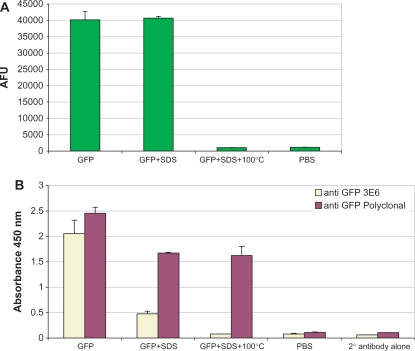
In the first experiments described here, we used an extremely stable form of GFP (86) (superfolder GFP – sfGFP), as well as two previously described derivatives, myc-GFP and clys-GFP (56,75). These are able to recognize the monoclonal antibody 9E10 and the antigen lysozyme, respectively. We adopted two criteria to determine whether sfGFP and the derivatives were correctly folded and displayed on phage. The first, based on the finding that GFP is not fluorescent unless it is correctly folded (62), was the observation of fluorescence in phage. Although this shows that correctly folded sfGFP is associated with phage, it does not show that it is in an accessible ‘displayed’ format, i.e. recognizable by antibodies or other targets. This was the basis for the second criterion, carried out with an anti-GFP monoclonal antibody that recognized a conformational epitope exquisitely sensitive to the presence of SDS and heat denaturation. As shown in Figure 2, 3E6 (Q-biogene) shows strong binding to GFP, which can be significantly reduced if the GFP is treated with 2% SDS, and abolished if heat treatment (10 min at 100°C) is additionally applied, a condition under which fluorescence is also lost. Since there is no loss of fluorescence, the reduction of 3E6 reactivity with SDS treatment may be associated with masking of the epitope by the binding of SDS rather than protein denaturation. It is important to note, that as the GFP fluorophore is formed after folding (62), it is possible that a non-fluorescent correctly folded GFP molecule is able to bind this antibody. The polyclonal antibody (Rockland Immunochemicals) binds GFP under all three conditions.
Figure 2.
3E6 recognizes a conformational epitope on GFP. (A) The fluorescence of GFP, and (B) its recognition by 3E6, and a polyclonal anti-GFP antibody under different conditions, including (i) native, (ii) treated with SDS and (iii) treated with SDS and 100°C heat treatment. Bound GFP was detected using polyclonal anti-GFP and m3E6 antibodies followed by HRP labeled corresponding secondary antibodies.
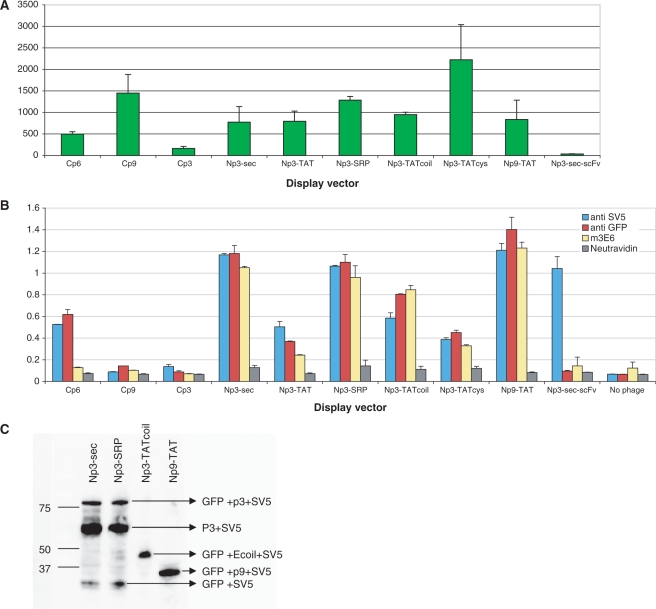
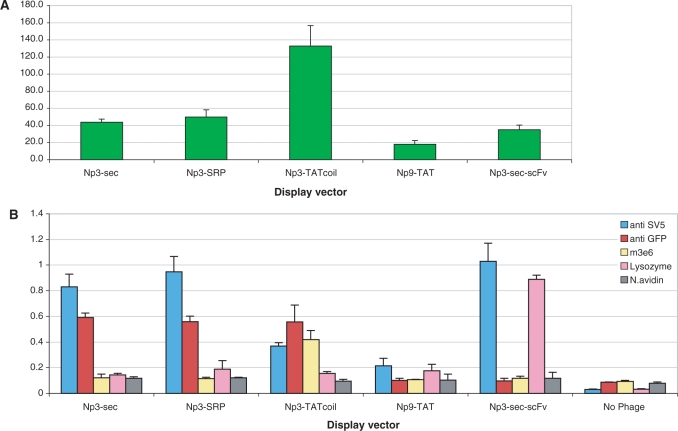
Bacteria containing the different constructs displaying sfGFP were infected with helper phage and assessed for phage production. Each of the constructs was able to make phage at similar titers (1012–13/ml) to those we have routinely observed with antibody phage display using pDAN5. The fluorescence of 1012 phagemid particles produced by these different vectors is shown in Figure 3A. The fluorescence of a standard p3 phage display vector displaying a scFv is shown for comparison. The least fluorescent phage were from the Cp6 and Cp3 constructs, while the remaining phage had fluorescent levels that were similar. The presence of fluorescence in a phage preparation gives no indication as to whether the GFP is accessible in a form suitable for display. This was studied by phage ELISA (7 × 1010 phage) using three different antibodies (Figure 3B): (i) anti-SV5 (79), a monoclonal antibody recognizing the SV5 peptide tag found at the C terminus of sfGFP; (ii) a polyclonal anti-GFP antibody able to recognize folded and unfolded GFP (Figure 2) and (iii) 3E6, the mAb that recognizes a GFP conformational epitope (Figure 2). Of these, only 3E6 will exclusively recognize GFP that is correctly folded, while the first two antibodies will recognize GFP or the attached peptide whether GFP is correctly folded or not (Figure 2). Surprisingly, there was little correlation between phage fluorescence and positive ELISA signals, especially with 3E6. In particular, Figure 3B shows that Cp9, Np3-TAT and Np3-TATcys produced fluorescent phage that were not, or only weakly, recognized by any of the antibodies, while one of the least fluorescent phage, those produced by Cp6, were relatively well recognized by SV5 and the polyclonal anti-GFP antibody, but not at all by 3E6. All the remaining phage constructs (Np3-Sec, Np3-SRP, Np3-TATcoil and Np9-TAT) produced phage particles that were fluorescent and well recognized by all three antibodies.
Figure 3.
Expression and display of GFP using different display vectors. (A) The fluorescence levels of 1012 phages displaying sfGFP using the different vector systems are compared. An Np3-sec vector carrying an scFv gene recognizing lysozyme is used as a negative control. (B) Assessment of GFP display with different vectors ELISA signals obtained with phage produced by different vectors. The SV5 antibody recognizes a linear peptide tag found at the C terminus of GFP, the polyclonal anti-GFP recognizes both folded and unfolded GFP, and 3E6 recognizes only correctly folded GFP. An Np3-sec vector carrying a scFv gene is used as positive control and measure of standard display. (C) Assessment of GFP display using western blot. The figure shows specific bands of display for Np3 sec and Np3 SRP vector (GFP + p3 + SV5), the p9 display is indicated by (GFP + p9 + SV5). In the case of the Np3 TATcoil vector, sfGFP is not covalently linked to p3, and the expected band comprises GFP + E coil + SV5 as indicated.
These four best vectors (Np3-Sec, Np3-SRP, Np3-TATcoil and Np9-TAT) were further analyzed by western blotting (Figure 3C) using the anti-SV5 antibody. As can be seen, the Np3-sec and Np3-SRP vectors incorporated the most SV5 epitope, however, most of this corresponded to a p3-SV5 truncation product with only 15–20% corresponding to the full length displayed p3-sfGFP protein. When only the amount of displayed sfGFP is taken into account, all four vectors had similar display levels.
Display of modified sfGFP
The sfGFP used in these studies is extremely stable, and folds rapidly (72), and as such, is probably not representative of most cytoplasmic proteins. In order to simulate less stable, less efficiently folding proteins, we investigated the display of two sfGFP derivatives: myc-GFP contains the linear myc peptide epitope recognized by the monoclonal antibody 9E10 (87), while clys-GFP (75) contains a CDR3 derived from a lysozyme binding VHH domain that is able to recognize lysozyme as a soluble protein. On the basis of the results obtained above, the four most promising vectors (Np3-Sec, Np3-SRP, Np3-TATcoil and Np9-TAT) were further studied with these modified GFPs.
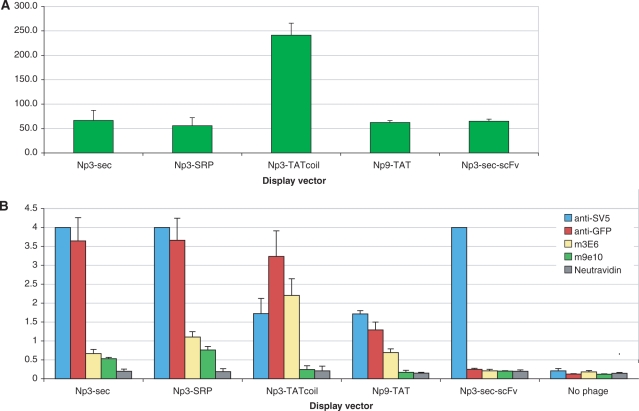
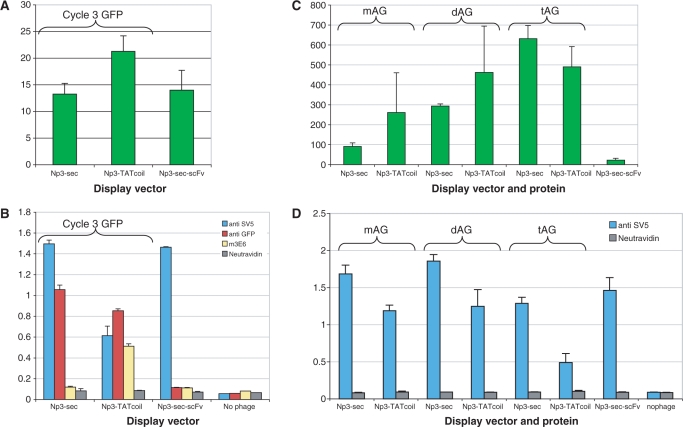
Figure 4A shows that for myc-GFP, the only vector able to produce phage particles more fluorescent than a phage displayed antibody (Np3-scFv) was Np3-TATcoil. The same was true for clys-GFP (Figure 5A). All the other vectors produced phages that were either less fluorescent, or only slightly more fluorescent than the antibody phage.
Figure 4.
Phage fluorescence and display of modifies GFP using different display vectors: myc-GFP. (A) Fluorescence of 1012 phage particles displaying myc-GFP using different vectors. (B) Analysis of myc-GFP display levels by ELISA.
Figure 5.
Fluorescence of 1012 phage particles displaying clys-GFP using different vectors. (A) Fluorescence from display of clys-GFP of 1012 phages. (B) Analysis of clys-GFP display levels by ELISA.
All four vectors, displaying either clys-GFP or myc-GFP, were able to produce phage particles that could be recognized by anti-SV5, and in most cases, the polyclonal anti-GFP antibody. However, only the Np3-TATcoil vector consistently produced phages that were well recognized by 3E6, the antibody recognizing the conformational epitope (Figures 4B and 5B). Interestingly, the SV5 signals did not correlate with either 3E6 or the polyclonal anti-GFP signals, with Np3-Sec and Np3-SRP having the highest SV5, but 3E6 values barely above the negative control scFv. When these constructs were tested for recognition by the 9E10 antibody that recognizes the linear myc epitope cloned within GFP, the Np3-SRP and Np3-Sec vectors gave the strongest signals, with the Np3-TATcoil and Np9-TAT vectors producing phages that were not recognized (Figure 4B).
clys-GFP is able to recognize lysozyme as a pure protein at a relatively low affinity [1.4 µM (75)]. When we carried out ELISA using phage displaying clys-GFP and lysozyme as the target (Figure 5B), all four vectors gave similar relatively low signals.
On the basis of the 3E6 results, the Np3-TATcoil vector appeared to be the most suitable for the display of modified GFP and therefore perhaps other proteins expressed in the cytoplasm. However, this result was contradicted by those obtained with 9E10 and lysozyme, in which Np3-TATcoil gave lower signals. In order to determine which of these vectors was the most effective within a selection context, two selection experiments were carried out. The best three display vectors (Np3-Sec, Np3-SRP and Np3-TATcoil) displaying myc-GFP were mixed in equal titers and selected on 9E10, the antibody that recognizes the linear myc epitope, 3E6, the antibody recognizing the conformational GFP epitope or lysozyme, as a negative control. A similar selection was carried out for the clys-GFP displayed on the three constructs, using lysozyme, 3E6 or myoglobin (used as a negative control). In both experiments the identity of the eluted phages was determined by PCR using specific primers to amplify 48 colonies from each selection. The results in Table 4, show clearly that the Np3-TATcoil vectors were preferentially selected when 3E6 was used to select either of the modified GFPs, as well as when lysozyme was used to select the clys-GFP constructs. Furthermore, whenever a non-specific selection was carried out, the Np3-TATcoil vector gave the lowest number of colonies. In contrast, the Np3-SRP vector gave the most colonies under non-specific selection conditions, where it constituted ∼50% of the output. In the case of selection on the linear myc epitope, the Np3-SRP vector gave the greatest number of clones, although the number of clones identified was only slightly higher than those with the non-specific target (lysozyme).
Table 4.
Competitive selection outcome
| Displayed protein | Target antigen | Display system: % of output phage |
||
|---|---|---|---|---|
| Np3–TATcoil | Np3-Sec | Np3-SRP | ||
| Myc-GFP | 9e10sp | 8 | 32 | 60 |
| lysozymensp | 13 | 26 | 49 | |
| 3E6sp | 82 | 6 | 14 | |
| clys-GFP | lysozymesp | 80 | 5 | 15 |
| myoglobin nsp | 7 | 38 | 58 | |
| 3E6sp | 95 | 4 | 1 | |
| GFP | Lysozymensp | 5 | 38 | 58 |
Equal titers of phagemid particles made with the Np3–TATcoil, Np3-Sec and Np3-SRP display vectors displaying the different proteins indicated, were mixed and a single round of selection carried out on the indicated targets. The designations sp and nsp indicate whether the target is specific (sp) or non-specific (nsp) for the displayed protein. For each selection the identity of 48 different clones was determined by PCR using vector specific primers. Each selection was carried out two or three times and the mean provided in the table (this explains why the total percentage for each selection does not always equal 100%).
Display of other fluorescent proteins
As described above, superfolder GFP has been evolved to be particularly stable, and the ability to display sfGFP, and modified variants, may therefore not be representative of other fluorescent or cytoplasmic proteins. In order to see whether this vector was suitable for other fluorescent proteins, wild type GFP (88), cycle 3 GFP (89), monomeric, dimeric and tetrameric Azami green (74) and DsRed-T1 (76,90) were cloned into the Np3-ATcoil and Np3-sec vectors and tested for display.
In the case of the two GFP constructs, only the fluorescence of phage displaying cycle 3 GFP using the Np3-TATcoil vector (Figure 6A) could be measured, while that of wild-type GFP, or cycle 3 GFP using the Np3-Sec vector, was no different to control phage displaying antibody. However, the fluorescence of phage displaying cycle 3 GFP was extremely low. Display was also assessed by ELISA, using 3E6, the polyclonal anti-GFP antibody and SV5. Figure 6B shows that only the Np3-TATcoil vector was able to give a detectable 3E6 signal, indicating that this was the only vector able to display cycle 3 GFP in a correctly folded form. Interestingly, as observed earlier, although the SV5 signals were high for all vectors, they tended to be lower in those with higher 3E6 signals.
Figure 6.
Phage fluorescence and display of different green fluorescent proteins. (A) Fluorescence of 1012 cycle3 GFP phages. (B) Display ELISA for cycle3 GFP phages. (C) Fluorescence of 1012 Azami green phages and (D) display ELISA for Azami green phages.
Unfortunately, we did not have a 3E6-like antibody for the other fluorescent proteins, and for this reason only SV5 could be used. The wild-type Azami green protein is a tetramer (74) that can be converted to a dimer by a single amino acid mutation, and a monomer by a further two mutations (74). Figure 6C shows that the Np3-TATcoil vector was most effective at producing fluorescent phage for the monomeric and dimeric Azami green proteins, while the Np3-Sec was slightly better than the Np3-TATcoil vector for the tetrameric protein. Figure 6D shows that SV5 recognized Azami green phage produced using either Np3-Sec or Np3-TATcoil, but that in general, the Np3-Sec vector gave higher signals than the Np3-TATcoil vector- similar to the results obtained with display of wild-type and cycle 3 GFP.
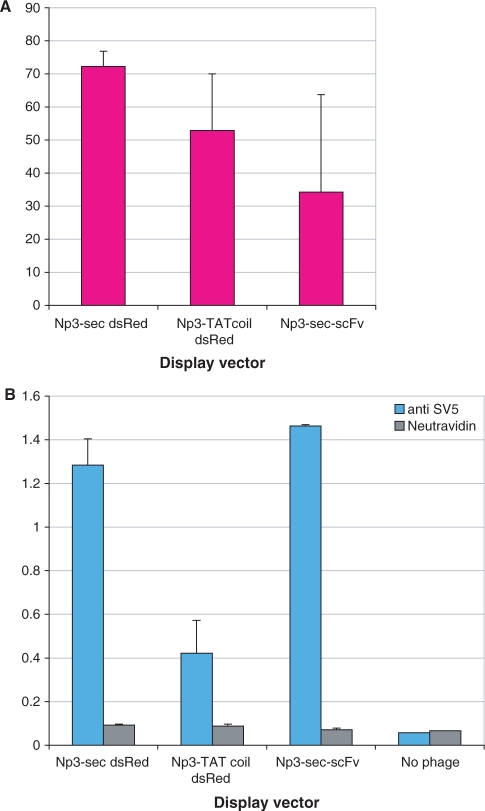
We also attempted to display DsRed T1 using the Np3-Sec and Np3-TATcoil vectors. As can be seen in Figure 7, neither of the two vectors was able to create phage particles significantly more fluorescent than the displayed scFv (Figure 7A), although a display ELISA using anti-SV5 (Figure 7B) showed that the Np3-sec DsRed display level was similar to that of the Np3-sec-scFv.
Figure 7.
Phage fluorescence and display of DsRed. (A) Fluorescence of 1012 dsRed phage. (B) Display ELISA of dsRed.
Mock selection
One concern with the use of the Np3-TATcoil vector is the possibility that there may be exchange of proteins between phage, as a result of the fact that displayed proteins are not covalently bound to g3p. In a final experiment to address this concern, and to determine whether the Np3-TATcoil vector could be used for selection, phage displaying either GFP or mAG using the Np3-TATcoil vector were mixed at three different ratios (Table 3). These phage mixtures were selected on: (i) 3E6, the antibody recognizing the conformational epitope on correctly folded GFP; or (ii) streptavidin beads as a negative control. The results in Table 3 show clear enrichment of phage displaying the GFP when selected on 3E6, but not on streptavidin. In the best case scenario (GFP:mAG 1:5000), enrichment after a single round of selection was over 2500-fold.
DISCUSSION
In this article we have examined a series of different vectors for their ability to display GFP or GFP variants that contain loop insertions. GFP was chosen for these experiments as it is a protein that is not fluorescent unless correctly folded (62), and so is able to serve as a surrogate to assess the effectiveness of cytoplasmic protein display using filamentous phage vectors under different conditions. Under normal conditions, functional fluorescent GFP can be folded in the bacterial cytoplasm, but not the bacterial periplasm (61). However, GFP that has folded in the cytoplasm can be translocated to the periplasm by the fusion of a TAT leader to the N terminus (39). Four of the vectors we constructed and tested were based on previously published vectors described as being suitable for the display of cytoplasmic proteins, either because the portion of the phage protein to which the GFP was fused is normally resident in the cytoplasm [Cp3 (19) and Cp6 (20,21)], or because the displayed protein is translocated from the cytoplasm to the periplasm by the TAT leader (43) (Np3-TATcys and Np3-TATcoil). In addition to these, we created additional vectors (see Figure 1), based on the use of the N and C termini of p9 (Np9-TAT and Cp9), and direct fusions between the three different leaders (Sec, SRP and TAT) to the N terminus of p3 (Np3-Sec, Np3-SRP and Np3-TAT).
In addition to using fluorescence as a measure of protein folding, we also identified a commercially available monoclonal antibody (3E6—Q-biogene) that recognized a conformational epitope destroyed by SDS and boiling (Figure 2). This allowed us to determine whether fluorescent GFP observed in phage preparations was actually displayed, i.e. accessible to external affinity reagents such as antibodies, or buried within the phage and consequently not available for binding.
In experiments with sfGFP, vectors that produced fluorescent phage (Cp9, Np3-Sec, Np3-TAT, Np3-TATcoil, Np3-TATcys and Np9-TAT) came from fluorescent bacteria, and vectors that produced phage that were no more fluorescent than control phage (Cp6, Cp3 and Np3-SRP) came from bacteria that had very low or no fluorescence (Figure 3A and data not shown). Np3-Sec was an exception: it produced the most fluorescent bacteria but phage of relatively low fluorescence. Interestingly, phage and bacteria produced by the two constructs (Cp3 and Cp6) based on previously described vectors (19–21) for the display of cytoplasmic proteins and libraries were essentially non-fluorescent. Two of the constructs (Cp9 and Np3-TATcys) produced very fluorescent phage, but were unable to bind 3E6, or only weakly so, indicating that the GFP in these phage was inaccessible, even if well expressed and correctly folded. The ability of the Np3-Sec vector to display sfGFP, as determined by the production of fluorescent phage that were able to bind 3E6, is in itself significant, since GFP has not been previously displayed using standard phage display vectors. This probably reflects the folding robustness of superfolder GFP compared to the other GFPs used for previous attempts.
The four vectors able to produce fluorescent phage that could also bind to 3E6 were further tested using an additional two GFP derivatives (clys-GFP and myc-GFP) that contain binding loops embedded within them conferring specific binding activity upon the GFP. The myc-GFP contains a linear epitope, recognized by the mAb 9E10 (87), that is functional in western blots. For this reason we believe that myc-GFP does not need to be folded to be recognized by 9E10. This is in contrast to clys-GFP that contains the CDR3 from a lysozme binding VHH. An examination of the structure of this VHH (91), and in particular the CDR3, in complex with lysozyme, would suggest that the CDR3 has to be in a particular orientation to be functional. Furthermore, we have previously shown, in a flow cytometry assay requiring both fluorescence and binding activity to reside in a single molecule, that clys-GFP binds to lysozyme with an affinity of ∼1.4 µM (75), indicating that correctly folded clys-GFP binds lysozyme. However, this does not exclude the possibility that poorly folded clys-GFP is also able to do so.
The insertion of these loops also affects folding, as manifested by reduced bacterial fluorescence and the need for longer time for fluorescence to develop. For both of these sfGFP derivatives, the Np3-TATcoil vector produced the most fluorescent phage, and was the only one that produced phage recognized by 3E6, indicating that it was the best at displaying correctly folded versions of these GFP derivatives. Similarly, in the competitive selection experiment (Table 4), in which all three vectors displaying the three different GFPs (GFP, clys-GFP and myc-GFP) competed with one another for binding to different selectors, the Np3-TATcoil vector was always specifically selected when 3E6 was used as the selector, or when lysozyme was used to select the clys-GFP constructs. This is in contrast to ELISA experiments using lysozyme and clys-GFP, where the Np3-Sec construct gave higher signals than the Np3-TATcoil. This may be due to the difference between the two assays: ELISA is carried out using polystyrene plates, which may denature the lysozyme, while the selection was carried out in solution which should preserve structural integrity. In the case of myc-GFP, tested with 9E10, the Np3-SRP construct gave both the highest ELISA signal, and was preferentially selected in the mock selection, reflecting the fact that this antibody recognizes a linear epitope that does not necessarily require folding (87). However, this result has to be tempered by the observation that phage produced using Np3-SRP were also the most commonly selected in all the remaining experiments, whether the selector was specific or not, suggesting that the poorly folded GFP derivatives displayed by this vector may have been relatively sticky, compared to the others.
On the basis of these results, we believe that the TAT based display vector, in which the g3p and the displayed protein are joined together by extremely high affinity [60 pM coils (84)], is the most efficient for the display of fluorescent proteins in particular, and perhaps cytoplasmic proteins in general. These results are confirmed in the experiments describing the display of the other fluorescent proteins (Figure 6). The TAT vector was able to produce fluorescent phage for all the fluorescent proteins except wild-type GFP (88), while the Sec based vector could only produce significantly fluorescent phage with sfGFP and the dimeric and tetrameric Azami greens (74). For the GFP based constructs, correct display could also be assessed using the monoclonal antibody 3E6, that recognizes a conformational epitope in the correctly folded protein. The TAT vector consistently gave high 3E6 signals for all the GFP based proteins (sfGFP, myc-GFP, clys-GFP and cycle 3 GFP), while a positive signal with the Sec vector was only obtained with the highly stable sfGFP protein. Display of the two tetrameric proteins (tetrameric Azami green and DsRed T1) was signicantly less efficient than display of the other fluorescent proteins. This could be because the size of these tetrameric proteins (∼110 kDa) approaches the size limit for the TAT pathway, or because of the slow folding (and assembly) kinetics of these proteins. Even though DsRed T1 was engineered for faster folding (76), in our hands it still took over 24 h for bacteria to become fluorescent. Given that phage are produced on a far shorter time scale (5–8 h), it is perhaps not surprising that correctly folded tetramers are not displayed when the TAT leader is used: phage production would be expected to be over by the time the proteins have folded and are competent for export. This is in contrast to the Np3-Sec vector in which unfolded protein can be exported and folding may occur on the phage after assembly.
One surprise of the work presented here, was the apparent lack of functionality of vectors that have been previously described as being suitable for the display of specific cytoplasmic proteins (19), or cDNA libraries (20,21). Although we attempted to replicate previously described vectors as closely as possible by using identical linker sequences, some elements, such as the peptide tag [SV5 (79)], and the leader sequence used, were different, and it is conceivable that such (or other) subtle differences, are responsible for the inability to display sfGFP and variants. It is also possible that the specific proteins displayed with these other vectors, were more suitable for these other vectors than the sfGFP we attempted to display. This may relate to particular folding properties, or the presence of longer natural peptide linkers that facilitated display with the Sec leader.
Although there is some overlap, in general, the Sec leader is suitable for secreted proteins, such as antibodies, the SRP leader for proteins or domains that fold rapidly, such as ankyrins and the TAT leader for proteins, which fold in the cytoplasm, such as fluorescent proteins. However, unlike the other two leaders, the TAT leader does not seem to be functional in phage display as a direct fusion, and has to be used in an alternative format, such as that described here. This mirrors results previously obtained with an intracellular carboxylesterase (85), in which direct fusion was also ineffective. The availability of display vectors using three different leaders, suggests that they should be used together to fully display complex proteomes. We would expect the combination of these three vectors to provide greater representation than any single vector alone. Such a combination of vectors should be readily applicable to cDNA selection approaches, in which cDNA, or open reading frame libraries are displayed on phage and selected with sera derived from patients with auto-immunity (21,92), viral/bacterial infections (55,93–98) or neoplasia (47,99–101) allowing the identification of proteins bearing antigenic epitopes, that may have utility in vaccination, treatment or diagnosis.
FUNDING
Funding for open access charge: LANL lab directed research funds.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
ARMB is grateful to Department of Energy, Genomes to Life program (DOE-GTL) and LANL lab directed research funds (LDRD-DR) for support.
REFERENCES
- 1.Bradbury A, Cattaneo A. The use of phage display in neurobiology. Trends Neurosci. 1995;18:243–249. [PubMed] [Google Scholar]
- 2.Hoogenboom HR, de Bruine AP, Hufton SE, Hoet RM, Arends JW, Roovers RC. Antibody phage display technology and its applications. Immunotechnology. 1998;4:1–20. doi: 10.1016/s1380-2933(98)00007-4. [DOI] [PubMed] [Google Scholar]
- 3.McGregor D. Selection of proteins and peptides from libraries displayed on filamentous bacteriophage. Nat. Biotechnol. 1996;6:155–162. doi: 10.1007/BF02740770. [DOI] [PubMed] [Google Scholar]
- 4.Szardenings M. Phage display of random peptide libraries: applications, limits, and potential. J. Recept. Signal. Transduct. Res. 2003;23:307–349. doi: 10.1081/rrs-120026973. [DOI] [PubMed] [Google Scholar]
- 5.Sblattero D, Bradbury A. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat. Biotechnol. 2000;18:75–80. doi: 10.1038/71958. [DOI] [PubMed] [Google Scholar]
- 6.Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 7.Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, McCafferty J, Hodits RA, Wilton J, Johnson KS. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat. Biotechnol. 1996;14:309–314. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 8.de Haard HJ, van Neer N, Reurs A, Hufton SE, Roovers RC, Henderikx P, de Bruine AP, Arends JW, Hoogenboom HR. A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J. Biol. Chem. 1999;274:18218–18230. doi: 10.1074/jbc.274.26.18218. [DOI] [PubMed] [Google Scholar]
- 9.Rothlisberger D, Pos KM, Pluckthun A. An antibody library for stabilizing and crystallizing membrane proteins – selecting binders to the citrate carrier CitS. FEBS Lett. 2004;564:340–348. doi: 10.1016/S0014-5793(04)00359-X. [DOI] [PubMed] [Google Scholar]
- 10.Hoet RM, Cohen EH, Kent RB, Rookey K, Schoonbroodt S, Hogan S, Rem L, Frans N, Daukandt M, Pieters H, et al. Generation of high-affinity human antibodies by combining donor-derived and synthetic complementarity-determining-region diversity. Nat. Biotechnol. 2005;23:344–348. doi: 10.1038/nbt1067. [DOI] [PubMed] [Google Scholar]
- 11.Zhang MY, Shu Y, Phogat S, Xiao X, Cham F, Bouma P, Choudhary A, Feng YR, Sanz I, Rybak S, et al. Broadly cross-reactive HIV neutralizing human monoclonal antibody Fab selected by sequential antigen panning of a phage display library. J. Immunol. Methods. 2003;283:17–25. doi: 10.1016/j.jim.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 13.Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 14.Cwirla SE, Peters EA, Barrett RW, Dower WJ. Peptides on phage: a vast library of peptides for identifying ligands. Proc. Natl Acad. Sci. USA. 1990;87:6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felici F, Castagnoli L, Musacchio A, Jappelli R, Cesareni G. Selection of antibody ligands from a large library of oligopeptides expressed on a multivalent exposition vector. J. Mol. Biol. 1991;222:301–310. doi: 10.1016/0022-2836(91)90213-p. [DOI] [PubMed] [Google Scholar]
- 16.Chappel JA, He M, Kang AS. Modulation of antibody display on M13 filamentous phage. J. Immunol. Methods. 1998;221:25–34. doi: 10.1016/s0022-1759(98)00094-5. [DOI] [PubMed] [Google Scholar]
- 17.Gao C, Mao S, Kaufmann G, Wirsching P, Lerner RA, Janda KD. A method for the generation of combinatorial antibody libraries using pIX phage display. Proc. Natl Acad. Sci. USA. 2002;99:12612–12616. doi: 10.1073/pnas.192467999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao C, Mao S, Lo CH, Wirsching P, Lerner RA, Janda KD. Making artificial antibodies: a format for phage display of combinatorial heterodimeric arrays. Proc. Natl Acad. Sci. USA. 1999;96:6025–6030. doi: 10.1073/pnas.96.11.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuh G, Sidhu SS. Efficient phage display of polypeptides fused to the carboxy-terminus of the M13 gene-3 minor coat protein. FEBS Lett. 2000;480:231–234. doi: 10.1016/s0014-5793(00)01946-3. [DOI] [PubMed] [Google Scholar]
- 20.Jespers LS, Messens JH, De Keyser A, Eeckhout D, Van den Brande I, Gansemans YG, Lauwereys MJ, Vlasuk GP, Stanssens PE. Surface expression and ligand-based selection of cDNAs fused to filamentous phage gene VI. Biotechnology (NY) 1995;13:378–382. doi: 10.1038/nbt0495-378. [DOI] [PubMed] [Google Scholar]
- 21.Hufton SE, Moerkerk PT, Meulemans EV, de Bruine A, Arends JW, Hoogenboom HR. Phage display of cDNA repertoires: the pVI display system and its applications for the selection of immunogenic ligands. J. Immunol. Methods. 1999;231:39–51. doi: 10.1016/s0022-1759(99)00139-8. [DOI] [PubMed] [Google Scholar]
- 22.Fransen M, Van Veldhoven PP, Subramani S. Identification of peroxisomal proteins by using M13 phage protein VI phage display: molecular evidence that mammalian peroxisomes contain a 2,4-dienoyl-CoA reductase. Biochem. J. 1999;340(Pt 2):561–568. [PMC free article] [PubMed] [Google Scholar]
- 23.Marvin DA. Filamentous phage structure, infection and assembly. Curr. Opin. Struct. Biol. 1998;8:150–158. doi: 10.1016/s0959-440x(98)80032-8. [DOI] [PubMed] [Google Scholar]
- 24.Held HA, Sidhu SS. Comprehensive mutational analysis of the M13 major coat protein: improved scaffolds for C-terminal phage display. J. Mol. Biol. 2004;340:587–597. doi: 10.1016/j.jmb.2004.04.060. [DOI] [PubMed] [Google Scholar]
- 25.Sidhu SS, Feld BK, Weiss GA. M13 bacteriophage coat proteins engineered for improved phage display. Methods Mol. Biol. 2007;352:205–219. doi: 10.1385/1-59745-187-8:205. [DOI] [PubMed] [Google Scholar]
- 26.Houbiers MC, Hemminga MA. Protein-lipid interactions of bacteriophage M13 gene 9 minor coat protein. Mol. Membr. Biol. 2004;21:351–359. doi: 10.1080/09687860400012918. [DOI] [PubMed] [Google Scholar]
- 27.Manting EH, Driessen AJ. Escherichia coli translocase: the unravelling of a molecular machine. Mol. Microbiol. 2000;37:226–238. doi: 10.1046/j.1365-2958.2000.01980.x. [DOI] [PubMed] [Google Scholar]
- 28.de Gier JW, Luirink J. Biogenesis of inner membrane proteins in Escherichia coli. Mol. Microbiol. 2001;40:314–322. doi: 10.1046/j.1365-2958.2001.02392.x. [DOI] [PubMed] [Google Scholar]
- 29.Driessen AJ, Manting EH, van der Does C. The structural basis of protein targeting and translocation in bacteria. Nat. Struct. Biol. 2001;8:492–498. doi: 10.1038/88549. [DOI] [PubMed] [Google Scholar]
- 30.Muller M, Koch HG, Beck K, Schafer U. Protein traffic in bacteria: multiple routes from the ribosome to and across the membrane. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:107–157. doi: 10.1016/s0079-6603(00)66028-2. [DOI] [PubMed] [Google Scholar]
- 31.Koch HG, Moser M, Muller M. Signal recognition particle-dependent protein targeting, universal to all kingdoms of life. Rev. Physiol. Biochem. Pharmacol. 2003;146:55–94. doi: 10.1007/s10254-002-0002-9. [DOI] [PubMed] [Google Scholar]
- 32.Mergulhao FJ, Summers DK, Monteiro GA. Recombinant protein secretion in Escherichia coli. Biotechnol. Adv. 2005;23:177–202. doi: 10.1016/j.biotechadv.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Berks BC. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 34.Berks BC, Palmer T, Sargent F. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr. Opin. Microbiol. 2005;8:174–181. doi: 10.1016/j.mib.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 35.Berks BC, Sargent F, Palmer T. The Tat protein export pathway. Mol. Microbiol. 2000;35:260–274. doi: 10.1046/j.1365-2958.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 36.Barrett CM, Ray N, Thomas JD, Robinson C, Bolhuis A. Quantitative export of a reporter protein, GFP, by the twin-arginine translocation pathway in Escherichia coli. Biochem. Biophys. Res. Commun. 2003;304:279–284. doi: 10.1016/s0006-291x(03)00583-7. [DOI] [PubMed] [Google Scholar]
- 37.Ize B, Gerard F, Wu LF. In vivo assessment of the Tat signal peptide specificity in Escherichia coli. Arch. Microbiol. 2002;178:548–553. doi: 10.1007/s00203-002-0488-1. [DOI] [PubMed] [Google Scholar]
- 38.DeLisa MP, Samuelson P, Palmer T, Georgiou G. Genetic analysis of the twin arginine translocator secretion pathway in bacteria. J. Biol. Chem. 2002;277:29825–29831. doi: 10.1074/jbc.M201956200. [DOI] [PubMed] [Google Scholar]
- 39.Thomas JD, Daniel RA, Errington J, Robinson C. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 2001;39:47–53. doi: 10.1046/j.1365-2958.2001.02253.x. [DOI] [PubMed] [Google Scholar]
- 40.Santini CL, Bernadac A, Zhang M, Chanal A, Ize B, Blanco C, Wu LF. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J. Biol. Chem. 2001;276:8159–8164. doi: 10.1074/jbc.C000833200. [DOI] [PubMed] [Google Scholar]
- 41.Fisher AC, Kim W, DeLisa MP. Genetic selection for protein solubility enabled by the folding quality control feature of the twin-arginine translocation pathway. Protein Sci. 2006;15:449–458. doi: 10.1110/ps.051902606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steiner D, Forrer P, Stumpp MT, Pluckthun A. Signal sequences directing cotranslational translocation expand the range of proteins amenable to phage display. Nat. Biotechnol. 2006;24:823–831. doi: 10.1038/nbt1218. [DOI] [PubMed] [Google Scholar]
- 43.Paschke M, Hohne W. A twin-arginine translocation (Tat)-mediated phage display system. Gene. 2005;350:79–88. doi: 10.1016/j.gene.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Stroud RM, Serwer P, Ross MJ. Assembly of bacteriophage T7. Dimensions of the bacteriophage and its capsids. Biophys. J. 1981;36:743–757. doi: 10.1016/S0006-3495(81)84763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehmann D, Sodoyer R, Leterme S. Characterization of BoHV-1 gE envelope glycoprotein mimotopes obtained by phage display. Vet. Microbiol. 2004;104:1–17. doi: 10.1016/j.vetmic.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 46.Houshmand H, Bergqvist A. Interaction of hepatitis C virus NS5A with La protein revealed by T7 phage display. Biochem. Biophys. Res. Commun. 2003;309:695–701. doi: 10.1016/j.bbrc.2003.08.054. [DOI] [PubMed] [Google Scholar]
- 47.Hansen MH, Ostenstad B, Sioud M. Identification of immunogenic antigens using a phage-displayed cDNA library from an invasive ductal breast carcinoma tumour. Int. J. Oncol. 2001;19:1303–1309. doi: 10.3892/ijo.19.6.1303. [DOI] [PubMed] [Google Scholar]
- 48.Kurakin A, Wu S, Bredesen DE. Target-assisted iterative screening of phage surface display cDNA libraries. Methods Mol. Biol. 2004;264:47–60. doi: 10.1385/1-59259-759-9:047. [DOI] [PubMed] [Google Scholar]
- 49.Nowak JE, Chatterjee M, Mohapatra S, Dryden SC, Tainsky MA. Direct production and purification of T7 phage display cloned proteins selected and analyzed on microarrays. Biotechniques. 2006;40:220–227. doi: 10.2144/000112099. [DOI] [PubMed] [Google Scholar]
- 50.Tan GH, Yusoff K, Seow HF, Tan WS. Antigenicity and immunogenicity of the immunodominant region of hepatitis B surface antigen displayed on bacteriophage T7. J. Med. Virol. 2005;77:475–480. doi: 10.1002/jmv.20479. [DOI] [PubMed] [Google Scholar]
- 51.Han Z, Xiong C, Mori T, Boyd MR. Discovery of a stable dimeric mutant of cyanovirin-N (CV-N) from a T7 phage-displayed CV-N mutant library. Biochem. Biophys. Res. Commun. 2002;292:1036–1043. doi: 10.1006/bbrc.2002.6741. [DOI] [PubMed] [Google Scholar]
- 52.Bukanov NO, Meek AL, Klinger KW, Landes GM, Ibraghimov-Beskrovnaya O. A modified two-step phage display selection for isolation of polycystin-1 ligands. Funct. Integr. Genomics. 2000;1:193–199. doi: 10.1007/s101420000024. [DOI] [PubMed] [Google Scholar]
- 53.Takakusagi Y, Kobayashi S, Sugawara F. Camptothecin binds to a synthetic peptide identified by a T7 phage display screen. Bioorg. Med. Chem. Lett. 2005;15:4850–4853. doi: 10.1016/j.bmcl.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 54.Krumpe LR, Atkinson AJ, Smythers GW, Kandel A, Schumacher KM, McMahon JB, Makowski L, Mori T. T7 lytic phage-displayed peptide libraries exhibit less sequence bias than M13 filamentous phage-displayed peptide libraries. Proteomics. 2006;6:4210–4222. doi: 10.1002/pmic.200500606. [DOI] [PubMed] [Google Scholar]
- 55.Santini C, Brennan D, Mennuni C, Hoess RH, Nicosia A, Cortese R, Luzzago A. Efficient display of an HCV cDNA expression library as C-terminal fusion to the capsid protein D of bacteriophage lambda. J. Mol. Biol. 1998;282:125–135. doi: 10.1006/jmbi.1998.1986. [DOI] [PubMed] [Google Scholar]
- 56.Dai M, Temirov J, Pesavento E, Kiss C, Velappan N, Pavlik P, Werner JH, Bradbury AR. Using T7 phage display to select GFP-based binders. Protein Eng Des Sel. 2008;21:413–424. doi: 10.1093/protein/gzn016. [DOI] [PubMed] [Google Scholar]
- 57.Hoffman R. Green fluorescent protein imaging of tumour growth, metastasis, and angiogenesis in mouse models. Lancet Oncol. 2002;3:546–556. doi: 10.1016/s1470-2045(02)00848-3. [DOI] [PubMed] [Google Scholar]
- 58.Kendall JM, Stubbs S. Fluorescent proteins in cellular assays. J. Clinical Ligand Assay. 2002;25:280–292. [Google Scholar]
- 59.Southward CM, Surette MG. The dynamic microbe: green fluorescent protein brings bacteria to light. Mol. Microbiol. 2002;45:1191–1196. doi: 10.1046/j.1365-2958.2002.03089.x. [DOI] [PubMed] [Google Scholar]
- 60.Miyawaki A. Visualization of the spatial and temporal dynamics of intracellular signaling. Dev. Cell. 2003;4:295–305. doi: 10.1016/s1534-5807(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 61.Feilmeier BJ, Iseminger G, Schroeder D, Webber H, Phillips GJ. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 2000;182:4068–4076. doi: 10.1128/jb.182.14.4068-4076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reid BG, Flynn GC. Chromophore formation in green fluorescent protein. Biochemistry. 1997;36:6786–6791. doi: 10.1021/bi970281w. [DOI] [PubMed] [Google Scholar]
- 63.Li L, Kang DG, Cha HJ. Functional display of foreign protein on surface of Escherichia coli using N-terminal domain of ice nucleation protein. Biotechnol. Bioeng. 2004;85:214–221. doi: 10.1002/bit.10892. [DOI] [PubMed] [Google Scholar]
- 64.Shi H, Wen Su W. Display of green fluorescent protein on Escherichia coli cell surface. Enzyme Microb. Technol. 2001;28:25–34. doi: 10.1016/s0141-0229(00)00281-7. [DOI] [PubMed] [Google Scholar]
- 65.Huang D, Shusta EV. Secretion and surface display of green fluorescent protein using the yeast Saccharomyces cerevisiae. Biotechnol. Prog. 2005;21:349–357. doi: 10.1021/bp0497482. [DOI] [PubMed] [Google Scholar]
- 66.Ye K, Shibasaki S, Ueda M, Murai T, Kamasawa N, Osumi M, Shimizu K, Tanaka A. Construction of an engineered yeast with glucose-inducible emission of green fluorescence from the cell surface. Appl. Microbiol. Biotechnol. 2000;54:90–96. doi: 10.1007/s002539900307. [DOI] [PubMed] [Google Scholar]
- 67.Kratz PA, Bottcher B, Nassal M. Native display of complete foreign protein domains on the surface of hepatitis B virus capsids. Proc. Natl Acad. Sci. USA. 1999;96:1915–1920. doi: 10.1073/pnas.96.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mottershead D, van der Linden I, von Bonsdorff CH, Keinanen K, Oker-Blom C. Baculoviral display of the green fluorescent protein and rubella virus envelope proteins. Biochem. Biophys. Res. Commun. 1997;238:717–722. doi: 10.1006/bbrc.1997.7372. [DOI] [PubMed] [Google Scholar]
- 69.Toivola J, Ojala K, Michel PO, Vuento M, Oker-Blom C. Properties of baculovirus particles displaying GFP analyzed by fluorescence correlation spectroscopy. Biol. Chem. 2002;383:1941–1946. doi: 10.1515/BC.2002.218. [DOI] [PubMed] [Google Scholar]
- 70.Rahman MM, Gopinathan KP. Bombyx mori nucleopolyhedrovirus-based surface display system for recombinant proteins. J. Gen. Virol. 2003;84:2023–2031. doi: 10.1099/vir.0.19225-0. [DOI] [PubMed] [Google Scholar]
- 71.Meulenbroek RA, Sargent KL, Lunde J, Jasmin BJ, Parks RJ. Use of adenovirus protein IX (pIX) to display large polypeptides on the virion–generation of fluorescent virus through the incorporation of pIX-GFP. Mol. Ther. 2004;9:617–624. doi: 10.1016/j.ymthe.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 72.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 73.Ayriss J, Woods T, Bradbury A, Pavlik P. High-throughput screening of single-chain antibodies using multiplexed flow cytometry. J. Proteome. Res. 2007;6:1072–1082. doi: 10.1021/pr0604108. [DOI] [PubMed] [Google Scholar]
- 74.Karasawa S, Araki T, Yamamoto-Hino M, Miyawaki A. A green-emitting fluorescent protein from Galaxeidae coral and its monomeric version for use in fluorescent labeling. J. Biol. Chem. 2003;278:34167–34171. doi: 10.1074/jbc.M304063200. [DOI] [PubMed] [Google Scholar]
- 75.Kiss C, Fisher H, Pesavento E, Dai M, Valero R, Ovecka M, Nolan R, Phipps ML, Velappan N, Chasteen L, et al. Antibody binding loop insertions as diversity elements. Nucleic Acids Res. 2006;34:e132. doi: 10.1093/nar/gkl681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat. Biotechnol. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 77.Mourez M, Collier RJ. Use of phage display and polyvalency to design inhibitors of protein-protein interactions. Methods Mol. Biol. 2004;261:213–228. doi: 10.1385/1-59259-762-9:213. [DOI] [PubMed] [Google Scholar]
- 78.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 79.Hanke T, Szawlowski P, Randall RE. Construction of solid matrix-antibody-antigen complexes containing simian immunodeficiency virus p27 using tag-specific monoclonal antibody and tag-linked antigen. J. Gen. Virol. 1992;73:653–660. doi: 10.1099/0022-1317-73-3-653. [DOI] [PubMed] [Google Scholar]
- 80.Crameri R, Suter M. Display of biologically active proteins on the surface of filamentous phages: a cDNA cloning system for selection of functional gene products linked to the genetic information responsible for their production. Gene. 1993;137:69–75. doi: 10.1016/0378-1119(93)90253-y. [DOI] [PubMed] [Google Scholar]
- 81.Crameri R, Jaussi R, Menz G, Blaser K. Display of expression products of cDNA libraries on phage surfaces. A versatile screening system for selective isolation of genes by specific gene-product/ligand interaction. Eur. J. Biochem. 1994;226:53–58. doi: 10.1111/j.1432-1033.1994.tb20025.x. [DOI] [PubMed] [Google Scholar]
- 82.Crameri R, Suter M. Display of biologically active proteins on the surface of filamentous phages: a cDNA cloning system for the selection of functional gene products linked to the genetic information responsible for their production [Gene 137 (1993) 69–75] Gene. 1995;160:139. doi: 10.1016/0378-1119(93)90253-y. [DOI] [PubMed] [Google Scholar]
- 83.Rothe C, Urlinger S, Lohning C, Prassler J, Stark Y, Jager U, Hubner B, Bardroff M, Pradel I, Boss M, et al. The human combinatorial antibody library HuCAL GOLD combines diversification of all six CDRs according to the natural immune system with a novel display method for efficient selection of high-affinity antibodies. J. Mol. Biol. 2008;376:1182–1200. doi: 10.1016/j.jmb.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 84.De Crescenzo G, Litowski JR, Hodges RS, O'C;onnor-McCourt MD. Real-time monitoring of the interactions of two-stranded de novo designed coiled-coils: effect of chain length on the kinetic and thermodynamic constants of binding. Biochemistry. 2003;42:1754–1763. doi: 10.1021/bi0268450. [DOI] [PubMed] [Google Scholar]
- 85.Droge MJ, Boersma YL, Braun PG, Buining RJ, Julsing MK, Selles KG, van Dijl JM, Quax WJ. Phage display of an intracellular carboxylesterase of Bacillus subtilis: comparison of Sec and Tat pathway export capabilities. Appl. Environ. Microbiol. 2006;72:4589–4595. doi: 10.1128/AEM.02750-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 87.Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 89.Crameri A, Whitehorn EA, Tate E, Stemmer WP. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 90.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 91.Desmyter A, Transue TR, Ghahroudi MA, Thi MH, Poortmans F, Hamers R, Muyldermans S, Wyns L. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat. Struct. Biol. 1996;3:803–811. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- 92.Bluthner M, Bautz EK, Bautz FA. Mapping of epitopes recognized by PM/Scl autoantibodies with gene-fragment phage display libraries. J. Immunol. Methods. 1996;198:187–198. doi: 10.1016/s0022-1759(96)00160-3. [DOI] [PubMed] [Google Scholar]
- 93.Wang LF, Du Plessis DH, White JR, Hyatt AD, Eaton BT. Use of a gene-targeted phage display random epitope library to map an antigenic determinant on the bluetongue virus outer capsid protein VP5. J. Immunol. Methods. 1995;178:1–12. doi: 10.1016/0022-1759(94)00235-o. [DOI] [PubMed] [Google Scholar]
- 94.Beghetto E, Pucci A, Minenkova O, Spadoni A, Bruno L, Buffolano W, Soldati D, Felici F, Gargano N. Identification of a human immunodominant B-cell epitope within the GRA1 antigen of Toxoplasma gondii by phage display of cDNA libraries. Int. J. Parasitol. 2001;31:1659–1668. doi: 10.1016/s0020-7519(01)00288-0. [DOI] [PubMed] [Google Scholar]
- 95.Beghetto E, Spadoni A, Buffolano W, Del Pezzo M, Minenkova O, Pavoni E, Pucci A, Cortese R, Felici F, Gargano N. Molecular dissection of the human B-cell response against Toxoplasma gondii infection by lambda display of cDNA libraries. Int. J. Parasitol. 2003;33:163–173. doi: 10.1016/s0020-7519(02)00256-4. [DOI] [PubMed] [Google Scholar]
- 96.Ludewig B, Krebs P, Metters H, Tatzel J, Tureci O, Sahin U. Molecular characterization of virus-induced autoantibody responses. J. Exp. Med. 2004;200:637–646. doi: 10.1084/jem.20040358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sahin U, Tureci O, Graf C, Meyer RG, Lennerz V, Britten CM, Dumrese C, Scandella E, Wolfel T, Ludewig B. Rapid molecular dissection of viral and bacterial immunomes. Eur. J. Immunol. 2006;36:1049–1057. doi: 10.1002/eji.200535538. [DOI] [PubMed] [Google Scholar]
- 98.Kim SH, Park SY. Selection and characterization of human antibodies against hepatitis B virus surface antigen (HBsAg) by phage-display. Hybrid Hybridomics. 2002;21:385–392. doi: 10.1089/153685902761022742. [DOI] [PubMed] [Google Scholar]
- 99.Somers VA, Brandwijk RJ, Joosten B, Moerkerk PT, Arends JW, Menheere P, Pieterse WO, Claessen A, Scheper RJ, Hoogenboom HR, et al. A panel of candidate tumor antigens in colorectal cancer revealed by the serological selection of a phage displayed cDNA expression library. J. Immunol. 2002;169:2772–2780. doi: 10.4049/jimmunol.169.5.2772. [DOI] [PubMed] [Google Scholar]
- 100.Chen YT, Scanlan MJ, Sahin U, Tureci O, Gure AO, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old LJ. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc. Natl Acad. Sci. USA. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Scanlan MJ, Chen YT, Williamson B, Gure AO, Stockert E, Gordan JD, Tureci O, Sahin U, Pfreundschuh M, Old LJ. Characterization of human colon cancer antigens recognized by autologous antibodies. Int. J. Cancer. 1998;76:652–658. doi: 10.1002/(sici)1097-0215(19980529)76:5<652::aid-ijc7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 102.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 103.McCafferty J, Jackson RH, Chiswell DJ. Phage-enzymes: expression and affinity chromatography of functional alkaline phosphatase on the surface of bacteriophage. Protein Eng. 1991;4:955–961. doi: 10.1093/protein/4.8.955. [DOI] [PubMed] [Google Scholar]
- 104.Iannolo G, Minenkova O, Petruzzelli R, Cesareni G. Modifying filamentous phage capsid: limits in the size of the major capsid protein. J. Mol. Biol. 1995;248:835–844. doi: 10.1006/jmbi.1995.0264. [DOI] [PubMed] [Google Scholar]
- 105.Weiss GA, Wells JA, Sidhu SS. Mutational analysis of the major coat protein of M13 identifies residues that control protein display. Protein Sci. 2000;9:647–654. doi: 10.1110/ps.9.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kretzschmar T, Geiser M. Evaluation of antibodies fused to minor coat protein III and major coat protein VIII of bacteriophage M13. Gene. 1995;155:61–65. doi: 10.1016/0378-1119(94)00897-2. [DOI] [PubMed] [Google Scholar]