Abstract
Purpose
PAX6 is a critical regulator of the developing lens, other ocular tissues, central nervous system, and pancreas. There are two alternatively spliced forms of the protein, PAX6 and PAX6(5a), that may have different regulatory functions. This study was designed to determine the amounts of PAX6 and PAX6(5a) transcripts present in adult human lens epithelium and fibers, human cornea and monkey retina.
Methods
PAX6 and PAX6(5a) transcript levels were monitored in microdissected lens epithelia, lens fibers, whole lens, cornea, and retina by competitive RT-PCR. The levels of TBP/TFIID were examined in adult human lens epithelium and fibers as control.
Results
PAX6 and PAX6(5a) were expressed at equal levels in lens epithelium and fibers. Ninety-five times more PAX6 transcripts were detected in the epithelial cells than in the fibers. Adult human cornea and monkey retina expressed less PAX6/PAX6(5a) than lens epithelium but more than lens fibers. Correspondingly, 40 fold higher levels of TBP transcripts were detected in lens epithelium than fibers, suggesting reduced overall expression of transcription factors in the adult lens fibers.
Conclusions
The presence of PAX6 and PAX6(5a) messages and proteins in adult lens epithelium suggest functions for both forms of PAX6 in the growth and maintenance of the adult human lens. The reduced levels of both forms of PAX6 in the lens fibers suggest down regulation of this gene during differentiation of epithelia into fibers. The lower level of TBP expression in lens fibers also suggests reduced transcriptional competence of adult lens fibers.
PAX6 is a critical regulatory gene located on human chromosome 11p13 and is expressed in many developing ocular tissues, brain, and pancreas [1]. PAX6 encodes a specific DNA-binding transcription factor capable of initiating lens [2] and eye [3] development. Heterozygous mutations in PAX6 induce a spectrum of ocular diseases including aniridia, Peters' anomaly, autosomal dominant keratitis, foveal hypoplasia, and early onset cataract [1]. Homozygous PAX6 mutations cause anophthalmia, severely damaged brain, and neonatal lethality [4]. Interestingly, a specific mutation in PAX6 may also be involved in subtle abnormalities of frontal lobe function [5]. Two major forms of the protein, PAX6 and PAX6(5a) (Figure 1A), result from alternate splicing [6]. Studies on the transcriptional regulation of crystallin genes in vertebrate lenses implicate PAX6 as an important regulatory factor [7,8]. The function of Pax-6(5a) has been less extensively studied; however, overexpression of PAX6(5a) relative to PAX6 was detected in human congenital cataract [4]. In addition, ectopic expression of PAX6(5a) in lens fiber cells of transgenic mice resulted in abnormal lens phenotype associated with changes in the levels of cell adhesion proteins [9].
Figure 1.
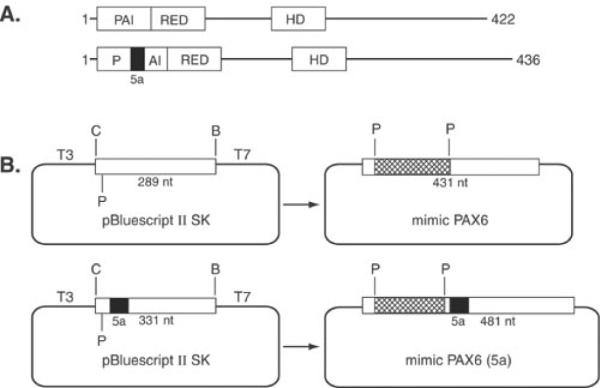
Schematic representations of PAX6 and PAX6(5a) and plasmids used as internal standards. A: Structure of PAX6, 422 amino acids and PAX6(5a), 436 amino acids. Exon 5a encodes a 14 amino acid insertion into the PAI subdomain of the paired domain disrupting its ability to bind DNA. RED subdomain and homeodomain (HD) are also shown. B: Internal standards (mimics) were created by subcloning three copies of a synthetic oligonucleotide into a unique Pst I site of the PAX6 and PAX6(5a) PCR generated fragments in pBluescript SK II as described in the Materials and Methods. The sizes of PCR products for PAX6, mimic PAX6, PAX6(5a) and mimic PAX6(5a) are 289, 431, 331, and 481 bases, respectively.
Expression of PAX6 in adult ocular tissues has not been well documented and the levels of expression of PAX6 and PAX6(5a) mRNA and protein in adult lens has not been investigated. Since PAX6 is likely to play a major role in adult lens growth and maintenance we have characterized the levels of PAX6 and PAX6(5a) expression in the adult human lens using a highly sensitive competitive RT-PCR assay and we have monitored the corresponding protein by immunohistochemistry. We demonstrate that PAX6 and PAX6(5a) are highly expressed at equal ratios in the adult lens epithelium, retina, and cornea. Correspondingly, the levels of PAX6 and PAX6(5a) were 95 times less abundant between lens epithelium and lens fibers. Similarly, TBP/TFIID expression also declined between lens epithelia and lens fibers. The present data confirm greater transcriptional competence of lens epithelium relative to lens fibers and provide the basis for understanding the role of PAX6 and PAX6(5a) in adult lens maintenance.
METHODS
Preparation of RNA
Whole normal human lenses were obtained from an eye bank and microdissected into epithelia and fiber cells as previously described [10]. Lens epithelial and corneal RNA preparations were prepared using the RNeasy kit as specified by the manufacturer (Qiagen, Valencia, CA). Whole lens and fiber RNAs were prepared using TRIZOL reagent as specified by the manufacturer (Life Technologies, Inc., Gaithersburg, MD). Monkey macula RNA was a gift from Dr. Ignacio Rodriguez (National Eye Institute, Bethesda, MD) and was used in lieu of human tissue to avoid potential RNA degradation that is common in human retinal RNA preparations.
Cloning of human PAX6, PAX6(5a), and TBP partial cDNAs
Reverse transcription-polymerase chain reaction (RT-PCR) was employed using a One-step Superscript II RT-PCR kit (Life Technologies, Inc., Gaithersburg, MD) to generate PAX6/PAX6(5a) and TBP products, using total RNA prepared from human lens epithelial cell line B3 [11]. The PAX6 and TBP sequences were obtained from Genbank under accession numbers M93650 and M34960, respectively. Oligonucleotides used for PAX6/PAX6(5a) (AGC TAT CGA TGG CAG AAG ATT GTA GAG and CGG GAT CCG ATG ACA CGC TTG GTA TG) and TBP (CGG GAT CCT TCC ACT CAC AGA CTC TCA C and CGG AAT TCG CTC TCT TAT CCT CAT GAT TAC C) amplification, were extended with non-complementary sequences (blue) with restriction sites ClaI, BamHI, and EcoRI (red) and subcloned as a ClaI-BamHI (PAX6) and a BamHI-EcoRI (TBP) fragments into a pBluescript SK II (Stratagene, LaJolla, CA). The PCR products were purified in 1% agarose gels, specific bands were isolated from the gel using the GFX system (Amersham-Pharmacia, Piscataway, NJ), the resulting fragments were digested with appropriate enzymes, and ligated into a pBluescript KS II. The resulting clones were verified by sequencing at the Albert Einstein College of Medicine DNA Sequencing Core Facility (AECOM).
Design of PCR Mimics
PAX6 and PAX6(5a) transcripts were monitored in the presence or absence of internal quantification standards called PCR mimics [12]. These mimics are plasmid DNAs containing the PAX6 or PAX6(5a) cDNA sequences with an insertion of three copies of a synthetic double stranded oligonucleotide at a unique PstI site of PAX6 or PAX6(5a) partial cDNAs as described above and therefore contain identical coding sequence and primer binding sites as unmodified PAX6 or PAX6(5a) cDNA. A diagrammatic summary of the PCR mimics is shown in Figure 1B.
Competitive Reverse Transcriptase-PCR
Indicated transcripts were reverse-transcribed and amplified using the One Step RT-PCR system (Life Technologies, Inc.) in the presence or absence of PCR mimics as internal standards. Where indicated, control reactions were performed identically in the absence of reverse transcriptase. The initial reverse transcriptase step was conducted at 50 °C for 30 min followed by 30 PCR cycles at annealing temperature of 55 °C. All primers used in this study were designed to cross intron-exon boundaries and were tested in the absence of reverse transcriptase. Control reactions were performed to ensure linearity of amplification over the concentrations of RNA in the present study. The sequences of the primers used for amplification of PAX6 and PAX6(5a) transcripts were 5′-CGG CAG AAG ATT GTA GAG-3′ and 5′-GAT GAC ACG CTT GGT ATG-3′. Where indicated, 0.13 pg of mimic DNA templates (0.65 pg each) were co-amplified with authentic PAX6 mRNA as internal standards. TBP was examined under identical conditions as a control. The sequences of the TBP primers were 5′-TTC CAC TCA CAG ACT CTC AC-3′ and 5′-GCT CTC TTA TCC TCA TGA TTA CC-3′. Where indicated, glyceraldehyde phosphate dehydrogenase (GAPDH) was also examined as a control using the primers 5′-TGT TCC AGT ATG ATT CCA CCC-3′ and 5′-TCC ACC ACC CTG TTG CTG TA-3′.
RESULTS
Quantification of PAX6 and PAX6(5a) transcripts in human lens epithelia and fibers
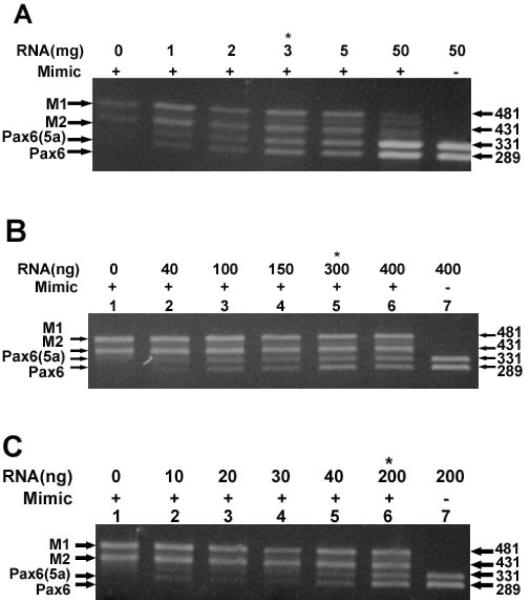
The relative levels of PAX6 and PAX6(5a) transcripts were measured in adult human lens epithelia, lens fibers and whole lenses by competitive RT-PCR [13–15] using PAX6 and PAX6(5a) mimics (Figure 1) as internal quantification standard. The data is shown in Figure 2. Equal amounts of two bands corresponding to PAX6 and PAX6(5a) transcripts were detected in lens epithelia (Figure 2A, lane 7), lens fibers (Figure 2B, lane 7) and whole lens (Figure 2C, lane 7) in the absence of the mimics. Purification and sequencing of these bands confirmed the PAX6 and PAX6(5a) transcripts. As control, the DNA mimics were amplified in the absence of RNA (Figure 2A–C, lane 1). In competitive RT-PCR, the amount of RNA required to compete with a fixed amount of mimic DNA is a quantitative approximation of the amount of target transcript [13–15]. Equal levels of PAX6 and PAX6(5a) transcripts and mimic transcripts were detected at a ratio of 3 ng of epithelial RNA to 0.065 pg of each mimic DNA (Figure 2A, lane 4). Thus, the amount of PAX6 and PAX6(5a) transcript in lens epithelial total RNA is approximately 2.0 × 10−2 pg transcript per ng of RNA. In contrast to the lens epithelium, equal levels of PAX6 and PAX6(5a) transcripts and mimic transcripts were not detected until at least 300 ng of lens fiber RNA was co-amplified with 0.065 pg of each mimic DNA (Figure 2B, lane 5). Based on these values we estimate that the amount of PAX6 and PAX6(5a) transcripts in lens fibers is approximately 2.1 × 10−4 pg transcript per ng of RNA which is 95 times less than that detected in lens epithelia. Using whole lens RNA, equal levels of PAX6 and PAX6(5a) transcripts and mimic transcripts were detected at a ratio of 200 ng of total lens RNA to 0.065 pg of each mimic DNA (Figure 2B, lane 6). Thus, the amount of PAX6 and PAX6(5a) transcript in total lens RNA is approximately 3.2 × 10−4 pg transcript per ng of RNA.
Figure 2.
Quantitation of PAX6 and PAX6(5a) transcripts. Quantitation of PAX6 and PAX6(5a) transcripts in (A) lens epithelia, (B) lens fibers, and (C) whole lens RNA preparations. Ethidium bromide stained gels showing the levels of PAX6 and PAX6(5a) indicated transcripts using increasing amounts of total RNAs. Also indicated are the RNA and mimic DNA concentrations and the positions of the M1 (PAX6(5a) mimic), M2 (PAX6 mimic), PAX6(5a), and PAX6 products. Asterisks indicate amounts of RNA that equally compete with 0.13 pg of mimic DNAs.
Relative levels of TBP in human lens epithelia and fibers
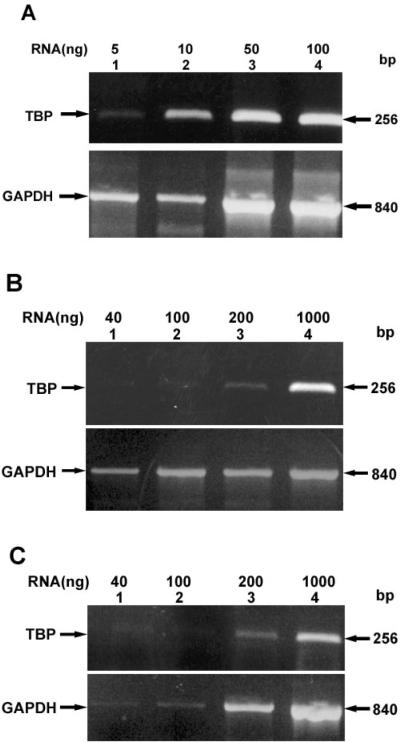
As a measure of the potential of PAX6 and PAX6(5a) to be transcriptionally active in lens epithelia or fibers, the level of TBP mRNA in each preparation was examined relative to GAPDH transcripts. Figure 3A shows the relative levels of TBP present in lens epithelial RNA. Significant amounts of TBP transcript were detected in as little as 5 ng of epithelial RNA (Figure 3A, lane 1). By contrast as much as 200 ng of lens fiber (Figure 3, lane 3) or whole lens (Figure 3B, lane 3) RNA was required to yield similar amounts of transcript. Shown as controls for each panel are the corresponding levels of GAPDH.
Figure 3.
Relative levels of TBP. Relative levels of TBP in (A) lens epithelia, (B) lens fibers, and (C) whole lens total RNA preparations. Ethidium bromide stained gels showing the levels of TBP relative to GAPDH control transcript with indicated amounts of RNA. Also shown are the positions of TBP and GAPDH transcripts.
Quantification of PAX6 and PAX6(5a) transcripts in monkey macula and human cornea
The levels of PAX6 and PAX6(5a) transcripts were also measured in monkey macula and human cornea RNA preparations. Equal levels of PAX6 and PAX6(5a) transcripts and mimic transcripts were obtained using 5 ng of the macula RNA (Figure 4, lane 5) or 10 ng of the cornea RNA preparation using 0.065 pg of each mimic DNA. The amount of each transcript is 1.3 × 10−2 pg transcript per ng of monkey macula RNA and 6.5 × 10−3 pg of transcript per ng of human cornea RNA.
Figure 4.
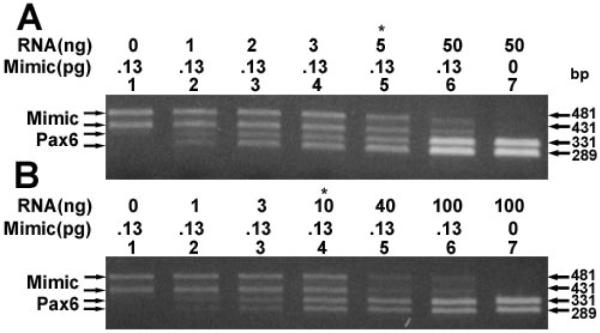
Quantification of PAX6 and PAX6(5a) transcripts. Quantification of PAX6 and PAX6(5a) transcripts in (A) monkey macula and (B) human cornea RNA preparations. Ethidium bromide stained gels showing the levels of PAX6 and PAX6(5a) transcripts using increasing amounts of indicated RNAs. Also indicated are the RNA concentrations and the positions of the M1 (PAX6(5a) mimic), M2 (PAX6 mimic), PAX6(5a), and PAX6 products. Asterisks indicate amounts of RNA that equally compete with 0.13 pg of mimic DNAs.
DISCUSSION
In the present study, we have characterized the expression levels of PAX6 and PAX6(5a) mRNAs in the adult human lenses and compared the absolute levels of these transcripts in lens epithelium and fibers. We also characterized RNA levels of these transcripts in retina and cornea. Our data indicate that lens fibers express lower levels of PAX6 mRNA while human cornea and lens epithelium expressed about 20 times and 95 times more of these mRNAs, respectively. In addition, we found 40 fold more PAX6/PAX6(5a) transcripts in monkey retina over the human lens fibers. The data suggest that while lens fiber cells contain an overwhelming majority of mRNAs encoding crystallins they still express PAX6 mRNAs at appreciable levels. To gain insight into the transcriptional competence of PAX6/PAX6(5a) in lens epithelium and lens fibers, we examined the corresponding levels of TBP, a general transcription factor required for transcription of all structural genes [16]. TBP has been used as a standard for determination of absolute levels of genes expressed in a homogenous population of eukaryotic cells [17,18]. In the present study, we observed about 40 fold reduction of TBP mRNA in lens fiber cells relative to lens epithelium. Thus, we propose that lower expression of PAX6 in lens fibers compared to epithelium can be attributed to two factors, (1) reduced expression as a result of cellular differentiation and (2) a general decrease of expression of other transcription factors represented by TBP/TFIID. Our data provide the first quantification of regulatory mRNAs in adult human lenses, an important step towards the mapping of human lens transcriptosome using cDNA microarrays [18].
Expression of Pax-6/PAX6 in developing lens and eye has been previously examined both using radioactive in situ hybridizations and immunohistochemistry. These studies revealed Pax-6/PAX6 expression and nuclear localization in the lens epithelium while reduced staining was observed in the differentiating lens fibers not distinguishing the two major forms, Pax-6 and Pax-6(5a). In mouse, those studies focused on E12.5 to 15.5 [19–21], and in human, data are available only for 6 and 10 week old human eyes [22]. Later stages were not examined since the denucleation of lens fiber cells make similar studies difficult to conduct. Here, we also performed an immunostaining with a Pax6 specific monoclonal antibody [23] and frozen sections of lens obtained from a 16 year old female donor. As expected [22], PAX6 proteins were detected in the nuclei of lens epithelium but not in the differentiating fiber nuclei (data not shown). In addition, western analysis (data not shown) failed to detect PAX6 in lens extracts obtained from dissected human lenses. This was likely due to the amount of PAX6 below the detection limit of the assay in agreement with previous western analysis using chicken [24] and mouse [9] lenses.
We found equal amounts of PAX6 and PAX6(5a) mRNAs in the adult lens epithelia and fibers. This is in contrast to previous studies on Pax-6 expression in embryonic and neonatal lenses and cultured lens cells. These studies [25–27] detected higher levels of Pax-6 over the Pax-6(5a) in samples examined. Quantitative RNase protections demonstrated constitutive splicing of Pax-6 and Pax-6(5a) at ratio 8:1 throughout mouse embryogenesis and in the adult mouse brain [25]. Similar data were observed using RT-PCR analysis and western blots of Pax-6 expression in cultured mouse (αTN4) and rabbit (N/N1003A) lens epithelial cells [26]. By contrast, a study of bovine lens, retina, cornea, and iris revealed possible tissue-specific fluctuation of Pax6 and Pax6(5a) ratio with the highest level of Pax6(5a) expression in the iris while Pax6 form dominated in the lens [27]. Our data indicate that the adult human lens expresses appreciable high levels of PAX6 and PAX6(5a) mRNAs in lens epithelium but not fibers. Moreover, the levels of PAX6 and PAX6(5a) are also high in the retina and cornea. Surprisingly, the ratio of PAX6 and PAX6(5a) was found to be equal in all tissues examined. This is potentially an important observation since PAX6 and PAX6(5a) may regulate different sets of target genes [4,6,9,28] and earlier studies focused almost exclusively on the role of the presumably more abundant form, PAX6. Not surprisingly, corresponding levels of TBP/TFIID declined in lens fibers indicating a loss of transcriptional competence during the differentiation of lens epithelia into lens fibers. These studies provide the groundwork for future studies to determine the role of PAX6 and PAX6(5a) in adult human lens maintenance and cataract.
ACKNOWLEDGEMENTS
We thank Drs. Ronald M. Burde for his support, Usha P. Andley for B3 lens cells, and Ignacio Rodriguez for monkey macula RNA. Pax-6 monoclonal antibody was obtained from the Developmental Studies Hybridoma Bank (The University of Iowa, Dept. of Biological Sciences, Iowa City, IA) maintained under contract N01-HD-7-3263 from the NICHD. Supported by grants from NIH, EY12200 to A.C., and EY13022 to M.K., Resnick Gerontology Center, Human Genome Program and an unrestricted grant from RPB to the Department of Ophthalmology and Visual Sciences of the Albert Einstein College of Medicine. A.C. is a recipient of a Research to Prevent Blindness Inc. (RPB) Career Development Award.
REFERENCES
- 1.Glaser T, Walton DS, Cai J, Epstein JA, Jepeal L, Maas RL. PAX6 gene mutations in aniridia. In: Wiggs JL, editor. Molecular genetics of ocular disease. Wiley-Liss; New York: 1995. pp. 55–81. [Google Scholar]
- 2.Altmann CR, Chow RL, Lang RA, Hemmati-Brivanlou A. Lens induction by Pax-6 in Xenopus laevis. Dev Biol. 1997;185:119–23. doi: 10.1006/dbio.1997.8573. [DOI] [PubMed] [Google Scholar]
- 3.Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–22. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- 4.Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–71. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 5.Heyman I, Frampton I, van Heyningen V, Hanson I, Teague P, Taylor A, Simonoff E. Psychiatric disorder and cognitive function in a family with an inherited novel mutation of the developmental control gene PAX6. Psychiatr Genet. 1999;9:85–90. doi: 10.1097/00041444-199906000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Epstein JA, Glaser T, Cai J, Jepeal L, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–34. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 7.Cvekl A, Piatigorsky J. Lens development and crystallin gene expression: many roles for Pax-6. Bioessays. 1996;18:621–30. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- 8.Kondoh H. Transcription factors for lens development assessed in vivo. Curr Opin Genet Dev. 1999;9:301–8. doi: 10.1016/s0959-437x(99)80045-8. [DOI] [PubMed] [Google Scholar]
- 9.Duncan MK, Kozmik Z, Cveklova K, Piatigorsky J, Cvekl A. Overexpression of PAX6(5a) in lens fiber cells results in cataract and upregulation of α5β1 integrin expression. J Cell Sci. 2000;113:3173–85. doi: 10.1242/jcs.113.18.3173. [DOI] [PubMed] [Google Scholar]
- 10.Horwitz J, Dovrat A, Straatsma BR, Revilla PJ, Lightfoot DO. Glutathione reductase in human lens epithelium: FAD-induced in vitro activation. Curr Eye Res. 1987;6:1249–56. doi: 10.3109/02713688709025235. [DOI] [PubMed] [Google Scholar]
- 11.Andley UP, Rhim JS, Chylack LT, Jr, Fleming TP. Propagation and immortalization of human lens epithelial cells in culture. Invest Ophthalmol Vis Sci. 1994;35:3094–102. [PubMed] [Google Scholar]
- 12.Bhat SP, Rayner SA, Huang CM, Ariyasu RG. Quantitative estimation of RNA transcripts suggests persistence of Pax-6 expression in the postembryonic chick retina. Dev Neurosci. 1999;21:140–6. doi: 10.1159/000017376. [DOI] [PubMed] [Google Scholar]
- 13.Gilliland G, Perrin S, Blanchard K, Bunn HF. Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction. Proc Natl Acad Sci U S A. 1990;87:2725–9. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouaboula M, Legoux P, Pessegue B, Delpech B, Dumont X, Piechaczyk M, Casellas P, Shire D. Standardization of mRNA titration using a polymerase chain reaction method involving co-amplification with a multispecific internal control. J Biol Chem. 1992;267:21830–8. [PubMed] [Google Scholar]
- 15.Diviacco S, Norio P, Zentilin L, Menzo S, Clementi M, Biamonti G, Riva S, Falaschi A, Giacca M. A novel procedure for quantitative polymerase chain reaction by coamplification of competitive templates. Gene. 1992;122:313–20. doi: 10.1016/0378-1119(92)90220-j. [DOI] [PubMed] [Google Scholar]
- 16.Carey M, Smale ST. Transcriptional regulation in eukaryotes: concepts, strategies, and techniques. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 2000. [Google Scholar]
- 17.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1996;93:5208–12. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lockhart DJ, Winzeler EA. Genomics, gene expression and DNA arrays. Nature. 2000;405:827–36. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- 19.Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–49. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 20.Davis JA, Reed RR. Role of Olf-1 and Pax-6 transcription factors in neurodevelopment. J Neurosci. 1996;16:5082–94. doi: 10.1523/JNEUROSCI.16-16-05082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West-Mays JA, Zhang J, Nottoli T, Hagopian-Donaldson S, Libby D, Strissel KJ, Williams T. AP-2alpha transcription factor is required for early morphogenesis of the lens vesicle. Dev Biol. 1999;206:46–62. doi: 10.1006/dbio.1998.9132. [DOI] [PubMed] [Google Scholar]
- 22.Nishina S, Kohsaka S, Yamaguchi Y, Handa H, Kawakami A, Fujisawa H, Azuma N. PAX6 expression in the developing human eye. Br J Ophthalmol. 1999;83:723–7. doi: 10.1136/bjo.83.6.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–80. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- 24.Duncan MK, Cvekl A, Li X, Piatigorsky J. Truncated forms of Pax-6 disrupt lens morphology in transgenic mice. Invest Ophthalmol Vis Sci. 2000;41:464–73. [PubMed] [Google Scholar]
- 25.Kozmik Z, Czerny T, Busslinger M. Alternatively spliced insertions in the paired domain restrict the DNA sequence specificity of Pax6 and Pax8. EMBO J. 1997;16:6793–803. doi: 10.1093/emboj/16.22.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson J, Cvekl A, Wistow G. Pax-6 is essential for lens-specific expression of zeta-crystallin. Proc Natl Acad Sci U S A. 1995;92:4676–80. doi: 10.1073/pnas.92.10.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaworski C, Sperbeck S, Graham C, Wistow G. Alternative splicing of Pax6 in bovine eye and evolutionary conservation of intron sequences. Biochem Biophys Res Commun. 1997;240:196–202. doi: 10.1006/bbrc.1997.7623. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi Y, Sawada J, Yamada M, Handa H, Azuma N. Auto-regulation of Pax6 transcriptional activation by two distinct DNA-binding subdomains of the paired domain. Genes Cells. 1997;2:255–61. doi: 10.1046/j.1365-2443.1997.1170315.x. [DOI] [PubMed] [Google Scholar]