Abstract
Study Objectives:
Contractile properties of upper airway muscles influence upper airway patency, an issue of particular importance for subjects with obstructive sleep apnea. Expression of genes related to cellular energetics is, in turn, critical for the maintenance of contractile integrity over time during repetitive activation. We tested the hypothesis that sternohyoid has lower expression of genes related to lipid and carbohydrate energetic pathways than the diaphragm.
Methods:
Sternohyoid and diaphragm from normal adult rats were examined with gene expression arrays. Analysis focused on genes belonging to Gene Ontology (GO) groups carbohydrate metabolism and lipid metabolism.
Results:
There were 433 genes with at least ± 2-fold significant differential expression between sternohyoid and diaphragm, of which 192 had sternohyoid > diaphragm and 241 had diaphragm > sternohyoid expression. Among genes with higher sternohyoid expression, there was over-representation of the GO group carbohydrate metabolism (P = 0.0053, n = 13 genes, range of differential expression 2.1- to 6.2-fold) but not lipid metabolism (P = 0.44). Conversely, among genes with higher diaphragm expression, there was over-representation of the GO group lipid metabolism (P = 0.0000065, n = 32 genes, range of differential expression 2.0- to 37.9-fold) but not carbohydrate metabolism (P = 0.23). Nineteen genes with diaphragm > sternohyoid expression were related to fatty acid metabolism (P = 0.000000058), in particular fatty acid β oxidation and biosynthesis in the mitochondria.
Conclusions:
Sternohyoid has much lower gene expression than diaphragm for mitochondrial enzymes that participate in fatty acid oxidation and biosynthesis. This likely contributes to the lower fatigue resistance of pharyngeal upper airway muscles compared with the diaphragm.
Citation:
van Lunteren E; Spiegler S; Moyer M. Differential expression of lipid and carbohydrate metabolism genes in upper airway versus diaphragm muscle. SLEEP 2010;33(3):363-370.
Keywords: Gene expression, upper airway, diaphragm, muscle, metabolism
OBSTRUCTIVE SLEEP APNEA IS A DISORDER WITH A HIGH PREVALENCE IN THE GENERAL POPULATION1 AND CONSIDERABLE ADVERSE CONSEQUENCES.2–4 The anatomic narrowing of the upper airway present in subjects with obstructive sleep apnea5,6 increases the mechanical load on the upper airway and thoracic muscles. As a result, muscle activation must increase above normal levels to maintain upper airway patency and ventilation, not only during wakefulness but also during the termination phase of the repetitive obstructive apneas.7–9 In turn, the structural and contractile properties of the upper airway and thoracic muscles10–13 influence the extent to which these muscle groups can maintain and/or restore upper airway patency and ventilation.
Maintenance of thoracic and upper airway respiratory muscle contractile integrity over time during repetitive contractions is dependent on a sufficient supply of ATP and other energetic intermediates, which in turn are derived from the metabolism of lipids and carbohydrates. The diaphragm is well suited to repetitive activation over long periods of time, being comprised of sizeable proportions of slow-twitch fibers.14–16 On the other hand, most muscles which dilate the pharyngeal upper airway have low proportions of slow-twitch fibers and low resistance to fatigue in relation to the diaphragm.14–20 Based on these differential fiber type distributions, the diaphragm is better suited to oxidative metabolism than are the pharyngeal upper airway muscles. Not well delineated is whether the upper airway muscles and diaphragm also differ with respect to patterns of lipid versus carbohydrate utilization. Patterns of substrate utilization are of interest because they have an important impact on muscle contractile performance,21,22 which further may be perturbed by diseases relevant to sleep apnea, such as diabetes.23–27 The present study tested the hypothesis that an upper airway dilator muscle, the sternohyoid, has lower expression of genes related to cellular lipid and carbohydrate energetic pathways than the diaphragm.
METHODS
Studies were performed on sternohyoid and diaphragm muscle from 6 normal young-adult male Sprague-Dawley rats (weight 391 ± 10 grams). All protocols were approved by the Institutional Animal Care and Use Committee and conformed to animal care guidelines established by the National Institutes of Health. The animals were well anesthetized with a rodent anesthetic cocktail consisting of ketamine, xylazine, and acepromazine. The sternohyoid and diaphragm muscles were removed surgically, placed in RNA-later, and stored at −80° C.
Gene expression array studies were performed as described previously.28–30 Trizol (GibcoBRL, Rockville, MD) was used to extract total RNA, and the RNA pellets were resuspended at 1 μg RNA/μL DEPC-treated water. This was followed by a cleanup protocol with a Qiagen (Valencia, CA) RNeasy Total RNA mini kit. Total RNA was prepared for use on Affymetrix (Santa Clara, CA) microarrays, according to the directions from the manufacturer. Briefly, 8 μg of RNA was used in a reverse transcription reaction (SuperScript II; Life Technologies, Rockville, MD) to generate first strand cDNA. After second strand synthesis, double strand cDNA was used in an in vitro transcription reaction to generate biotinylated cRNA. This was purified and fragmented, following which 15 μg of biotin-labeled cRNA was used in a 300 μL hybridization cocktail, which included spiked transcript controls. Subsequently, 200 μL of the cocktail was loaded onto Affymetrix RAE 230A microarrays (Santa Clara, CA) and hybridized for 16 h at 45oC with agitation. Standard post-hybridization washes and double-stain protocols used an Affymetrix GeneChip Fluidics Station 400. Arrays were scanned using a Hewlett Packard Gene Array scanner and analyzed with Affymetrix MAS 5.0 software.
Bayesian analysis of variance for microarrays (BAM) was chosen for statistical analysis of the microarray data, using BAMarray software (http://www.bamarray.com).28–31 Genes with significantly changed expression identified by BAM were then further selected based on consistent and appropriate present and absent calls per Affymetrix software. Signals were then averaged for diaphragm and sternohyoid muscle, and fold differential expression was calculated based on average values from each group. Analysis focused on genes whose expression was at least ± 2-fold different in the 2 muscles, unless indicated otherwise. To assign biological meaning to the group of genes with differential expression, the subset of genes which met the above criteria was analyzed with the Gene Ontology (GO) classification system, using DAVID software (http://apps1.niaid.nih.gov/david/).32,33 This software determines over-representation of genes with altered expression within specific GO categories by using the one-tailed Fisher exact probability test; this is modified by the addition of a jackknifing procedure, which penalizes the significance of categories with very few genes and favors more robust categories with larger numbers of genes.
Real-time PCR (RT-PCR) was used to confirm changes in gene expression, as described previously.28–30 Testing was done with the same tissue that had been used for gene expression arrays, using an Applied Biosystems ABI 7900HT unit with automation attachment (Foster City, CA). To execute the first step and make archival cDNA, 3 μg of total RNA was reverse transcribed in a 100 μL reaction using Applied Biosystems enzymes and reagents in accordance with the manufacturer's protocols. RNA samples were accurately quantitated using a Nanodrop Technologies ND-1000 spectrophotometer (Wilmington, DE). Equal amounts of total RNA were reverse transcribed and then used in PCR amplifications. GAPDH had very little variation in expression across the sample set and therefore was chosen as the endogenous control. The cDNA reaction from above was diluted by a factor of 10. For the PCR step, 9 μL of this diluted cDNA was used for each of 3 replicate 15 μL reactions carried out in a 384-well plate. Standard PCR conditions were used for the Applied Biosystems assays: 50°C for 2 min, followed by 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec alternating with 60°C for 1 min each. Values for RNA abundance were normalized for each gene with respect to the endogenous control in that sample, mean values for fold changes were calculated for each gene, and statistical testing was performed with the unpaired t-test.
RESULTS
A total of 433 genes had ± 2-fold differential expression between the sternohyoid and diaphragm muscles. There were more genes that had higher expression in diaphragm than sternohyoid (241) compared with the other way around (192). This finding was even more prominent among the 131 genes with > 3-fold differential expression (88 vs 43 genes), and particularly for the 42 genes with > 5-fold differential expression (36 vs 6 genes). Analysis focused on those genes with at least 2-fold differential expression between muscles which belong to the GO terms lipid metabolism, carbohydrate metabolism, and related terms.
For the 241 genes with higher expression in diaphragm than sternohyoid, there were 32 genes assigned to the GO term lipid metabolism, and this was highly significant by over-representation analysis (Table 1). There were also 5 other lipid-related GO terms for genes with higher diaphragm than sternohyoid expression which achieved statistical significance. In contrast, the 11 genes assigned to the GO group carbohydrate metabolism did not meet statistical criteria for over-representation.
Table 1.
Sternohyoid versus Diaphragm GO groups
| GO Term | Number of Genes | P Value for Degree of Over-Representation |
|---|---|---|
| Diaphragm > Sternohyoid Expression | ||
| Lipid Metabolism | 32 | 0.0000065 |
| Cellular Lipid Metabolism | 27 | 0.000030 |
| Fatty Acid Metabolism | 19 | 0.000000058 |
| Lipid Binding | 11 | 0.016 |
| Fatty Acid Binding | 6 | 0.000065 |
| Carbohydrate Metabolism | 11 | NS (P = 0.23) |
| Sternohyoid > Diaphragm Expression | ||
| Carbohydrate Metabolism | 13 | 0.0053 |
| Cellular Carbohydrate Metabolism | 10 | 0.016 |
| Cellular Polysaccharide Metabolism | 5 | 0.000190 |
| Polysaccharide Metabolism | 5 | 0.00022 |
| Polysaccharide Binding | 4 | 0.067 |
| Regulation of Glucose Import | 2 | 0.0086 |
| Lipid Metabolism | 12 | NS (P = 0.44) |
Assignment of genes with differential expression between sternohyoid (SH) and diaphragm (DIA) muscle to the gene ontology (GO) groups lipid metabolism, carbohydrate metabolism and related terms.
Among the 192 genes with higher expression in sternohyoid than diaphragm, there were 13 genes assigned to the GO term carbohydrate metabolism, which was significant by over-representation analysis (Table 1). There were also 5 other carbohydrate-related GO terms which achieved statistical significance among genes with higher sternohyoid than diaphragm expression. In contrast, the 12 genes assigned to the GO group lipid metabolism did not meet statistical criteria for over-representation.
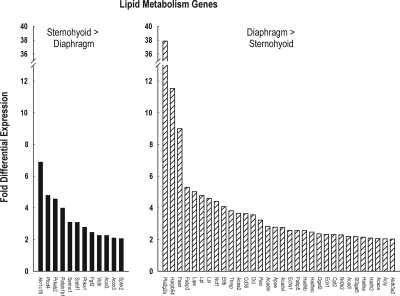
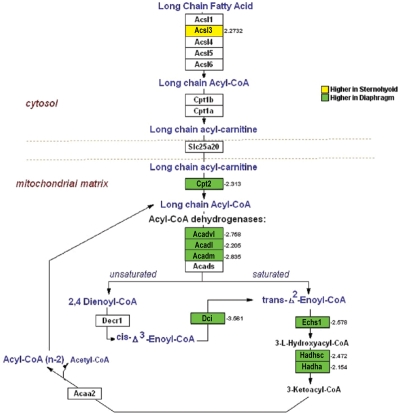
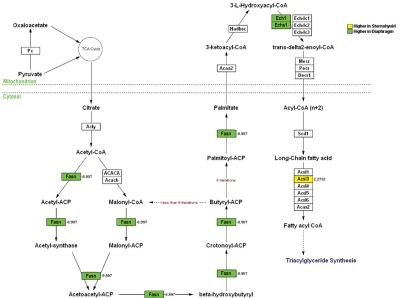
Figure 1 depicts fold-differential expression for each of the genes which were assigned to the GO group lipid metabolism. There were 32 genes with diaphragm > sternohyoid lipid gene expression, the magnitude of which ranged from 2.0- to 37.9-fold, with a mean of 4.6 and a median of 2.8-fold. Conversely the magnitude of the differential expression for the 12 lipid genes with a sternohyoid > diaphragm pattern ranged from 2.1- to 6.9-fold, with a mean of 3.8- and a median of 2.9-fold. Of the 32 lipid metabolism genes, 11 are involved directly in fatty acid energetics pathways, and 10 of the 11 genes had greater diaphragm than sternohyoid expression. Among these 11 genes, 9 participate in specific steps of fatty acid β-oxidation (Figure 2), and 4 participate in specific steps of fatty acid synthesis (Figure 3), with 2 of the 11 genes involved in both (Acsl3 and Echs1). Nine of these 11 genes are involved in mitochondrial rather than cytosolic steps of fatty acid metabolism (Figure 2 and 3).
Figure 1.
Extent of differential expression between sternohyoid and diaphragm muscle of lipid metabolism genes. Left panel depicts data for 12 genes with higher expression in sternohyoid than diaphragm muscle; right panel depicts data for 32 genes with higher expression in diaphragm than sternohyoid muscle.
Figure 2.
Genes with differential expression between sternohyoid and diaphragm muscle that are involved in specific steps of fatty acid β-oxidation. Genes with higher expression in sternohyoid than diaphragm are indicated in red; genes with higher expression in diaphragm than sternohyoid are indicated in green. Numbers indicate fold-differential expression.
Figure 3.
Genes with differential expression between sternohyoid and diaphragm muscle that are involved in specific steps of fatty acid synthesis. Genes with higher expression in sternohyoid than diaphragm are indicated in red; genes with higher expression in diaphragm than sternohyoid are indicated in green. Numbers indicate fold-differential expression.
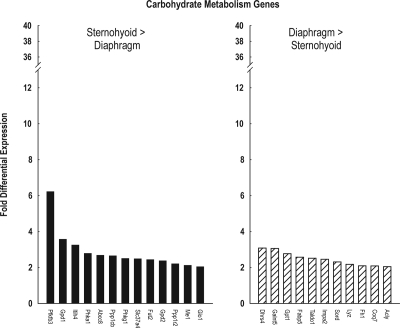
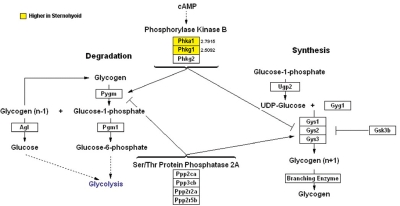
Figure 4 depicts fold-differential expression for each of the genes which were assigned to the GO group carbohydrate metabolism. The magnitude of differential sternohyoid > diaphragm carbohydrate metabolism gene expression ranged from 2.1- to 6.2-fold, with a mean of 2.9- and a median of 2.5-fold. Conversely the magnitude of differential diaphragm > sternohyoid carbohydrate metabolism gene expression ranged from 2.0- to 3.1-fold, with a mean of 2.5- and a median of 2.5-fold. Only 2 of these genes participate directly in cellular energetics pathways, and both had greater sternohyoid than diaphragm expression. Both of these genes are involved in glycogen metabolism (Figure 5).
Figure 4.
Extent of differential expression between sternohyoid and diaphragm muscle of carbohydrate metabolism genes. Left panel depicts data for 13 genes with higher expression in sternohyoid than diaphragm muscle; right panel depicts data for 11 genes with higher expression in diaphragm than sternohyoid muscle.
Figure 5.
Genes with differential expression between sternohyoid and diaphragm muscle that are involved in specific steps of glycogen metabolism. Genes with higher expression in sternohyoid than diaphragm are indicated in red; genes with higher expression in diaphragm than sternohyoid are indicated in green. Numbers indicate fold-differential expression.
RT-PCR studies were performed for 31 genes (Table 2). In all instances the direction of differential expression delineated by PCR was the same as that delineated by gene expression microarrays. Furthermore, all PCR results were statistically significant.
Table 2.
Confirmation by RT-PCR
| Gene Symbol | Fold Change by Array | Fold Change by PCR | P Value by PCR |
|---|---|---|---|
| Sternohyoid > Diaphragm Expression | |||
| Pfkfb3 | 6.2 | 2.6 | 0.028 |
| Phka1 | 2.8 | 2.5 | < 0.001 |
| Abcc8 | 2.7 | 6.1 | < 0.001 |
| Ppp1cb | 2.7 | 1.5 | 0.035 |
| Phkg1 | 2.5 | 1.1 | 0.048 |
| Gpd2 | 2.4 | 4.1 | < 0.001 |
| Ppp1r2 | 2.2 | 1.3 | 0.037 |
| Diaphragm > Sternohyoid Expression | |||
| Pla2g2a | 37.9 | 13.5 | 0.002 |
| Harpb64 | 11.5 | 5.2 | < 0.001 |
| Fasn | 9.0 | 20.6 | 0.003 |
| Fabp3 | 5.3 | 8.0 | < 0.001 |
| Lipe | 5.0 | 9.0 | < 0.001 |
| Lpl | 4.8 | 8.6 | < 0.001 |
| Lsr | 4.6 | 37.7 | < 0.001 |
| Ncf1 | 4.4 | 16.2 | < 0.001 |
| Etfb | 4.1 | 4.5 | < 0.001 |
| Thrsp | 3.8 | 8.7 | 0.009 |
| Acaa2 | 3.7 | 6.5 | < 0.001 |
| Dci | 3.6 | 5.6 | < 0.001 |
| Peci | 3.2 | 3.5 | < 0.001 |
| Acadm | 2.8 | 1.9 | 0.003 |
| Apoe | 2.8 | 5.0 | < 0.001 |
| Cpt2 | 2.3 | 2.5 | < 0.001 |
| Nr0b2 | 2.3 | 10.8 | < 0.001 |
| Acadl | 2.2 | 3.5 | < 0.001 |
| St3gal5 | 2.2 | 2.5 | < 0.001 |
| Hadha | 2.2 | 3.9 | < 0.001 |
| Hadh2 | 2.1 | 4.7 | < 0.001 |
| Acaca | 2.1 | 2.8 | < 0.001 |
| Acly | 2.0 | 3.5 | 0.005 |
| Aldh3a2 | 2.0 | 2.1 | < 0.001 |
RT-PCR confirmation of differential lipid metabolism gene expression between sternohyoid and diaphragm muscles.
DISCUSSION
The present study found a large number of genes with differential expression between the diaphragm and sternohyoid muscle, 68 of which were involved in lipid and carbohydrate metabolism. Many GO terms related to lipid and carbohydrate metabolism were over-represented among the genes with differential expression, with lipid-related GO terms being prominent among genes with diaphragm > sternohyoid expression and carbohydrate-related GO terms being prominent among genes with sternohyoid > diaphragm expression. Thirteen of these differentially-expressed metabolism genes are involved directly in cellular energy pathways, most notably a higher expression of genes related to lipid energetics in diaphragm compared with sternohyoid muscle, most of which are involved with intra-mitochondrial portions of metabolic pathways.
The pharyngeal upper airway muscles differ importantly from the diaphragm with respect to both fiber type composition and contractile properties.14–20 The former are comprised predominantly of fast-twitch fibers whereas the latter are comprised of large proportions of both slow- and fast-twitch fibers. Furthermore, pharyngeal upper airway muscles have faster isometric twitch kinetics and lower fatigue resistance than the diaphragm.
Previous comparisons of pharyngeal and thoracic respiratory muscle biochemical properties have focused on the oxidative vs glycolytic enzyme spectrum or the fast vs slow myosin spectrum rather than the lipid vs carbohydrate enzyme spectrum. LaFramboise et al.14 found that genioglossus muscle had predominantly type 2 (2B > 2A > 2X) but little β /slow myosin heavy chain isoforms, whereas diaphragm had a mixture of β /slow, 2A and 2X, but relative little 2B, myosin heavy chain isoforms. Brozanski et al.34 found that adult genioglossus muscle had a predominance of 2X and 2B with a smaller proportion of 2A myosin heavy chain isoforms whereas diaphragm had approximately equal proportions of slow, 2A, and 2X with a small proportion of 2B myosin heavy chain isoforms. Pae et al.15 found that geniohyoid muscle fibers had only the type 2A myosin heavy chain isoform, sternohyoid muscle fibers had type 2A and 2B myosin heavy chain isoforms, and diaphragm muscle fibers had a mixture of type 1, 2A, 2B, and 2X myosin heavy chain isoforms. With respect to the specific muscles examined in the present study, in rat sternohyoid the proportion of fibers is FG > FOG > SO, irrespective of whether the proportions are quantified by fiber number or fiber cross-sectional area.16 Compared with diaphragm, the SH has a higher proportion of FG fibers and a substantially lower proportion of SO fibers. With respect to FOG fibers, when quantified with respect to fiber number the two muscles have similar proportions, whereas when quantified with respect to fiber cross-sectional area the diaphragm has a higher FOG proportion than the SH.
Data comparing upper airway muscles and diaphragm with regards to specific enzymes involved in cellular energetics are limited. Enzyme activity for citrate synthase is lower for rat sternohyoid and geniohyoid muscles than the diaphragm, whereas enzyme activity of lactate dehydrogenase is higher for the two upper airway muscles than the diaphragm.36 Citrate synthase is a key enzyme of the Krebs (or citric acid) cycle and is involved in aerobic metabolism, whereas lactate dehydrogenase catalyzes the interconversion of lactate and pyruvate and is especially important in anaerobic metabolism. However neither enzyme provides insight into lipid versus carbohydrate metabolism.
We are not aware of previous studies that have compared genome-wide expression patterns among different muscles involved in breathing. However, several previous gene expression array studies have compared diaphragm with limb muscle, albeit without a specific focus on genes involved in metabolism. Porter et al.37 compared normal diaphragm and mixed limb muscle (gastrocnemius/soleus) as part of a study examining dystrophin-deficient muscular dystrophy in mdx mice, and found 726 differentially expressed transcripts. Only a small number of these genes could be attributed to differences in relative proportions of fiber types between diaphragm and pooled limb muscle. Eight slow fiber-related genes were more prominent in diaphragm, whereas 3 fast fiber-related genes were more prominent in pooled limb muscle. In addition four myogenesis-related genes were identified among the differentially expressed genes. Subsequently Haslett et al.38 compared mouse diaphragm with 5 limb muscles, also as part of a study of dystrophin-deficient mdx mice. They found 44 to 50 genes with differential expression each for the normal diaphragm compared with the normal extensor digitorum longus, gastrocnemius, quadriceps, and tibialis anterior muscles, but less than 15 genes for the diaphragm compared with the normal soleus. Genes differentially expressed for the diaphragm were identified as involved in transcriptional regulation, mitochondrial metabolism and fiber type-specific isoforms, but further details about the metabolism group of genes were not provided. Sanoudou et al.39 examined mice with nemaline myopathy, and as part of the study compared normal diaphragm, tibialis anterior, gastrocnemius, plantaris, and extensor digitorum longus. The greatest numbers of genes with differential expression among the normal muscles were seen between the diaphragm and each of the limb muscles rather than among various limb muscles, with values ranging from 1435 to 2350 probe sets. However, no further details were provided about the cellular processes in which these genes participate. Thus there clearly are considerable differences between normal diaphragm and limb muscles in gene expression, but previous studies did not address what extent these differences involve lipid and carbohydrate metabolism. The present study concurs with previous findings indicating considerable heterogeneity among skeletal muscles in patterns of gene expression, and furthermore indicates that heterogeneity in gene expression among muscles involves those related to lipid and carbohydrate metabolism.
One limitation of the present study is that rats are not natural models of obstructive sleep apnea. However, much of what is known about the pharyngeal upper airway respiratory muscles is derived from normal rodents.14–17,19,20,34,36,40 In addition, rodents are frequently used as a model to study the effects of chronic intermittent hypoxia, an important consequence of the episodic upper airway closure that occurs in obstructive sleep apnea.41–44 Finally, there are many rodent models of obesity, diabetes, and hypertension, disorders which in humans frequently coexist with obstructive sleep apnea—and rodent models of these diseases have been used to study thoracic and upper airway respiratory muscles.18,23–26,45–47 Thus the present study using rat muscle is in line with extensive other data from the literature using rodent models to study various aspects of obstructive sleep apnea.
Kimoff48 recently reviewed the literature examining the evidence that abnormalities of the upper airway muscle structure and function play important roles in the pathophysiology of obstructive sleep apnea. This includes alterations in contractile properties, histological perturbations, myopathic changes, evidence for muscle denervation, and a high prevalence of obstructive sleep apnea in primary muscle diseases which involve the pharyngeal muscles. Regarding specifically whether muscle fatigue plays a role in obstructive sleep apnea, data from previous studies that address this include the following. First, there are changes in fiber type distributions in upper airway muscles from slow-twitch type I fibers to fast-twitch type II fibers in sleep apnea, which are in a direction which should increase muscle fatigability. Series et al.49,50 found increased proportions of fast-twitch type IIa fibers in musculus uvulae and genioglossus muscle of humans with OSA compared with snorers. In addition, the musculus uvulae has increased levels of enzymes associated with anaerobic metabolism in subjects with OSA compared with snorers. Subsequently Carrera et al.11,51 found that the type II fiber increase in the genioglossus with OSA is also present when comparisons are made with normal control subjects, and this is not affected by the presence or absence of obesity. An increase in the type II fiber proportion is also present in the sternohyoid muscle of bulldogs that have sleep apnea compared with control animals.13 Second, there are some direct data which indicate that there indeed is worsened muscle fatigability in upper airway muscles of humans with sleep apnea. Carrera et al.11 found that the genioglossus muscle of subjects with obstructive sleep apnea had greater force loss during repetitive stimulation compared with control subjects. Interestingly, in a subsequent study the worsened genioglossus fatigability with obstructive sleep apnea was found in non-obese but not obese subjects.51 Third, hypoxia is an important consequence of the periodic airway closure that occurs in obstructive sleep apnea, and this worsens upper airway muscle fatigability. Acute hypoxia has been demonstrated to impair the fatigue resistance of the geniohyoid muscle52 as well as tongue protruder and retractor muscles.40 In addition, chronic episodic hypoxia increases sternohyoid and geniohyoid muscle fatigue and reduces recovery from fatigue.53,54
Patterns of lipid versus carbohydrate metabolism in both skeletal and cardiac muscle change with disease. One example that is particularly relevant to subjects with sleep apnea is diabetes mellitus. For the diaphragm there is a diabetes-induced shift in cellular energetics away from carbohydrate metabolism23,25,26 and towards lipid metabolism.24,26,27 Similar changes occur in the heart.22,24,26 This appears to be mediated at least in part at a gene expression level, based on findings of increased expression of lipid metabolism genes with reciprocally decreased expression of carbohydrate metabolism genes with diabetes in both limb skeletal muscle55,56 and the heart.30,38,57 Future studies are needed to delineate the impact of obstructive sleep apnea, as well as diseases associated with obstructive sleep apnea, on the expression of metabolism genes in the diaphragm and sternohyoid muscle to determine how this might affect the balance of gene expression in upper airway versus thoracic respiratory muscles.
DISCLOSURE STATEMENT
This was not an industry supported study. The authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
These studies were supported by grants from the Department of Veterans Affairs (Veterans Health Administration). We would like to thank Dr. Patrick Leahy from the Gene Expression Array Core Facility of Case Western Reserve University for his valuable assistance.
REFERENCES
- 1.Young T, Palta M, Dempsey J, Skatrud J, Weber S, Badr S. The occurrence of sleep-disordered breathing among middle-aged adults. New Engl J Med. 1993;32:1230–5. doi: 10.1056/NEJM199304293281704. [DOI] [PubMed] [Google Scholar]
- 2.Flemons WW, Tsai W. Quality of life consequences of sleep-disordered breathing. J Allergy Clin Immunol. 1997;99:S750–6. doi: 10.1016/s0091-6749(97)70123-4. [DOI] [PubMed] [Google Scholar]
- 3.Peppard P, Young T, Palta M, Skatrud J. Prospective study of the association between sleep disordered breathing and hypertension. N Engl J Med. 2000;342:1378–84. doi: 10.1056/NEJM200005113421901. [DOI] [PubMed] [Google Scholar]
- 4.Teran-Santos J, Jimenez-Gomez A, Cordero-Guevara J, Cooperative Group Burgos-Santander The association between sleep apnea and the risk of traffic accidents. N Engl J Med. 1999;340:847–51. doi: 10.1056/NEJM199903183401104. [DOI] [PubMed] [Google Scholar]
- 5.Haponik E, Smith P, Bohlman M, Allan R, Goldman S, and Bleecker E. Computerized tomography in obstructive sleep apnea: correlation of airway size with physiology during sleep and wakefulness. Am Rev Respir Dis. 1983;127:221–6. doi: 10.1164/arrd.1983.127.2.221. [DOI] [PubMed] [Google Scholar]
- 6.Schwab RJ, Gupta KB, Gefter WB, Metzger LJ, Hoffman EA, Pack AI. Upper airway and soft tissue anatomy in normal subjects and patients with sleep-disordered breathing. Significance of the lateral pharyngeal walls. Am J Respir Crit Care Med. 1995;152:1673–89. doi: 10.1164/ajrccm.152.5.7582313. [DOI] [PubMed] [Google Scholar]
- 7.Fogel RB, Trinder J, White DP, et al. The effect of sleep onset on upper airway muscle activity in patients with sleep apnoea versus controls. J Physiol. 2005;564:549–62. doi: 10.1113/jphysiol.2005.083659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mezzanotte WS, Tangel DJ, White DP. Waking genioglossal electromyogram in sleep apnea patients versus normal controls (a neuromuscular compensatory mechanism) J Clin Invest. 1992;89:1571–9. doi: 10.1172/JCI115751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suratt PM, McTier RF, Wilhoit SC. Upper airway muscle activation is augmented in patients with obstructive sleep apnea compared with that in normal subjects. Am Rev Respir Dis. 1988;137:889–94. doi: 10.1164/ajrccm/137.4.889. [DOI] [PubMed] [Google Scholar]
- 10.Boyd JH, Petrof BJ, Hamid Q, Fraser R, Kimoff RJ. Upper airway muscle inflammation and denervation changes in obstructive sleep apnea. Am J Respir Crit Care Med. 2004;170:541–6. doi: 10.1164/rccm.200308-1100OC. [DOI] [PubMed] [Google Scholar]
- 11.Carrera M, Barbé F, Sauleda J, Tomés M, Gómez C, Agustí AGN. Patients with obstructive sleep apnea exhibit genioglossus dysfunction that is normalized after treatment with continuous positive airway pressure. Am J Respir Crit Care Med. 1999;159:1960–6. doi: 10.1164/ajrccm.159.6.9809052. [DOI] [PubMed] [Google Scholar]
- 12.Lindman R, Stal PS. Abnormal palatopharyngeal muscle morphology in sleep-disordered breathing. J Neurol Sci. 2002;195:11–23. doi: 10.1016/s0022-510x(01)00676-1. [DOI] [PubMed] [Google Scholar]
- 13.Petrof BJ, Pack AI, Kelly AM, Eby J, Hendricks JC. Pharyngeal myopathy of loaded upper airway in dogs with sleep apnea. J Appl Physiol. 1994;76:1746–52. doi: 10.1152/jappl.1994.76.4.1746. [DOI] [PubMed] [Google Scholar]
- 14.LaFramboise WA, Watchko JF, Brozanski BS, Daood MJ, Guthrie RD. Myosin heavy chain expression in respiratory muscles of the rat. Am J Respir Cell Mol Biol. 1992;6:335–9. doi: 10.1165/ajrcmb/6.3.335. [DOI] [PubMed] [Google Scholar]
- 15.Pae EK, Wu J, Nguyen D, Monti R, Harper RM. Geniohyoid muscle properties and myosin heavy chain composition are altered after short-term intermittent hypoxic exposure. J Appl Physiol. 2005;98:889–94. doi: 10.1152/japplphysiol.00978.2004. [DOI] [PubMed] [Google Scholar]
- 16.van Lunteren E, Vafaie H, Salomone RJ. Comparative effects of aging on pharyngeal and diaphragm muscles. Respir Physiol. 1995;99:113–25. doi: 10.1016/0034-5687(94)00077-d. [DOI] [PubMed] [Google Scholar]
- 17.Cobos AR, Segade LA, Fuentes I. Muscle fibre types in the suprahyoid muscles of the rat. J Anat. 2001;198:283–94. doi: 10.1046/j.1469-7580.2001.19830283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Lunteren E. Effects of genetic obesity on rat upper airway muscle and diaphragm contractile properties. Eur Respir J. 1996;9:2139–44. [PubMed] [Google Scholar]
- 19.van Lunteren E, Vafaie H. Force potentiation in respiratory muscles: comparison of diaphragm and sternohyoid. Am J Physiol. 1993;264:R1095–100. doi: 10.1152/ajpregu.1993.264.6.R1095. [DOI] [PubMed] [Google Scholar]
- 20.Volz LM, Mann LB, Russell JA, Jackson MA, Leverson GE, Connor NP. Biochemistry of anterior, medial, and posterior genioglossus muscle in the rat. Dysphagia. 2007;22:210–14. doi: 10.1007/s00455-006-9075-y. [DOI] [PubMed] [Google Scholar]
- 21.Dyck JR, Cheng JF, Stanley WC, et al. Malonyl coenzyme a decarboxylase inhibition protects the ischemic heart by inhibiting fatty acid oxidation and stimulating glucose oxidation. Circ Res. 2004;94:e78–84. doi: 10.1161/01.RES.0000129255.19569.8f. [DOI] [PubMed] [Google Scholar]
- 22.Lopaschuk GD. Metabolic abnormalities in the diabetic heart. Heart Fail Rev. 2002;7:149–59. doi: 10.1023/a:1015328625394. [DOI] [PubMed] [Google Scholar]
- 23.Beloff-Chain A, Rookledge KA. The metabolism of glucose in diaphragm muscle from normal rats, from streptozotocin-treated diabetic rats and from rats treated with anti-insulin serum. Biochem J. 1968;110:529–32. doi: 10.1042/bj1100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garland PB, Randle PJ. Regulation of glucose uptake by muscles. 10. Effects of alloxan-diabetes, starvation, hypophysectomy and adrenalectomy, and of fatty acids, ketone bodies and pyruvate, on the glycerol output and concentrations of free fatty acids, long-chain fatty acyl-coenzyme A, glycerol phosphate and citrate-cycle intermediates in rat heart and diaphragm muscles. Biochem J. 1964;93:678–87. doi: 10.1042/bj0930678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ianuzzo CD, Noble EG, Hamilton N, Dabrowski B. Effects of streptozotocin diabetes, insulin treatment, and training on the diaphragm. J Appl Physiol. 1982;52:1471–5. doi: 10.1152/jappl.1982.52.6.1471. [DOI] [PubMed] [Google Scholar]
- 26.Randle PJ, Newsholme EA, Garland PB. Regulation of glucose uptake by muscle. 8. Effects of fatty acids, ketone bodies and pyruvate, and of alloxan-diabetes and starvation, on the uptake and metabolic fate of glucose in rat heart and diaphragm muscles. Biochem J. 1964;93:652–65. doi: 10.1042/bj0930652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stearns SB, Tepperman HM, Tepperman J. Studies on the utilization and mobilization of lipid in skeletal muscles from streptozotocin-diabetic and control rats. J Lipid Res. 1979;20:654–62. [PubMed] [Google Scholar]
- 28.van Lunteren E, Moyer M, Leahy P. Gene expression profiling of diaphragm muscle in alpha2-laminin (merosin)-deficient dy/dy dystrophic mice. Physiol Genomics. 2006;25:85–95. doi: 10.1152/physiolgenomics.00226.2005. [DOI] [PubMed] [Google Scholar]
- 29.van Lunteren E, Spiegler S, Moyer M. Contrast between cardiac left ventricle and diaphragm muscle in expression of genes involved in carbohydrate and lipid metabolism. Respir Physiol Neurobiol. 2008;161:41–53. doi: 10.1016/j.resp.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 30.van Lunteren E, Moyer M. Oxidoreductase, morphogenesis, extracellular matrix and calcium ion binding gene expression in streptozotocin-induced diabetic rat heart. Am J Physiol Endocrinol Metab. 2007;293:E759–68. doi: 10.1152/ajpendo.00191.2007. [DOI] [PubMed] [Google Scholar]
- 31.Ishwaran H, Rao JS. Detecting differentially expressed genes in microarrays using Bayesian model selection. J Am Stat Assoc. 2003;98:438–55. [Google Scholar]
- 32.Dennis G, Sherman BT, Hosack DA, et al. DAVID: data base for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- 33.Hosack DA, Dennis G, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brozanski BS, Daood MJ, Watchko JF, LaFramboise WA, Guthrie RD. Postnatal expression of myosin isoforms in the genioglossus and diaphragm muscles. Pediatr Pulmonol. 1993;15:212–19. doi: 10.1002/ppul.1950150406. [DOI] [PubMed] [Google Scholar]
- 35.Dick TE, van Lunteren E. Fiber subtype distribution of pharyngeal dilator muscles and diaphragm in the cat. J Appl Physiol. 1990;68:2237–40. doi: 10.1152/jappl.1990.68.5.2237. [DOI] [PubMed] [Google Scholar]
- 36.van Lunteren E, Brass EP. Metabolic profiles of cat and rat pharyngeal and diaphragm muscles. Respir Physiol. 1996;105:171–77. doi: 10.1016/0034-5687(96)00043-6. [DOI] [PubMed] [Google Scholar]
- 37.Porter JD, Merriam AP, Leahy P, et al. Temporal gene expression profiling of dystrophin-deficient (mdx) mouse diaphragm identifies conserved and muscle-group specific mechanisms in the pathogenesis of muscular dystrophy. Hum Mol Genet. 2004;13:257–69. doi: 10.1093/hmg/ddh033. [DOI] [PubMed] [Google Scholar]
- 38.Haslett JN, Kang PB, Han M, et al. The influence of muscle type and dystrophin deficiency on murine expression profiles. Mamm Genome. 2005;16:739–48. doi: 10.1007/s00335-005-0053-8. [DOI] [PubMed] [Google Scholar]
- 39.Sanoudou D, Corbett MA, Han M, et al. Skeletal muscle repair in a mouse model of nemaline myopathy. Hum Mol Genet. 2006;15:2603–12. doi: 10.1093/hmg/ddl186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuller DD, Fregosi RF. Fatiguing contractions of tongue protrudor and retractor muscles: influence of systemic hypoxia. J Appl Physiol. 2000;88:2123–30. doi: 10.1152/jappl.2000.88.6.2123. [DOI] [PubMed] [Google Scholar]
- 41.Bradford A, McGuire M, O'Halloran KD. Does episodic hypoxia affect upper airway dilator muscle function? Implications for the pathophysiology of obstructive sleep apnoea. Respir Physiol Neurobiol. 2005;147:223–34. doi: 10.1016/j.resp.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Dematteis M, Godin-Ribuot D, Arnaud C, et al. Cardiovascular consequences of sleep-disordered breathing: contribution of animal models to understanding the human disease. ILAR J. 2009;50:262–81. doi: 10.1093/ilar.50.3.262. [DOI] [PubMed] [Google Scholar]
- 43.Gauda EB. Introduction: Sleep-disordered breathing across the life span: exploring a human disorder using animal models. ILAR J. 2009;50:243–7. doi: 10.1093/ilar.50.3.243. [DOI] [PubMed] [Google Scholar]
- 44.Mahamed S, Mitchell GS. Is there a link between intermittent hypoxia-induced respiratory plasticity and obstructive sleep apnoea? Exp Physiol. 2007;92:27–37. doi: 10.1113/expphysiol.2006.033720. [DOI] [PubMed] [Google Scholar]
- 45.O'Halloran KD, McGuire M, O'Hare T, MacDermott M, Bradford A. Upper airway EMG responses to acute hypoxia and asphyxia are impaired in streptozotocin-induced diabetic rats. Respir Physiol Neurobiol. 2003;14(138):301–8. doi: 10.1016/j.resp.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 46.van Lunteren E, Moyer M. Streptozotocin-diabetes alters action potentials in rat diaphragm. Respir Physiol Neurobiol. 2003;135:9–16. doi: 10.1016/s1569-9048(03)00038-7. [DOI] [PubMed] [Google Scholar]
- 47.van Lunteren E, Moyer M. Altered diaphragm action potentials in Zucker diabetic fatty (ZDF) rats. Respir Physiol Neurobiol. 2006;153:157–165. doi: 10.1016/j.resp.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 48.Kimoff RJ. Upper airway myopathy is important in the pathophysiology of obstructive sleep apnea. J Clin Sleep Med. 2007;3:567–9. [PMC free article] [PubMed] [Google Scholar]
- 49.Series F, Cote C, Simoneau J, et al. Physiologic, metabolic, and muscle fiber type characteristics of musculus uvulae in sleep apnea hypopnea syndrome and in snorers. J Clin Invest. 1995;95:20–5. doi: 10.1172/JCI117640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Series F, Simoneau J, St. Pierre S, Marc I. Characteristics of the genioglossus and musculus uvulae in sleep apnea hypopnea syndrome and in snorers. Am J Respir Crit Care Med. 1996;153:1870–4. doi: 10.1164/ajrccm.153.6.8665048. [DOI] [PubMed] [Google Scholar]
- 51.Carrera M, Barbé F, Sauleda J, et al. Effects of obesity upon genioglossus structure and function in obstructive sleep apnoea. Eur Respir J. 2004;23:425–9. doi: 10.1183/09031936.04.00099404. [DOI] [PubMed] [Google Scholar]
- 52.Salomone RJ, van Lunteren E. Effects of hypoxia and hypercapnia on geniohyoid contractility and endurance. J Appl Physiol. 1991;71:709–15. doi: 10.1152/jappl.1991.71.2.709. [DOI] [PubMed] [Google Scholar]
- 53.McGuire M, MacDermott M, Bradford A. Effects of chronic episodic hypoxia on rat upper airway muscle contractile properties and fiber-type distribution. Chest. 2002;122:1012–7. doi: 10.1378/chest.122.3.1012. [DOI] [PubMed] [Google Scholar]
- 54.Dunleavy M, Bradford A, O'Halloran KD. Oxidative stress impairs upper airway muscle endurance in an animal model of sleep-disordered breathing. Adv Exp Med Biol. 2008;605:458–62. doi: 10.1007/978-0-387-73693-8_80. [DOI] [PubMed] [Google Scholar]
- 55.Knoll KE, Pietrusz JL, Liang M. Tissue-specific transcriptome responses in rats with early streptozotocin-induced diabetes. Physiol Genomics. 2005;21:222–9. doi: 10.1152/physiolgenomics.00231.2004. [DOI] [PubMed] [Google Scholar]
- 56.Lecker SH, Jagoe RT, Gilbert A, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 57.Gerber LK, Aronow BJ, Matlib MA. Activation of a novel long-chain free fatty acid generation and export system in mitochondria of diabetic rat hearts. Am J Physiol. 2006;291:C1198–207. doi: 10.1152/ajpcell.00246.2006. [DOI] [PubMed] [Google Scholar]