Abstract
A large body of evidence indicates that metazoan innate immunity is regulated by the nervous system, but the mechanisms involved in the process and the biological significance of such control remain unclear. We show that a neural circuit involving npr-1, which encodes a G-protein-coupled receptor related to mammalian neuropeptide Y receptors, functions to suppress innate immune responses. The inhibitory function of NPR-1 requires a cyclic GMP-gated ion channel encoded by tax-2 and tax-4 as well as the soluble guanylate cyclase GCY-35. Furthermore, we show that npr-1- and gcy-35-expressing sensory neurons actively suppress immune responses of non-neuronal tissues. A full-genome microarray analysis on animals with altered neural function due to mutation in npr-1 shows an enrichment in genes that are markers of innate immune responses, including those regulated by a conserved PMK-1/P38 MAPK signaling pathway. These results present evidence that neurons directly control innate immunity in C. elegans, suggesting that G-protein coupled receptors may participate in neural circuits that receive inputs from either pathogens or infected sites and integrate them to coordinate appropriate immune responses.
Innate immune defense comprises a variety of mechanisms used by metazoans to prevent microbial infections. Activation of the innate immune system upon pathogen recognition results in a rapid and definitive microbicidal response to invading microorganisms that is fine-tuned to prevent deleterious deficiencies or excesses in the response. The nervous system, which can respond in milliseconds to many types of nonspecific environmental stimuli, has several characteristics that make it an ideal partner with the innate immune system to regulate nonspecific host defenses (1–3). However, even though a large body of evidence indicates that metazoan innate immunity is under the control of the nervous system, the mechanisms involved in the process and the biological significance of such control remain unclear. To provide insights into the neural mechanisms that regulate innate immunity, we have taken advantage of the simple and well studied nervous and innate immune systems of Caenorhabditis elegans.
The powerful genetic approaches available to C. elegans research have been used to address central questions concerning the functions of the nervous system (4). With its 302 neurons and 56 glial cells, which represent 37% of all somatic cells in a hermaphrodite, the nervous system is perhaps the most complex organ of C. elegans. Ablation of different neurons has demonstrated that sensory neurons regulate a variety of physiological processes, including dauer formation and adult lifespan (5–8). In addition, C. elegans neurons are known to express numerous secreted peptides of the TGF beta family, the insulin family, and neuropeptide families (6, 9–13). This myriad of secreted factors has the potential to act at a distance to modulate various physiological processes by regulating the function of neuronal and non-neuronal cells throughout the animal.
Like other free-living nematodes, C. elegans lives in soil environments where it is in contact with soil-borne microbes, including human microbial pathogens; it has evolved physiological mechanisms to respond to different pathogens by activating the expression of innate immune response genes that are conserved across metazoans (14–19). C. elegans also has behavioral responses to pathogenic bacteria such as Bacillus thuringiensis (20, 21), Microbacterium nematophilum (22), Photorhabdus luminescens (23), Pseudomonas aeruginosa (24–26) and Serratia marcescens (24, 27, 28). Animals infected with these pathogens avoid lawns of the pathogen, or migrate away from pathogen odors. It is currently unknown how the nematode can sense pathogenic bacteria, though mutants in sensory transduction molecules such as the Gi-like protein ODR-3 and the G protein receptor kinase GRK-2 are incapable of S. marcescens lawn avoidance (28). These results suggest that G-protein coupled receptors may participate in neural circuits that receive inputs from either pathogens or infected sites and integrate them to coordinate appropriate defense responses.
To study the role of GPCRs in the regulation of innate immune response, we first determined the susceptibility of forty C. elegans strains carrying mutations in GPCRs to the human opportunistic pathogen Pseudomonas aeruginosa strain PA14, a clinical isolate capable of rapidly killing C. elegans at 25°C (29, 30) (Table 1). Out of the 40 mutants studied, three mutants exhibited enhanced resistance to P. aeruginosa and only one mutant exhibited enhanced susceptibility to P. aeruginosa (Fig. 1, A and B). Interestingly, the strain exhibiting enhanced susceptibility to P. aeruginosa-mediated killing carries a loss-of-function mutation in npr-1, which encodes a G protein-coupled receptor related to mammalian neuropeptide Y receptors (31).
Table 1.
GPCRs are involved in innate immunity to P. aeruginosa
| Strain | Gene | Description | Closest human homologa | Expression Pattern | TD50 Mean±SEM, N | P-value | PA14 phenotypeb |
|---|---|---|---|---|---|---|---|
| N2 | --- | --- | --- | --- | 57.20 ± 5.748 N=5 | --- | WT |
| RB799 | C25G6.5 | Putative GPCR | Prolactin-releasing peptide receptor | AIA, AIY, PVQ | 68.00 ± 5.115 N=4 | 0.2147 | WT |
| RB1284 | C30F12.6 | Putative GPCR | Thyrotropin-releasing hormone receptor | pharynx, intestine | 68.33 ± 5.925 N=3 | 0.2528 | WT |
| RB1289 | C43C3.2 | Putative GPCR | Melanin-concentrating hormone receptor 1 | unknown | 68.00 ± 7.000 N=2 | 0.3427 | WT |
| RB1288 | C48C5.1 | Putative GPCR | Neuromedin U receptor 2 | unknown | 81.50 ± 9.500 N=2 | 0.0753 | ERP |
| RB1423 | C49A9.7 | Putative GPCR | Substance P receptor | unknown | 70.00 ± 7.572 N=3 | 0.2244 | WT |
| RB1321 | C56G3.1 | Putative GPCR | Isoform B of Somatostatin receptor | unknown | 78.33 ± 1.764 N=3 | 0.0347* | ERP |
| RB1162 | cfz-2 | Frizzled family of membrane receptors | Frizzled-8 precursor | pharyngeal neurons | 51.25 ± 4.820 N=4 | 0.4689 | WT |
| RB665 | dop-1 | D1-like dopamine receptor | D(1B) dopamine receptor | head support cells, RIS, AVM, ALM, ALN, PLN, PVQ, PLM, PHC, ALM, AUA, RIB, RIM | 68.75 ± 3.945 N=4 | 0.1616 | WT |
| LX702 | dop-2 | D2-like dopamine receptor | Isoform 3 of D(2) dopamine receptor | RIA, SIA, SIB, RID, PDA | 55.00 ± 3.606 N=3 | 0.7951 | WT |
| BZ873 | dop-3 | D2-like dopamine receptor | Isoform 2 of D(2) dopamine receptor | neurons of the head, ventral cord and tail, GABAergic neurons | 39.50 ± 0.5000 N=2 | 0.1252 | ESP |
| RB761 | F35G8.1 | Putative GPCR | Isoform 2 of Neuropeptide FF receptor 2 | unknown | 60.00 ± 6.245 N=3 | 0.7642 | WT |
| RB509 | gnrr-1 | GoNadotropin-Releasing hormone Receptor | Isoform 1 of Gonadotropin- releasing hormone receptor | unknown | 74.33 ± 6.173 N=3 | 0.1023 | WT |
| RB1349 | F57H12.4 | Putative GPCR | Isoform 1A of Growth hormone secretagogue receptor type 1 | unknown | 89.33 ± 3.283 N=3 | 0.0071** | ERP |
| RB896 | gar-1 | G-protein-linked acetylcholine receptor | Muscarinic acetylcholine receptor M1 | ciliated head neurons, PVM | 72.00 ± 4.000 N=3 | 0.1212 | WT |
| RB756 | gar-2 | G-protein-linked acetylcholine receptor | Muscarinic acetylcholine receptor M2 | sensory, ventral cord neurons, HSN | 64.00 ± 5.033 N=3 | 0.4542 | WT |
| JD217 | gar-3 | G-protein-linked acetylcholine receptor | Muscarinic acetylcholine receptor M1 | pharyngeal muscle, I3, extrapharyngeal neurons | 75.50 ± 7.500 N=2 | 0.1389 | WT |
| VC158 | lat-2 | Latrophilin receptor | Uncharacterized protein LPHN2 | g1 gland cells, arcade cells | 56.50 ± 3.279 N=4 | 0.9245 | WT |
| DA609 | npr-1 | G-protein coupled neuropeptide receptor | Isoform 2 of Neuropeptide FF receptor 2 | AQR, ASE, ASG, ASH, URX, IL2L/R OLQ, AUA, SAAD, RMG, SMBD, M3, VD, DD PQR, PHA, PHB, RIV, RIG, SDQ | 37.80 ± 4.067 N=5 | 0.0246* | ESP |
| XA3702 | npr-2 | G-protein coupled neuropeptide receptor | Isoform 2 of Neuropeptide FF receptor 2 | unknown | 62.67 ± 2.186 N=3 | 0.5111 | WT |
| CX3410 | odr-10 | Odorant receptor | Olfactory receptor 5B17 | AWA | 62.00 ± 2.517 N=3 | 0.5648 | WT |
| RB1141 | R13H7.2 | Putative GPCR | Neuromedin U receptor 2 | Intestine, head neurons | 64.67 ± 2.028 N=3 | 0.3757 | WT |
| DA1814 | ser-1 | Serotonin/octopamine receptor | 5-hydroxytryptamine 2A receptor | RMH, RMF, RMD, pharyngeal muscles | 63.00 ± 8.505 N=3 | 0.578 | WT |
| OH313 | ser-2 | Serotonin/octopamine receptor | 5-hydroxytryptamine receptor 1A | AIY, AVH, AUA, RIC, SAB, RID, RIA, SDQ, CAN, DA9, LUA, ALN, PVC, NSM, AIZ, DVA, BDU, SIA, PVT, RME, OLL, PVD | 54.00 ± 8.963 N=3 | 0.7616 | WT |
| RB1622 | ser-3 | Serotonin/octopamine receptor | Isoform 2 of Alpha-1A adrenergic receptor | Head, tail neurons | 64.25 ± 4.956 N=4 | 0.3983 | WT |
| AQ866 | ser-4 | Serotonin/octopamine receptor | 5-hydroxytryptamine receptor 1B | PVT, RIB, DVA, RIS, DVC | 59.00 ± 2.517 N=3 | 0.8269 | WT |
| DA2100 | ser-7 | Serotonin/octopamine receptor | Isoform D of 5- hydroxytryptamine receptor 7 | Pharyngeal neurons MC, M4, I2, I3, M5, M3, I4, I6 and M2 | 61.00 ± 2.000 N=3 | 0.6435 | WT |
| CB5414 | srd-1 | Serpentine Receptor, class D | Melanin-concentrating hormone receptor 2 | ASI | 58.00 ± 2.082 N=3 | 0.9218 | WT |
| VC459 | srd-2 | Serpentine Receptor, class D | G protein-coupled receptor MRGX1 | unknown | 62.67 ± 6.839 N=3 | 0.572 | WT |
| RB1526 | srd-44 | Serpentine Receptor, class D | DRG kappa 1 splice variant KOR 1A | unknown | 59.00 ± 6.494 N=4 | 0.8413 | WT |
| RB1419 | srw-140 | Serpentine Receptor, class W | Isoform 1A of Growth hormone secretagogue receptor type 1 | unknown | 58.67 ± 8.192 N=3 | 0.885 | WT |
| RB1306 | str-182 | 7-transmembrane olfactory receptor | none | unknown | 83.33 ± 7.860 N=3 | 0.0342* | ERP |
| VC342 | str-31 | 7-transmembrane olfactory receptor | Frizzled-8 precursor | unknown | 64.25 ± 6.945 N=4 | 0.4556 | WT |
| RB785 | T02E9.3 | Putative GPCR | Isoform 2 of D(2) dopamine receptor | Head, tail neurons | 57.00 ± 2.646 N=3 | 0.9806 | WT |
| VC125 | tag-126. | Tyramine receptor | beta-1-adrenergic receptor | Head, tail neurons, vulva | 68.67 ± 0.3333 N=3 | 0.1854 | WT |
| VC224 | tag-24 | Biogenic amine receptor | Alpha-2A adrenergic receptor | Head, tail neurons | 76.67 ± 6.839 N=3 | 0.0774 | ERP |
| VC270 | tag-49 | Putative GPCR | Neuromedin-K receptor | Intestine, renal gland cells, nervous system | 67.67 ± 5.840 N=3 | 0.2778 | WT |
| VC273 | tag-89 | Putative GPCR | Thyrotropin-releasing hormone receptor | unknown | 70.00 ± 3.937 N=4 | 0.1264 | WT |
| RB1365 | uvt-6 | Vitellogenin-linked GPCR | Somatostatin receptor type 3 | Head, tail neurons, ventral nerve cord, anal depressor cell, VM1 | 53.33 ± 7.311 N=3 | 0.6933 | WT |
| RB1393 | Y58G8A.4 | Putative GPCR | Prolactin-releasing peptide receptor | unknown | 59.00 ± 4.619 N=3 | 0.8369 | WT |
| RB1405 | Y59H11AL.1 | Putative GPCR | Substance-K receptor | unknown | 62.33 ± 1.202 N=3 | 0.5305 | WT |
Best BLASTP matches to longest protein product (www.wormbase.org)
ERP: enhanced resistance to P. aeruginosa; ESP: enhanced susceptibility to P. aeruginosa; Strains were considered to be significantly ERP or ESP when TD50 was significantly different from wild-type using Student’s exact t-test (bold). Additional strains are designated ERP or ESP due to significant differences (p<0.0001) in survival compared to wild-type in two independent experiments using PRISM to apply a logrank test.
Fig. 1.
C. elegans G-protein coupled receptor NPR-1 is involved in immunity to P. aeruginosa. (A) C. elegans strains carrying mutations in GPCRs were screened for altered survival on P. aeruginosa. C56G3.1(ok1439) (P=0.0347), F57H12.4 (ok1504) (P=0.0071), and str-182(ok1419) (P=0.0342) had enhanced resistance to P. aeruginosa and npr-1(ad609) (P=0.0246) had enhanced susceptibility to P. aeruginosa. Shown is the time required for 50% of the nematodes to die (TD50) as mean +/− SEM corresponding to at least three independent experiments, each of which used at least 40 adult nematodes per strain. (B) Wild-type N2 and npr-1(ad609) (P=0.0001) nematodes were exposed to P. aeruginosa and scored for survival over time. The graph represents combined results of four independent experiments, N≥40 adult nematodes per strain. (C) Wild-type N2 and npr-1(ad609) (P=0.1411) nematodes were exposed to heat-killed P. aeruginosa and scored for survival over time. The graph represents the combined results of two independent experiments, N=100 adult nematodes per strain. (D) Wild-type N2, npr-1(ad609) (P=0.0001), npr-1(ky13) (P=0.0001), npr-1(n1353) (P=0.0001), npr-1(ur89) (P=0.0001), npr-1(g320) (P=0.0001), and the wild isolate (WI) npr-1(g320) (P=0.0922) were exposed to P. aeruginosa and scored for survival over time. Shown is a representative assay of at least 3 independent experiments, N=48 adult nematodes per strain.
In order to determine whether the enhanced susceptibility to P. aeruginosa exhibited by npr-1(ad609) animals (Fig. 1A) was due to a reduction in lifespan or a deficient response to potentially pathogenic bacteria, npr-1(ad609) nematodes were fed heat-killed P. aeruginosa on plates supplemented with ampicillin. No difference in survival was seen between npr-1(ad609) and wild-type nematodes under these conditions, suggesting that the npr-1 mutation affects the immune response to living pathogenic bacteria without altering the basic lifespan of the animals (Fig. 1C and Fig. S1).
We confirmed that NPR-1 is required for C. elegans defense to P. aeruginosa by exposing five additional npr-1 mutants to the pathogen and comparing their survival to that of wild-type animals (Fig. 1D). Strains carrying loss-of-function alleles npr-1(ky13), npr-1(n1353), or npr-1(ur89) or the reduced-function allele npr-1(g320) were more susceptible to P. aeruginosa than wild-type, confirming that NPR-1 is required for the defense response to this pathogen. Interestingly, while the German wild isolate RC301, which contains the npr-1(g320) allele (31), is not significantly susceptible to P. aeruginosa compared to wild type, the npr-1(g320) allele confers susceptibility to P. aeruginosa in an N2 background (Fig. 1D). These results suggest that the German isolate may have evolved a mechanism to compensate for the increased susceptibility to pathogens due to reduced NPR-1 activity.
To determine whether the immune deficiency due to mutation in the npr-1 gene is specific for P. aeruginosa infection, we exposed npr-1(ad609) nematodes to Salmonella enterica and Enterococcus faecalis, two human pathogens known to kill C. elegans (32–34). As shown in Figures S3A and S3B, npr-1(ad609) nematodes exhibited enhanced susceptibility to these pathogens, suggesting that NPR-1 is required for immune responses to pathogens in general.
NPR-1 is involved in a neural circuit that integrates behavioral responses to environmental oxygen, food, and other animals. In nature, NPR-1 is found in two allelic forms that differ in a single amino acid at position 215, NPR-1(215V) and NPR-1(215F) (31). The NPR1(215V) allele, which is found the in standard laboratory strain, has high activity whereas the NPR-1(215F) allele has low activity (35, 36). Wild-type npr-1(215V) animals avoid oxygen levels above 10% when food is absent, but fail to avoid high oxygen in the presence of E. coli bacteria, the food provided to C. elegans in the laboratory. By contrast, npr-1(215F) and npr-1animals carrying loss-of-function (lf) alleles have strong hyperoxia avoidance in the absence or presence of E. coli (37). As a result, npr-1(215F) and npr-1(lf) show a preference for the thickest part of a bacterial lawn, the region in which oxygen levels are the lowest (35). In addition, as nematode aggregation into feeding groups decreases local oxygen concentrations, npr-1(215F) and npr-1(lf) form aggregates of nematodes when the animals are grown at densities high enough to allow this behavioral response (37).
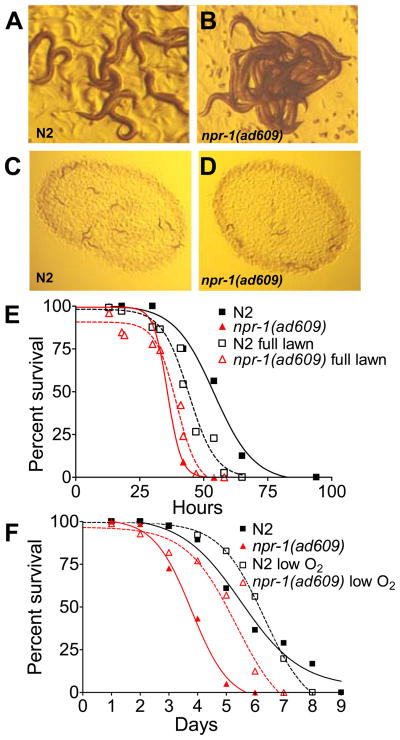
One potential explanation for the reduced lifespan of npr-1(lf) mutants grown on bacterial pathogens is that aggregation increases nematode susceptibility to pathogen infection. However, the animal density in the assays where the susceptibility to pathogens is tested was not sufficient to elicit aggregation, making this possibility unlikely (Fig. 2D). Even though npr-1(ad609) animals did not aggregate, they still exhibited a preference for the thickest part of the lawn where oxygen concentrations are lower (Fig. 2, C and D). In addition, long-term exposure to P. aeruginosa caused wild-type animals to leave the bacterial lawn, a potentially protective behavioral response, but leaving was not observed in npr-1(ad609) animals. Although the number of bacterial cells in npr-1(ad609) animals was not found to be greater than that in wild-type animals (Fig. S2) at early stages of the infection, suggesting that the bacterial dose received by the two animals is comparable, we asked whether the behavior of npr-1(ad609) animals could affect susceptibility to pathogens. Thus, we grew animals on agar plates that were completely covered in P. aeruginosa, a condition that eliminates both the lawn border (favored by npr-1 animals) and the ability to leave the lawn (favored by wild-type animals). As shown in Figure 2E, wild-type animals grown on plates completely covered by P. aeruginosa died at a higher rate than animals grown on plates containing a small lawn of P. aeruginosa in the center of the plate. npr-1(ad609) animals were equally susceptible to P. aeruginosa when grown on full or center lawns. Together, these results indicate that the lawn-leaving behavior of wild-type animals contributes to their increased survival. However, npr-1(ad609) animals still exhibited enhanced susceptibility to P. aeruginosa compared to wild type when the infections were performed in plates containing full lawns (Fig. 2E). These results indicate that lawn avoidance is part of C. elegans defense response to P. aeruginosa, but cannot account for all of the difference between wild-type and npr-1(ad609) animals.
Fig. 2.
Hyperoxia avoidance of NPR-1-deficient animals increases susceptibility to P. aeruginosa. (A) C. elegans wild-type N2 animals and (B) npr-1(ad609) mutants were propagated at 20°C as hermaphrodites on modified NG agar plates seeded with E. coli strain OP50 and then visualized using a Leica MZ FLIII stereomicroscope. The characteristic aggregate of npr-1(ad609) nematodes shown here is at the edge of the bacterial lawn. (C) Twelve wild-type N2 and (D) twelve npr-1(ad609) nematodes were exposed to P. aeruginosa for 24 hours under standard killing assay conditions and visualized using a Leica MZ FLIII stereomicroscope. Under these conditions, npr-1(ad609) nematodes do not form characteristic aggregates of the strain. (E) Wild-type N2 and npr-1(ad609) nematodes were exposed to either a full lawn or a center lawn of P. aeruginosa on a 3.5 cm in diameter plate and scored for survival over time. Under both conditions npr-1(ad609) animals were more susceptible to P. aeruginosa-mediated killing (P=0.0001). Wild-type animals on full lawns were more susceptible to P. aeruginosa-mediated killing than animals on center lawns (P=0.0001); npr-1(ad609) animals were equally susceptible (P=0.07). The graph represents combined results of three independent experiments, N≥40 adult nematodes per strain. (F) Wild-type N2 and npr-1(ad609) nematodes at exposed to P. aeruginosa at either 21% or 8% oxygen and scored for survival over time. Under both conditions npr-1(ad609) animals were more susceptible to P. aeruginosa-mediated killing (P=0.0001). npr-1(ad609) animals at 21% oxygen were more susceptible to P. aeruginosa-mediated killing than animals at 8% oxygen (P=0.0001); wild-type animals were equally susceptible (P=0.95). The graph represents combined results of two independent experiments, N=40 adult nematodes per strain.
To ask whether other elements of the oxygen response contribute to the enhanced susceptibility of npr-1(ad609) nematodes, animals grown at 21% oxygen were compared to those grown at 8% oxygen, a favorable oxygen environment that suppresses most behavioral phenotypes of npr-1 mutants. Under 8% oxygen, npr-1(ad609) animals do not exhibit a preference for the bacterial border, and are capable of leaving the P. aeruginosa lawn. As shown in Figure 2F, npr-1(ad609) animals were more resistant to P. aeruginosa-mediated killing at 8% oxygen than at 21% oxygen, but were still more susceptible than wild-type animals at 8% oxygen. These results indicate that animals deficient in NPR-1 activity are more susceptible to P. aeruginosa due to two factors: decreased pathogen avoidance and decreased innate immune responses.
The increased susceptibility of npr-1(ad609) to S. enterica (Fig. S3A), a pathogen that does not elicit an avoidance behavior (38), is consistent with a role of NPR-1 in the regulation of immune responses that are independent of pathogen avoidance. Since a small amount of S. enterica that passes through the pharyngeal grinder proliferates and colonizes the intestine in a process that is independent of the dose (32), and the pumping rates of npr-1(ad609) animals are comparable to those of wild type (Fig. S4), the results further support the function of NPR-1 in the regulation on immune responses.
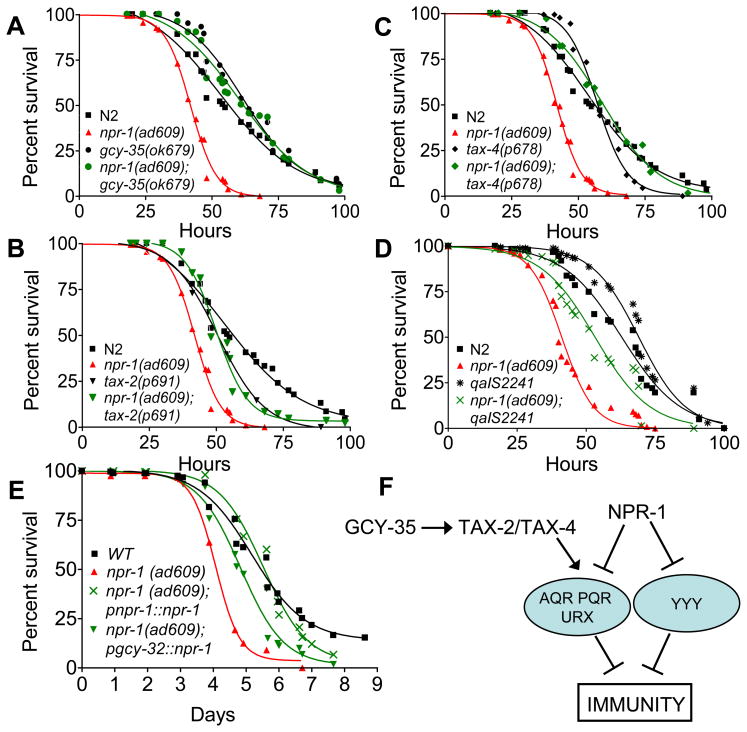
Genetic studies have identified a chemosensory circuit that coordinates oxygen preference and aggregation in npr-1 mutants (35, 37, 39–42). Aggregation and bordering of npr-1(ad609) nematodes depend on functional gcy-35, tax-2, or tax-4 genes (31, 40, 43). GCY-35 is a soluble guanylyl cyclase (sGC) that binds directly to molecular oxygen, and TAX-2 and TAX-4 are two subunits of a cGMP-gated-ion-channel (31, 40, 43). Through the activity of GCY-35 and other guanylate cyclases and the subsequent activation of TAX-2/TAX-4, AQR, PQR, and URX sensory neurons drive avoidance of high oxygen; these neurons are thought to be hyperactive in npr-1 mutants (40). To determine whether this part of the NPR-1 neural circuit regulates innate immune response, we studied the pathogen susceptibility of npr-1(ad609) animals carrying loss-of-function mutations in gcy-35, tax-2, or tax-4. As shown in Figure 3, the enhanced susceptibility to P. aeruginosa of npr-1(ad609) animals was rescued by mutations in gcy-35, tax-2, or tax-4. Similar results were obtained when the infections were performed in plates containing full lawns of P. aeruginosa (Fig. S5).
Fig. 3.
The NPR-1 neural circuit regulates innate immunity. (A) Wild-type N2, npr-1(ad609) (P=0.0001), gcy-35(ok769) (P=0.0125), and gcy-35(ok769);npr-1(ad609) (P=0.0639) were exposed to P. aeruginosa. (B) Wild-type N2, npr-1(ad609) (P=0.0001), tax-4(p678) (P=0.1673), tax-4(p678);npr-1(ad609) (P=0.3611), were exposed to P. aeruginosa. (C) Wild-type N2, npr-1(ad609) (P=0.0001), tax-2(p691) (P=0.0930), tax-2(p691);npr-1(ad609) (P=0.0031) were exposed to P. aeruginosa. (D) Wild-type N2, npr-1(ad609) (P=0.0001), qaIS2241 (P=0.0042), a strain which lacks AQR, PQR, and URX neurons, and npr-1(ad609); qaIS2241 (P=0.0001) were exposed to P. aeruginosa. The graphs represent combined results of at least three independent experiments, N≥40 adult nematodes per strain. (E) Wild-type N2, npr-1(ad609) (P=0.0001), pgcy-32::npr-1; npr-1(ad609) (P=0.0001), and pnpr-1::npr-1; npr-1(ad609) (P=0.1939) were exposed to P. aeruginosa. The graphs represent combined results of at least two independent experiments, N≥100 adult nematodes per strain. Killing assays were performed at 17°C, as low temperatures are known to increase the resolution of killing assays involving P. aeruginosa. (F) Model of the neural control of innate immunity in C. elegans: NPR-1 inhibits the activity of AQR, PQR, URX and additional neuron(s) designated YYY that suppress innate immunity, while GCY-35, TAX-2, and TAX-4 are required for the activation of AQR, PQR and URX neurons.
NPR-1 is expressed in at least twenty different neurons, including the gcy-35-expressing sensory neurons AQR, PQR, and URX (35). To confirm that at least AQR, PQR, and URX neurons are part of a neural network that inhibits innate immunity, we studied the susceptibility to P. aeruginosa of a strain in which these neurons were genetically ablated by expressing the cell-death activator gene egl-1 under the control of the gcy-36 promoter (42). The strain lacking AQR, PQR and URX neurons exhibited a significantly increased survival on P. aeruginosa (Fig. 3D), indicating that AQR, PQR and URX neurons suppress innate immunity. In addition, lack of AQR, PQR and URX neurons partially rescued the enhanced susceptibility to P. aeruginosa of npr-1(ad609) animals (Fig. 3D). Expression of npr-1 under the control of the gcy-32 promoter, which drives the expression of npr-1 to AQR, PQR and URX neurons, also rescued the enhanced susceptibility to P. aeruginosa of npr-1(ad609) animals (Fig. 3E), providing additional support to the role of these neurons in the regulation of innate immunity. Consistent with the idea that additional NPR-1 expressing neurons regulate innate immunity (Fig. 3F), npr-1 expression under the regulation of its own promoter fully rescued the enhanced susceptibility to P. aeruginosa phenotype of npr-1(ad609) animals (Fig. 3E). Taken together, these results indicate that genes and cells involved in the NPR-1 neural circuit modulate innate immune responses.
As in mammals, peristalsis, low pH, and antimicrobial substances prevent microbial colonization of the C. elegans intestine. In addition, accumulating evidence indicates that different genetic pathways regulate the expression of C. elegans genes that are markers of immune response (14–19). To provide insight into the immune function of the NPR-1 neural circuit, we utilized gene expression microarrays to find clusters of genes upregulated or downregulated in npr-1(ad609) mutants relative to wild-type animals grown on live P. aeruginosa (Tables 2 and 3). Interestingly, there is a significant enrichment in NPR-1-regulated genes that have at least one of three features: they are upregulated by P. aeruginosa infection in wild-type animals, expressed in the intestine, and/or have already been linked to the C. elegans P38 MAP kinase, PMK-1, which plays a crucial role in innate immunity (17, 44–47) (Table 4). Further analysis revealed that five of the genes most highly downregulated by NPR-1 are found in a cluster on chromosome V that appears to have been duplicated further downstream on that chromosome (Table 3). Of these five genes, three are also known to be downregulated by the C. elegans PMK-1/P38 pathway. Overall, most of the genes regulated by pathways linked to innate immunity correspond to PMK-1-regulated genes (Fig. 4K). In addition, these genes are similarly misregulated in animals deficient in NPR-1 or PMK-1 function (Tables 2 and 3). Since pmk-1 is not transcriptionally regulated by NPR-1 (Tables 2 and 3), we studied whether NPR-1 regulates PMK-1 at the post-transcriptional level. As shown in Figure 4L, npr-1(ad609) nematodes exhibit lower levels of active PMK-1 than wild-type nematodes, suggesting that the NPR-1 neural circuit modulates the activation of PMK-1. Inhibition of pmk-1 gene expression by RNAi in npr-1(ad609) nematodes results in increased susceptibility (Fig. S6), indicating that while the NPR-1 mediated immune pathway has overlapping targets with the PMK-1 mediated immune pathway, NPR-1 regulates both PMK-1-dependent and independent immune responses.
Table 2.
Genes upregulated by NPR-1
| Microarray | qRT-PCR | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Description | Intestinal expressiona | ↑ on PA14b | Known immune pathwayc | mean ± SEMd | mean ± SEMd | p-value |
| dct-17 | Germline tumor affecting | Y | daf-16 | 0.176±0.023 | 0.5411±0.0505 | 0.0003 | |
| F15D4.5 | Similarity to human synaptonemal complex protein | 0.303±0.029 | ND | ||||
| dod-21 | Lifespan abnormal (RNAi) | daf-16 | 0.3385±0.0325 | 0.4256±0.0775 | 0.0007 | ||
| F36F12.8 | Zinc finger protein | 0.3575±0.0115 | 0.6271±0.1184 | 0.0346 | |||
| F46F2.3 | None | Y | dbl-1 | 0.364±0.054 | 0.1904±0.0356 | 0.0001 | |
| F13H8.3 | Predicted inosine-uridine nucleoside hydrolase | 0.3845±0.0595 | ND | ||||
| gst-24 | Glutathione S-transferase | Y | Y | 0.3945±0.0285 | 0.4442±0.0437 | 0.0001 | |
| stdh-2 | Steroid dehydrogenase, lifespan abnormal(RNAi) | Y | 0.3995±0.0805 | 0.4456±0.0713 | 0.0015 | ||
| T10D4.6 | None | 0.4065±0.0505 | ND | ||||
| Y69A2AR.25 | Similarity to human neurogenic locus notch | 0.4135±0.0345 | ND | ||||
| gst-20 | Glutathione S-transferase | 0.4165±0.0405 | ND | ||||
| T24B8.5 | Similarity to roundworm mucin MUC-5 | Y | daf-16, pmk-1 | 0.426±0.028 | ND | ||
| T28F2.2 | Slow growth, decreased brood size (RNAi) | Y | 0.4275±0.0045 | 0.1441±0.0231 | 0.0001 | ||
| F36G9.12 | Predicted transcription factor, transferase activity | Y | 0.436±0.027 | ND | |||
| col-101 | Cuticle collagen | 0.467±0.013 | ND | ||||
| C14C6.5 | None | Y | Y | pmk-1 | 0.468±0.008 | 0.5562±0.0869 | 0.0070 |
| clec-85 | C-type lectin | Y | Y | dbl-1, pmk-1 | 0.6025±0.0995 | 0.5661±0.0549 | 0.0042 |
| dod-24 | CUB like region, lifespan abnormal(RNAi) | Y | daf-16 | 0.7555±0.0185 | 0.5546±0.0201 | 0.0001 | |
| lec-11 | Galectin family, binds sugar in vitro | Y | Y | 0.7655±0.0875 | 0.6171±0.0401 | 0.0024 | |
| abf-1 | Antibacterial factor | Y | Y | 0.8015±0.0225 | 0.682±0.0157 | 0.0001 | |
| lys-8 | Putative lysozyme, lifespan abnormal(RNAi) | Y | Y | daf-16, dbl-1, pmk-1 | 0.832±0.003 | 0.7331±0.0648 | 0.0092 |
| lys-2 | Putative lysozyme | Y | Y | pmk-1 | 0.851±0.044 | 0.5561±0.0257 | 0.0004 |
Table 3.
Genes downregulated by NPR-1
| Microarray | qRT-PCR | ||||
|---|---|---|---|---|---|
| Gene | Description | Known immune pathwaya | mean ± SEMb | mean ± SEMb | p-value |
|
Y19D10A.7 F56A4.9c |
Receptor with similarity to human insulin-like growth factor 1 precursor | pmk-1 | 14.295±0.845 | 13.84±2.208 | 0.0021 |
|
Y19D10A.4 C01B4.7 c |
Permease of the major facilitator superfamily, similar to human sialin | pmk-1 | 4.305±0.1677 | 3.346±0.2054 | 0.0003 |
|
Y19D10A.16 C01B4.6 c |
Similar to human Aldose 1-epimerase | pmk-1 | 4.272±0.4356 | 5.941±0.3604 | 0.0001 |
| nspb-1-5 | Nematode Specific Peptide family, group B | 4.246±1.256 | ND | ||
| F43C11.3 | None | 3.6555±0.4285 | ND | ||
| nlp-25 | Neuropeptide-like protein | 3.5235±0.6755 | ND | ||
| C42D4.3 | Fibronectin | 3.4845±0.1205 | ND | ||
|
Y19D10A.5 C01B4.8 c |
Permease of the major facilitator superfamily, similar to human sialin | 3.4±0.106 | 2.247±0.3004 | 0.0142 | |
|
Y19D10A.11 F56A4.12 c |
Permease of the major facilitator superfamily, similar to human sialin | 3.171±0.263 | 2.826±0.2391 | 0.0016 | |
| nspa-9 | Nematode Specific Peptide family, group A | 2.9805±0.5015 | ND | ||
| grl-21 | Grl domain, intercellular signalling | 2.9605±0.0925 | ND | ||
| col-97 | Cuticle collagen | 2.9565±0.4775 | ND | ||
| col-39 | Cuticle collagen | 2.9505±0.5555 | ND | ||
| ZK180.5 | Similar to human Diacylglycerol kinase kappa | 2.883±0.17 | ND | ||
| col-62 | Cuticle collagen | 2.5785±0.2495 | ND | ||
| clec-72 | C-type lectin | 2.51±0.439 | ND | ||
Table 4.
Over-represented gene sets among NPR-1-regulated genes
| Gene Set | Genes in Set | Genes in Common | Representation Factora | p-value |
|---|---|---|---|---|
| Pseudomonas aeruginosa-induced genes (7) | 197 | 12 | 36.1 | 3.21 × 10−16 |
| Intestinally-expressed genes (6) | 1947 | 12 | 3.6 | 5.46 × 10−05 |
| PMK-1-regulated genes (8) | 110 | 8 | 43.1 | 1.09 × 10−11 |
The representation factor is the number of overlapping genes divided by the expected number of overlapping genes drawn from the group of NPR-1-regulated genes and the group corresponding to a given gene set. For details, see http://elegans.uky.edu/MA/progs/representation.stats.html.
Fig. 4.
The NPR-1 neural circuit regulates expression of innate immune genes. (A–J) Quantitative reverse transcription–PCR analysis of C01B4.6/Y19D10A.16, F56A4.9/Y19D10A.7, C01B4.7/Y19D10A.4, F56A4.12/Y19D10A.11, abf-1, dod-24, F36F12.8, F46F2.3, gst-24, and T28F2.2 expression in npr-1(ad609) and gcy-35(ok769);npr-1(ad609) nematodes relative to wild-type nematodes exposed to P. aeruginosa. Data were analyzed by normalization to pan-actin (act-1,-3,-4) and relative quantification using the comparative cycle threshold method. Student’s exact t-test indicates differences among the groups are significantly different; bar graphs correspond to mean ± SEM. Point graphs correspond to gene quantification in independent isolations of npr-1(ad609)(N=6) and gcy-35(ok769);npr-1(ad609)(N=3). (K) The Venn diagram lists the genes identified by microarray analysis to be regulated by both NPR-1 and one or more known innate immune pathways in C. elegans. Genes that lie within two or three circles are regulated by multiple innate immune pathways in addition to NPR-1. Twenty-six genes have not been previously connected to any of the innate immune pathways and are depicted in the solitary circle. (L) Immunological detection of active PMK-1. Active PMK-1 was detected in wild-type N2, npr-1(ad609) and gcy-35(ok769);npr-1(ad609). Animals were grown at 20°C until 1 day old adult and whole worm lysates were used to detect active PMK-1 by Western blotting using an anti-human p38 antibody from Promega, Inc. Actin was detected using a polyclonal antibody from SIGMA. BioRad Quantity One Analysis Software was used to scan and analyze the Western blot.
To obtain insight into the mechanism by which gcy-35 mutation rescues the enhanced susceptibility to P. aeruginosa of npr-1(ad609) animals (Fig. 3A), we used quantitative RT-PCR (qRT-PCR) to compare the expression levels of selected genes of npr-1(ad609) to that of npr-1(ad609);gcy-35(ok769) animals. As shown in Figure 4, a gcy-35 mutation in npr-1(ad609) animals rescues the altered expression of 10 out of 19 genes tested that are markers of C. elegans immune response. These results indicate that the NPR-1 neural circuit modulates the expression of immune-related genes, many of which are known to be expressed in tissues that are in direct contact with pathogens during infection.
In summary, our results provide evidence that specific genes and neurons in the nervous system are responsible for effective innate immune responses that are independent of behavioral phenotypes and may take place in tissues that are in direct contact with pathogens. It has recently been postulated that cell non-autonomous signals from different neurons may act on non-neural tissues to regulate processes such as fat storage (48) and longevity (8). C. elegans neurons can regulate physiological processes through conserved neuroendocrine signals including insulin-related peptides, TGF-beta peptides, and neuropeptides. The URX, AQR, and PQR neurons that are part of the NPR-1 neural circuit that regulates innate immunity are exposed to the pseudocoelomic body fluid, which could communicate neuroendocrine signals to non-neural tissues involved in defense responses. The identification and characterization of the specific neuroendocrine signals that regulate innate immune responses in C. elegans should yield significant insights into the mechanisms used by the nervous system to regulate similar processes across metazoans.
Supplementary Material
Acknowledgments
We thank Caenorhabditis Genetics Center (University of Minnesota) for strains used in this study. A.A. is funded by The Whitehead Scholars Program, NIH SERCEB (U54 AI057157), and NIH GM070977.
References
- 1.Sternberg EM. Nat Rev Immunol. 2006 Apr;6:318. doi: 10.1038/nri1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson J. J Intern Med. 2005 Feb;257:122. doi: 10.1111/j.1365-2796.2004.01440.x. [DOI] [PubMed] [Google Scholar]
- 3.Tracey KJ. Nature. 2002 Dec 19–26;420:853. doi: 10.1038/nature01321. [DOI] [PubMed] [Google Scholar]
- 4.Sattelle DB, Buckingham SD. Invert Neurosci. 2006 Mar;6:1. doi: 10.1007/s10158-005-0014-7. [DOI] [PubMed] [Google Scholar]
- 5.Bargmann CI, Horvitz HR. Science. 1991 Mar 8;251:1243. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- 6.Schackwitz WS, Inoue T, Thomas JH. Neuron. 1996 Oct;17:719. doi: 10.1016/s0896-6273(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 7.Alcedo J, Kenyon C. Neuron. 2004 Jan 8;41:45. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 8.Bishop NA, Guarente L. Nature. 2007 May 31;447:545. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 9.Li C, Nelson LS, Kim K, Nathoo A, Hart AC. Ann N Y Acad Sci. 1999;897:239. doi: 10.1111/j.1749-6632.1999.tb07895.x. [DOI] [PubMed] [Google Scholar]
- 10.Li W, Kennedy SG, Ruvkun G. Genes Dev. 2003 Apr 1;17:844. doi: 10.1101/gad.1066503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nathoo AN, Moeller RA, Westlund BA, Hart AC. Proc Natl Acad Sci U S A. 2001 Nov 20;98:14000. doi: 10.1073/pnas.241231298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierce SB, et al. Genes Dev. 2001 Mar 15;15:672. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren P, et al. Science. 1996 Nov 22;274:1389. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- 14.Mallo GV, et al. Curr Biol. 2002;12:1209. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- 15.Kerry S, Tekippe M, Gaddis NC, Aballay A. PLoS ONE. 2006 Dec 20;1:e77. doi: 10.1371/journal.pone.0000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shapira M, et al. Proc Natl Acad Sci U S A. 2006 Sep 19;103:14086. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Troemel ER, et al. PLoS Genet. 2006 Nov 10;:2. [Google Scholar]
- 18.Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank JJ. Genome Biol. 2007 Sep 17;8:R194. doi: 10.1186/gb-2007-8-9-r194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. Genome Res. 2006 Aug;16:1005. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulenburg H, Muller S. Parasitology. 2004 Apr;128:433. doi: 10.1017/s003118200300461x. [DOI] [PubMed] [Google Scholar]
- 21.Hasshoff M, Bohnisch C, Tonn D, Hasert B, Schulenburg H. Faseb J. 2007 Jun;21:1801. doi: 10.1096/fj.06-6551com. [DOI] [PubMed] [Google Scholar]
- 22.Yook K, Hodgkin J. Genetics. 2007 Feb;175:681. doi: 10.1534/genetics.106.060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sicard M, Hering S, Schulte R, Gaudriault S, Schulenburg H. Environ Microbiol. 2007 Jan;9:12. doi: 10.1111/j.1462-2920.2006.01099.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Lu H, Bargmann CI. Nature. 2005 Nov 10;438:179. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
- 25.Beale E, Li G, Tan MW, Rumbaugh KP. Appl Environ Microbiol. 2006 Jul;72:5135. doi: 10.1128/AEM.00611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laws TR, Atkins HS, Atkins TP, Titball RW. Microb Pathog. 2006 Jun;40:293. doi: 10.1016/j.micpath.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Pujol N, et al. Curr Biol. 2001;11:809. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- 28.Pradel E, et al. Proc Natl Acad Sci U S A. 2007 Feb 13;104:2295. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan MW, Mahajan-Miklos S, Ausubel FM. Proc Natl Acad Sci U S A. 1999;96:715. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Cell. 1999;96:47. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 31.de Bono M, Bargmann CI. Cell. 1998 Sep 4;94:679. doi: 10.1016/s0092-8674(00)81609-8. [DOI] [PubMed] [Google Scholar]
- 32.Aballay A, Yorgey P, Ausubel FM. Curr Biol. 2000;10:1539. doi: 10.1016/s0960-9822(00)00830-7. [DOI] [PubMed] [Google Scholar]
- 33.Labrousse A, Chauvet S, Couillault C, Kurz CL, Ewbank JJ. Curr Biol. 2000;10:1543. doi: 10.1016/s0960-9822(00)00833-2. [DOI] [PubMed] [Google Scholar]
- 34.Garsin DA, et al. Proc Natl Acad Sci U S A. 2001;98:10892. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coates JC, de Bono M. Nature. 2002 Oct 31;419:925. doi: 10.1038/nature01170. [DOI] [PubMed] [Google Scholar]
- 36.Rogers C, et al. Nat Neurosci. 2003 Nov;6:1178. doi: 10.1038/nn1140. [DOI] [PubMed] [Google Scholar]
- 37.Gray JM, et al. Nature. 2004 Jul 15;430:317. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 38.Tenor JL, Aballay A. EMBO Rep. 2008 Jan;9:103. doi: 10.1038/sj.embor.7401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Bono M, Tobin DM, Davis MW, Avery L, Bargmann CI. Nature. 2002 Oct 31;419:899. doi: 10.1038/nature01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheung BH, Cohen M, Rogers C, Albayram O, de Bono M. Curr Biol. 2005 May 24;15:905. doi: 10.1016/j.cub.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 41.Rogers C, Persson A, Cheung B, de Bono M. Curr Biol. 2006 Apr 4;16:649. doi: 10.1016/j.cub.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 42.Chang AJ, Chronis N, Karow DS, Marletta MA, Bargmann CI. PLoS Biol. 2006 Sep;4:e274. doi: 10.1371/journal.pbio.0040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheung BH, Arellano-Carbajal F, Rybicki I, de Bono M. Curr Biol. 2004 Jun 22;14:1105. doi: 10.1016/j.cub.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 44.Kim DH, et al. Science. 2002;297:623. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 45.Aballay A, Drenkard E, Hilbun LR, Ausubel FM. Curr Biol. 2003;13:47. doi: 10.1016/s0960-9822(02)01396-9. [DOI] [PubMed] [Google Scholar]
- 46.Kim DH, et al. Proc Natl Acad Sci U S A. 2004 Jul 27;101:10990. doi: 10.1073/pnas.0403546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huffman DL, et al. Proc Natl Acad Sci U S A. 2004 Jul 27;101:10995. doi: 10.1073/pnas.0404073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mak HY, Nelson LS, Basson M, Johnson CD, Ruvkun G. Nat Genet. 2006 Mar;38:363. doi: 10.1038/ng1739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.