Abstract
Toll-like receptors (TLR) are highly conserved pattern recognition receptors of the innate immune system. Toll-like receptor 2 (TLR2) recognizes bacterial lipopeptides in a heterodimeric complex with TLR6 or TLR1, thereby discriminating between di- or triacylated lipopeptides, respectively. Previously, we found that HEK293 cells transfected with bovine TLR2 (boTLR2) were able to respond to diacylated lipopeptides but did not recognize triacylated lipopeptides, even after cotransfection with the so far published sequence of boTLR1. In this study we now could show that primary bovine cells were in general able to detect triacylated lipopetides. A closer investigation of the boTLR1 gene locus revealed an additional ATG 195 base pairs upstream from the published start codon. Its transcription would result in an N-terminus with high identity to human and murine TLR1 (huTLR1, muTLR1). Cloning and cotransfection of this longer boTLR1 with boTLR2 now resulted in the recognition of triacylated lipopeptides by HEK293 cells, thereby resembling the ex vivo observation. Analysis of the structure-activity relationship showed that the ester-bound acid chains of these lipopeptides need to consist of at least 12 carbon atoms to activate the bovine heterodimer showing similarity to the recognition by huTLR2/huTLR1. In contrast, HEK293 cell cotransfected with muTLR2 and muTLR1 could already be activated by lipopeptides with shorter fatty acids of only 6 carbon atoms. Thus, our data indicate that the additional N-terminal nucleotides belong to the full length and functionally active boTLR1 (boTLR1-fl) which participates in a species-specific recognition of bacterial lipopeptides.
Keywords: toll-like receptor, bacterial lipopeptide, species-specificity
1. INTRODUCTION
A prerequisite for the induction of an immune response against invading pathogens is the recognition of their presence. To discriminate between self and non-self the immune system has evolved a repertoire of germline-encoded so called pattern recognition receptors (PRR) which recognize highly conserved pathogen associated molecular patterns (PAMP) present on all microorganisms [16]. A prominent family of PRR is that of toll-like receptors (TLR) which have been found in most invertebrates and all vertebrate species [24]. In mammals there are so far 13 different TLR identified. With the exception of TLR10, 12 and 13 natural ligands are known for all TLR allowing the immune system to detect nearly all kinds of invading microorganisms [2, 22].
TLR are type I transmembrane receptors composed of an extracellular domain consisting of 19–25 leucine-rich repeats (LRR), a transmembrane domain and an intracellular toll/interleukin 1 receptor (TIR) domain [12]. The TIR domain is highly conserved between TLR and different species and mediates the induction of signal transduction cascades via MyD88 and/or as found with TLR3 and TLR4 via TRIF [7]. Ligand binding occurs via the LRR of the ectodomain. For this reason this domain highly varies between different TLR but shares great similarity between the same TLR of different species [24]. Nevertheless there are some species-specific differences in the ligand recognition [31].
While all other TLR are active as homodimers TLR2 is unique in its ability to form functional active heterodimers with either TLR1 or TLR6 [27]. Although this receptor is thought to recognize the most diverse repertoire of structural different ligands like lipoteichoic acids, peptidoglycan and endogenous ligands like heat shock proteins and hyaluronic acid, the best characterized (and maybe only) ligands are bacterial lipoproteins [34]. Lipoproteins represent an important constituent of the cell wall of Gram-positive and Gram-negative bacteria and are also present on the membrane of mycoplasma [1]. All lipoproteins are composed of a conserved N-terminal dihydroxypropyl cysteine which can be acylated with 2 or 3 fatty acids that anchor the protein to the cell membrane [32]. By generating synthetic lipopeptides the structure-activity relationship has extensively been studied in human and murine cells. Beside the number and length of the fatty acids also the N-terminal amino acids of lipoproteins are responsible for the ligand-receptor interaction on host cells [4, 21]. Taken together, most diacylated lipopeptides activate TLR2/6 heterodimers, while most triacylated lipopeptides are recognized by TLR2/1 heterodimers [3, 21, 23, 28, 29]. However, some di- and triacylated lipopeptides seem to be recognized by both, TLR2/1 and TLR2/6 heterodimers [5, 21]. The activation of TLR2/1 and/or TLR2/6 heterodimers then leads to similar or even identical signalling-pathways indicating that this heterodimerization broadens the ligand spectrum (di- and triacylated lipopeptides) rather than to mediate different immune responses [8].
Species-specific recognition of lipopeptides has been found using murine and human cells where lipopeptides with short-length fatty acids activate muTLR2 heterodimers whereas human receptors require longer acid chains like palmitic acids [13]. Recently we have established HEK293 cell lines stably transfected with bovine TLR (boTLR) – one with boTLR4 and boMD2 and one with boTLR2 – to examine their contribution to the pathology of bacteria causing diseases in cattle [9, 26]. Concerning boTLR2, we noticed that HEK293 cells transfected only with this receptor were able to recognize diacylated lipopeptides but could not be activated by triacylated lipopeptides.
In this study we therefore have investigated whether primary bovine cells in general and HEK293-boTLR2 cells additionally transfected with boTLR1 are able to recognize triacylated lipopeptides. Furthermore, we have analyzed the structural prerequisites of synthetic lipopeptides to activate bovine TLR2/TLR1 in comparison to human and murine heterodimers to help improving our understanding of the action of species-specific pathogens.
2. MATERIALS AND METHODS
2.1. Reagents and plasmids
Dulbecco’s modified Eagle’s medium GIBCO-LG (DMEM), penicillin-streptomycin (PS), L-glutamine (LG), sodium pyruvate, and Hepes buffer were obtained from Invitrogen (Karlsruhe, Germany). Fetal calf serum (FCS) (Linaris, Wertheim-Bettingen, Germany) was heat-inactivated. The synthetic LP were synthesized and analyzed by EMC microcollections GmbH (Tübingen, Germany) and show strict TLR2 dependency as determined by the use of TLR2-deficient mice and TLR2 transfected HEK293 cells (data not shown). The chemical structures, denotations and abbreviations of LP and fatty acids used in this study are represented in Supplementary Figure S1 1. Human recombinant TNF-α was kindly provided by Prof. D. Maennel (University of Regensburg, Regensburg, Germany). Plasmids of human and murine TLR were obtained from InvivoGen (San Diego, USA). cDNA of bovine TLR2 and short form of TLR1 was cloned into pCR 2.1-TOPO vector (Stratagene, La Jolla, USA) and subcloned into pUNO (InvivoGen) and pTarget (Promega, Mannheim, Germany), respectively. Coding sequences for generation of cDNA were obtained from NCBI GenBank. Identified full length bovine TLR1 covers nucleotides 871727–874106 on chromosome 6 (NW_001495180.2) thereby starting with the ATG 195 base pairs upstream of the published start codon (EU006652). cDNA was cloned into pENTR/D-TOPO and subcloned into pDEST47 using GATEWAY cloning system according to the provider’s manual (Invitrogen, Leek, Netherlands). The sequence of full length boTLR1 is now available at NCBI GenBank databases under accession number FJ147090.
2.2. Cell lines and cell culture
The human embryonic kidney cell line HEK293 (ECACC) was cultured in DMEM supplemented with 10% FCS, 1% PS and 1% LG. Cells were seeded at 0.5 × 106 and 0.3 × 106 cells/10 mL in 75 cm3 flasks and cultured at 37 °C, 5% CO2 for 3 or 4 days, respectively. The bovine B lymphocyte cell line BL3 was obtained from the Collection of Cell Lines in Veterinary Medicine (Friedrich-Loeffler-Institut, Greifswald-Insel-Riems). Cells were cultured in DMEM supplemented with 10% FCS, 1% PS and 1% LG and passaged twice a week with 1 × 106 and 0.7 × 106 cells/10 mL in 75 cm3 flasks and cultured at 37 °C, 5% CO2 for 3 or 4 days, respectively.
2.3. Transient transfection and stimulation of cell lines
HEK293 cells were seeded at 0.2 × 105 cells/150 μL in 96-well culture dishes. After 24 h transfection was performed using Polyfect reagent according to the provider’s instructions (Qiagen, Hilden, Germany). Cells were then stimulated for 24 h with indicated concentrations of LP ranging from 1 to 100 nM. Supernatants were analyzed for IL-8 production using commercial enzyme-linked immunoabsorbant assays (ELISA, Biosource, CA, USA). BL3 cells were seeded at 1 × 105 cells/400 μL in 24-well culture dishes and triplicates were stimulated after 24 h with 100 nM LP for 6 h. Cells were washed once with cold PBS and combined for RNA-isolation and cDNA-synthesis.
2.4. Preparation of bovine mononuclear cells (MNC) and spleen cells
Peripheral blood and spleen were obtained from two healthy Holstein cows. MNC were separated from heparinized blood diluted two-fold with phosphate-buffered saline (PBS) by gradient centrifugation (30 min, 4 °C, 1 100 × g) in lymphocyte separation medium (PAA Laboratories, Pasching, Austria). The layer of MNC was recovered, washed thrice in PBS and resuspended in DMEM supplemented with 10% FCS, 1% LG, 0.5% PS, 1% Hepes and 1% sodium pyruvate. Spleen cells were mechanically isolated using forceps and scalpel. Single cells were separated from tissue fragments by sedimentation and washed thrice with DMEM containing 10% FCS, 1% LG, 0.5% PS, 1% Hepes and 1% sodium pyruvate. All steps were performed on ice.
2.5. Stimulation of bovine MNC and spleen cells
MNC were seeded at 4 × 105 cells/300 μL per 48-well culture dish in supplemented DMEM. Triplicates were stimulated with 100 nM LP for 6 h. Cells were washed once with cold PBS before RNA-isolation and cDNA-synthesis.
Isolated spleen cells were stimulated in triplicates at 4 × 105/150 μL DMEM with 1, 10 and 100 nM LP for 24 h and pulsed with [3H] thymidine ([3H]TdR), 0.9 μCi/mL) for another 24 h of culture. Cells were harvested and [3H]TdR incorporation determined in a liquid scintillation spectrometer. The results are expressed in counts per minute per culture [6].
2.6. RNA-isolation, cDNA-synthesis and PCR
RNA was isolated using Absolutely RNA Miniprep Kit including DNase digestion on column as recommended by the manufacturer (Stratagene, Amsterdam, Netherlands). One microgram of total RNA was taken for cDNA-synthesis using SuperScript III Reverse Transcriptase from Invitrogen (Leek, Netherlands). Promega-PCR Mix was used for amplification of short (sense: CACCATGCCTGACATCCTCTCAC; antisense: ATGTATTTCTGCTGCTTTTTCC and full length boTLR1 (sense: CACCATGACTAAAAAAAATTCTAGC; antisense: ATGTATTTCTGCTGCTTTTTCC). Real-time PCR was performed with the Light Cycler 480 System (Roche Diagnostics) using LightCycler® Fast Start DNA MasterPLUS according to the manufacturer’s instructions. Relative cDNA amounts of IL-8 (sense: CCTTTCCACCCCAAATTTATC, antisense: ACCCACTTTTCCTTGG) were calculated compared to the expression of the house-keeping gene HPRT (sense: GGACTAATTATGGACAGG, antisense: CCTTGAGCACACAGAGGG).
2.7. Sequence alignment and computation
Multiple alignment of DNA sequences and calculations of percentage identities were made using ClustalW method in MegAline program of DNASTAR software (DNASTAR, Inc., Madison, USA).
3. RESULTS
3.1. Recognition of di- and triacylated lipopeptides by boTLR2-transfected HEK293 cells
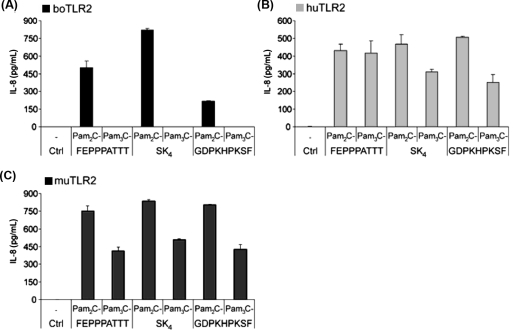
To examine which lipopeptide structures are recognized by boTLR2 we stimulated HEK293 cells transfected with boTLR2 (HEK293-boTLR2) with di- and triacylated forms of different lipopeptides (for structural details see Supplementary Fig. S11). The responsiveness was measured by IL-8 production (Fig. 1A) and compared with HEK293 cells transfected with human or murine TLR2 (Figs. 1B and C). HEK293-boTLR2 cells only recognized diacylated lipopeptides as shown for Pam2C-FEPPPATTT (representing a synthetic LP of Rhodopseudomonas viridis), Pam2C-SK4 and Pam2C-GDPKHPKSF (representing a synthetic LP of Mycoplasma salivarium; also named FSL-1). The triacylated forms did not induce any IL-8 release. In contrast, cells transfected with huTLR2 or muTLR2 responded to both, di- and triacylated lipopeptides. It should be noted that the HEK293 cells used in this study express native human TLR1 and TLR6 (Supplementary Fig. S21). Therefore transfection with huTLR2 leads to the formation of TLR2/1 and TLR2/6 heterodimers and the recognition of tri- and diacylated lipopeptides, respectively. MuTLR2 also seems to form active heterodimers with the native human coreceptors. However, when transfected with boTLR2 only diacylated lipopeptides were recognized indicating a heterodimerization with native human TLR6 but not with human TLR1.
Figure 1.
HEK293 cells transfected with boTLR2 do not recognize triacylated LP while human or murine heterodimers were activated by these stimuli. HEK293 cells were transfected with (A) boTLR2 (B) huTLR2 or (C) muTLR2 and stimulated after 24 h with 100 nM of di- and triacylated LP with the peptide moieties FEPPPATTT, SK4 or GDPKHPKSF. After 24 h IL-8 production was detected in the supernatant. Values are means ± SD of n = 2 and represent the result of at least 3 independent experiments.
3.2. Contribution of TLR1 to the recognition of triacylated lipopeptides by boTLR2
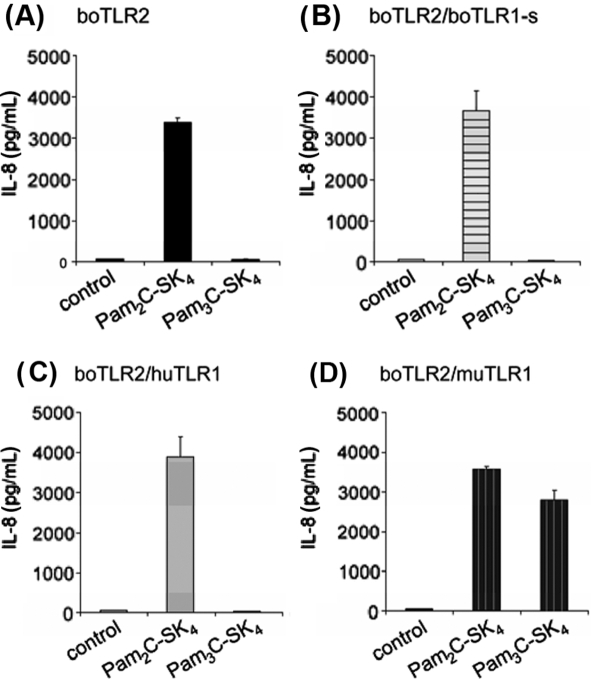
Most triacylated lipopeptides activate the cell through TLR2/1 heterodimers. For that reason we cotransfected HEK293 cells with boTLR2 and human or murine TLR1, or with a vector containing the published sequence of the boTLR1 (NCBI GenBank databases under accession number EU006652). With 2 184 bp its sequence is 183 and 204 bp shorter than the human and murine ortholog, respectively. Therefore, in the following it is termed boTLR1-short (boTLR1-s, see Sect. 3.4).
Cotransfection of boTLR2 with boTLR1-s did not result in an IL-8 release after stimulation with Pam3C-SK4 (Fig. 2B). Also overexpression of huTLR1 had no influence on the recognition of the triacylated stimulus (Fig. 2C). Interestingly, in the presence of muTLR1 stimulation with Pam3C-SK4 resulted in nearly the same IL-8 release compared to stimulation with Pam2C-SK4 did (Fig. 2D). This indicates that boTLR2 forms a functional complex with muTLR1 which can be activated via triacylated lipopeptides.
Figure 2.
Cotransfection of boTLR2 with muTLR1 but not with bovine or human TLR1 resulted in the recognition of triacylated LP. HEK293 cells were transfected with (A) boTLR2 alone or in combination with (B) boTLR1-s, (C) huTLR1 or (D) muTLR1. Cells were stimulated for 24 h with 100 nM of Pam2C-SK4 or Pam3C-SK4. IL-8 production was measured in the supernatant. Values are means ± SD of n = 2 and represent the result of at least 3 independent experiments.
3.3. Recognition of di- and triacylated lipopeptides by bovine primary cells and BL3 cell line
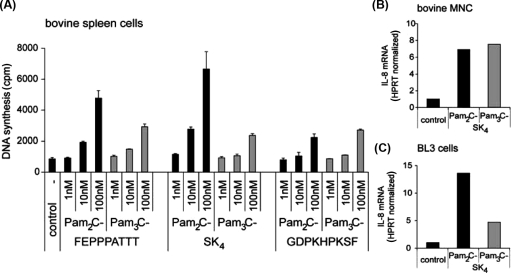
The recombinant boTLR1-s may differ from native boTLR1. We therefore investigated, whether triacylated lipopeptides are recognized by native bovine cells. Bovine spleen cells, MNC as well as the bovine B-cell line BL3 were stimulated with different di- and triacylated lipopeptides. As shown in Figure 3A all lipopeptides induced a dose-dependent B cell proliferation in freshly isolated spleen cells. While di- and triacylated lipopeptides with FEPPPATTT and SK4 peptide tails were active between 10 nM and 100 nM, the mitogenic activity of Pam2C- and Pam3C-GDPKHPKSF started at a concentration of 100 nM. Pam2C-FEPPPATTT and Pam2C-SK4 induced slightly stronger proliferation compared to their triacylated analogues. Nevertheless all triacylated structures were able to activate these cells. Also in primary MNC and the BL3 cell line, di- and triacylated lipopeptides were recognized as shown for Pam2C-SK4 and Pam3C-SK4 by induction of IL-8 mRNA (Figs. 3B and C). Thus, bovine cells have in principle the ability to react to triacylated lipopeptides.
Figure 3.
Primary bovine spleen cells, MNC and BL3 cells respond to both, di- and triacylated LP. (A) Bovine spleen cells were stimulated for 24 h with 1, 10 and 100 nM of di- and triacylated LP with the peptide moieties FEPPPATTT, SK4 or GDPKHPKSF. Activation was measured by [3H]TdR incorporation into the B-lymphocytes and results are expressed in counts per minute per culture. Values are means ± SD of n = 3 and represent the result of 3 independent experiments. (B) Primary bovine MNC and (C) BL3 cells were stimulated for 24 h with 100 nM of Pam2C-SK4 or Pam3C-SK4. IL-8 induction was determined on mRNA level by real-time PCR normalized to the house-keeping gene HPRT. The results are representative for at least 3 independent experiments.
3.4. Sequence comparison of bovine, human and murine TLR1
The published nucleotide sequence of boTLR1-s encompassed 2 184 bp (NCBI GenBank databases under accession number EU006652). Interestingly, 195 bp prior to the published start codon we found another ATG in the gene locus of boTLR1 (now published in NCBI GenBank databases under accession number NW_001495180.2, Nucleotides 871727–874106). An alignment with the sequence of human and murine TLR1 revealed that the boTLR1-s exhibits an N-terminus that is 183 to 204 bp shorter (Supplementary Fig. S31). However, usage of the other ATG would result in a longer receptor (boTLR1-full length) with a nucleotide sequence that shares high homology with the N-terminus of the human and murine TLR1 (Supplementary Fig. S21, boTLR1-fl). Also on protein level the first 100 amino acids would be 77% identical to huTLR1 and 57% identical to muTLR1 (data not shown) indicating that this sequence could be part of a full length boTLR1 (boTLR1-fl).
3.5. Amplification of full length boTLR1 in primary bovine cells
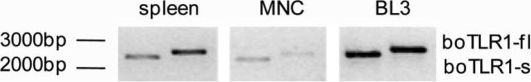
Both, full length boTLR1 of 2 379 bp as well as boTLR1-s (2 184 bp) could be detected via RT-PCR in the cDNA of primary spleen cells, MNC and BL3 cells (Fig. 4). Both products were of comparable intensity, leading to the assumption that both primer pairs use the cDNA of full length boTLR1 as template. This postulation could be confirmed using real-time PCR where CT values for boTLR1-s and boTLR1-fl were equal (data not shown).
Figure 4.
Long form of boTLR1 can be detected in primary bovine spleen cells, MNC and BL3 cells. RT-PCR with primer against the published boTLR1 (boTLR1-s) and the full length form (boTLR1-fl) amplified both TLR1 variants in cDNA from primary bovine spleen cells, MNC and BL3 cells.
3.6. Recognition of triacylated lipopeptides by boTLR2-transfected HEK293 cells cotransfected with full length boTLR1
BoTLR1-fl was cloned and cotransfected with boTLR2 into HEK293 cells and compared with cells cotransfected with boTLR2 and boTLR1-s. BoTLR2 in combination with boTLR1-s did not recognize triacylated lipopeptides as shown by the inability of Pam3C-SK4 to induce IL-8, while stimulation with the diacylated Pam2C-SK4 induced a cytokine release (Fig. 5, black bars). In contrast, HEK293 cells that were cotransfected with boTLR2 and boTLR1-fl, could be activated by both di- and triacylated lipopeptides (Fig. 5, grey bars). We therefore concluded that boTLR1-f1 is the true full length TLR1 which is able to form functional heterodimers with boTLR2. This boTLR2/1-fl is responsible for the recognition of triacylated lipopeptides like in other species.
Figure 5.
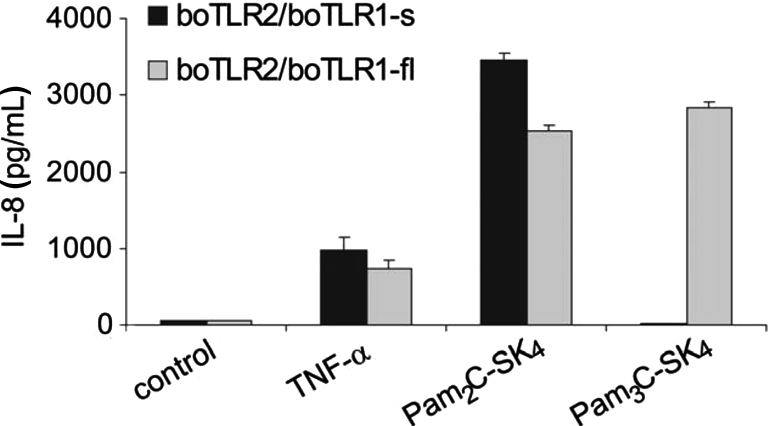
Cotransfection of boTLR2 with full length boTLR1 resulted in the recognition of triacylated LP. HEK293 cells were transfected with boTLR2 in combination with short boTLR1 (black bars) or full length boTLR1 (grey bars). Cells were stimulated for 24 h with 10 ng/mL TNF-α and 100 nM of the lipopeptides Pam2C-SK4 and Pam3C-SK4. IL-8 production was measured in the supernatant. Values are means ± SD of n = 2 and represent the result of at least 3 independent experiments.
3.7. Impact of ester-bound fatty acids of triacylated lipopeptides on their recognition by bovine, human and murine TLR2/1 heterodimers
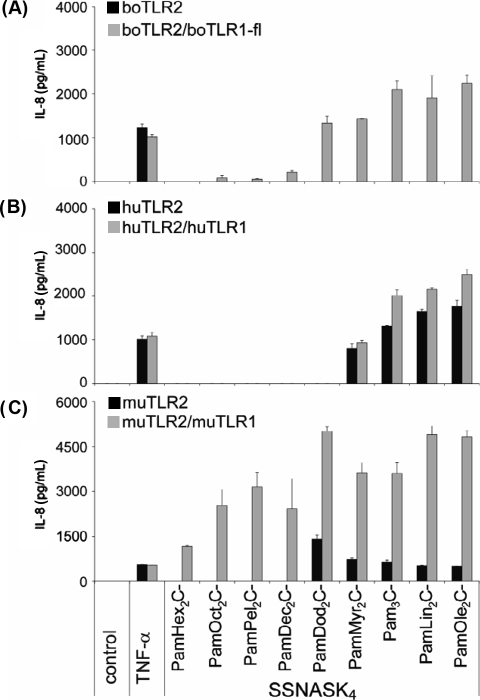
By using a lipopeptide collection with different acylation patterns we compared the structural requirements of triacylated lipopeptides to activate the bovine TLR2/1-fl heterodimer in comparison to the human and murine receptors. All lipopeptides consist of the conserved dihydroxypropyl cystein backbone conjugated with an amid-bound palmitic acid and the peptide sequence SSNASK4. The chain length of the ester-bound fatty acids increased from 6 to 18 carbon atoms (hexanoic acids to oleic acids; for details see on line Supplementary Fig. S11). Again, boTLR2 alone was not able to recognize any of the triacylated lipopeptides (Fig. 6A, black bars). Cotransfection of boTLR1-fl resulted in a reasonable recognition of lipopeptides with ester-bound fatty acids of at least 12 carbon atoms (dodecanoic acids). Lipopeptides with shorter fatty acids were not able to induce an IL-8 release (Fig. 6A, grey bars).
Figure 6.
Structure-activity relationship of triacylated lipopeptides of HEK293 cells transfected with bovine, human or murine TLR2 alone or in combination with respective TLR1. HEK293 cells were transfected with (A) bovine, (B) human or (C) murine TLR2 alone (black bars) or in combination with the respective TLR1 (grey bars). Cells were stimulated for 24 h with 10 ng/mL TNF-α and 100 nM of the indicated lipopeptides. Length of the ester-bound fatty acids increase from left to right. IL-8 production was measured in the supernatant. Values are means ± SD of n = 2 and represent the result of at least 3 independent experiments.
In contrast, the huTLR2/1 heterodimer could only be activated by lipopeptides with ester-bound myristic acids (14 carbon atoms, Fig. 6B, black bars) or fatty acids with a higher number of carbon atoms. Additional overexpression of huTLR1 did not considerably increase the IL-8 release after stimulation (Fig. 6B, grey bars).
When transfected alone, muTLR2 was able to recognize lipopeptides with ester-bound dodecanoic acids (Fig. 6C, black bars). Cotransfection with muTLR1 resulted in the response to lipopeptides with short ester-bound hexanoic acids as obtained with PamHex2C-SSNASK4. Additionally, it should be noted, that the cotransfection markedly enhanced the response towards lipopeptides that were recognized by cells transfected with muTLR2 alone (Fig. 6C, grey bars). The longer the chain length the higher was the induction of IL-8 with a maximal stimulation with lipopeptides possessing dodecanoic acids. Lipopeptides with longer fatty acids had no stronger stimulatory activity.
4. DISCUSSION
In our previous study we could show, that HEK293 cells stably transfected with boTLR2 were able to respond to Staphylococcus aureus, heat-killed Listeria monocytogenes (HKLM), Escherichia coli causing mastitis (strain JF 4.037) and to different diacylated lipopeptides [9]. The aim of the present study now was to understand in more detail the requirements of the boTLR2-coreceptor boTLR1 for the recognition of triacylated lipopeptides.
The HEK293 cells used in this study express native human TLR1 and TLR6 but no TLR2 (Supplementary Fig. S31). Hence, transfection of huTLR2 enables the cells to recognize all kinds of biologically active lipopeptides (Fig. 1B). The fact that muTLR2 transfected cells are also able to respond to the tested di- and triacylated lipopeptides (Fig. 1C) indicates that the murine receptor forms functionally active heterodimers with huTLR6 and at least partially with huTLR1.
In contrast to muTLR2, our data indicate that boTLR2 forms only active heterodimers with huTLR6 but not with huTLR1, as determined by the responsiveness to diacylated but not to triacylated lipopeptides (Figs. 1A and 2C). However, HEK293 cells cotransfected with boTLR2 and muTLR1 induced an IL-8 response when stimulated with Pam3C-SK4. This response was comparable to that of Pam2C-SK4 stimulated cells (Fig. 2D). The overall sequence identity of human and murine TLR1 and TLR6 amount 70.8% and 73.4%, respectively. The identity between human and bovine TLR1 and TLR6 is even higher (78.2% and 78.7%, respectively) and also TLR2 between these species shares 77% identity compared to 70.6% between huTLR2 and muTLR2. However, to explain the observation that boTLR2 forms functionally active heterodimers with muTLR1 but not with huTLR1, more detailed comparisons of the sequences and structures of the TLR of the different species are necessary. There are already studies applying site-directed mutagenesis, chimeras or crystal structures and modeling showing that distinct amino acids sequences are involved in the direct interaction between TLR2 and its coreceptors [10, 17, 25].
Because boTLR2/muTLR1 heterodimers were functionally active, it was surprising that the cotransfection of HEK293 cells with boTLR2 and a vector containing the so far published sequence of boTLR1 did not result in recognition of triacylated lipopeptides, like Pam3C-SK4 (Fig. 2B). On mRNA level we detected the transcript of this boTLR1 in transfected HEK293 cells (data not shown). Unfortunately, there are no boTLR1 antibodies available to investigate the protein expression, hence, its cellular presence and localization could not be confirmed. To follow the hypothesis that the so far published boTLR1 sequence may be incomplete, we analyzed the TLR1 sequences of the different species and figured out that the published sequence of the bovine receptor is 59 and 68 amino acids shorter than the human and murine TLR1, respectively, thereby possessing a shortened N-terminus (boTLR1-s, short). This led us to the assumption that the missing N-terminus might be responsible for the finding that triacylated ligands, like Pam3C-SK4, could not activate boTLR2/1-s transfected HEK293 cells.
We and others already showed that Gram-negative bacteria, which express triacylated lipoproteins, are recognized by native bovine cells in a TLR2-dependent manner [9, 33]. We, therefore, expected also triacylated synthetic lipopeptides to activate native bovine cells. And indeed, we could show that primary bovine cells and bovine cell lines could respond to di- as well as to triacylated ligands (Fig. 3). This again led us to the assumption that the published sequence of boTLR1 is incomplete and does not match the sequence of native boTLR1.
In fact, by investigating the sequence and the gene locus of boTLR1 more closely we found an additional ATG 195 base pairs upstream from the so far published start codon that was further proven to be the transcriptional start of the full length boTLR1 (boTLR1-fl; Supplementary Fig. S41).
After cloning the boTLR1-fl and transfecting this construct together with boTLR2, HEK293 cells were now able to induce an IL-8 production in response to triacylated lipopeptides (Fig. 5). From this we can conclude that the N-terminal amino acids are indispensable for a functionally active boTLR1. Even though we transfected the HEK293 cells with the same amounts of boTLR2 and boTLR1-fl plasmids we have no indication whether boTLR2 and boTLR1-fl need to be expressed in similar amounts by the cells, because there are no antibodies available to check the expression on protein level.
Although different studies showed that the amino acids which are involved in ligand binding, receptor interaction, and cellular distribution are mainly located at LRR 9–14, the transmembrane domain, the TIR domain, and the N-terminal part can also be important for the shape of the ectodomain, thereby influencing the ligand binding or the cellular targeting of the receptor [11, 18–20]. For example, mutation of four glycosylation sites of huTLR2 leads to its intracellular retention [30]. Also exchange of two cysteines at position 30 and 36 into serines caused an intracellular localization of this receptor thereby disabling the cell to recognize bacterial lipopeptides [19]. Preliminary data from computer-aided molecular modeling of TLR1-s and TLR1-fl indicate that beside the extracellular part of TLR1 also the transmembrane and the TIR domain express a modified conformation2. This finding indicates that all parts of the TLR1-molecule are affected by the deletion of the N-terminus in TLR1-s, which results in a not active molecule.
So far, lipopeptides used in this study were all di- or tripalmitoylated. This modification represents one of the most abundant forms of acylation found in lipoproteins of bacterial membranes [15]. However, the composition of bacterial cell walls is very diverse and many kinds of acylation patterns of lipoproteins can be found as described for the acylation pattern of LPS [14]. Therefore, we used the lipopeptide collection shown in Supplementary Figure S1A1 to evaluate the activation of boTLR2/1-fl by different structures in comparison to the human and murine heterodimers in HEK293 cells. Our data show clear species-specific differences in the recognition of these lipopeptides by the different heterodimers (Fig. 6). While the muTLR2/1 could already be activated with lipopeptides with ester-bound hexanoic acid residues the bovine and human receptor complex recognized lipopeptides with longer ester-bound dodecanoic and myristic acids, respectively. Therefore, it is likely that muTLR2 does form functionally less active complexes with huTLR1 than with huTLR6 and cotransfection with muTLR1 resulted in the marked enhancement of the response towards the triacylated ligands. Our data show that the bovine TLR2/1-heterodimer discriminates between diverse acylated lipopeptides in a different way compared to the human- and more pronounced to murine TLR2/1. On this basis it could be possible to explain species-specific susceptibilities and to get hints on the co-evolution of hosts and their species-specific environment that lead to the discrimination between pathogens and commensals [31].
Taken together, we firstly identified the full length functionally active boTLR1 and determined some structural prerequisites of triacylated lipopeptides to stimulate the boTLR2/1-fl heterodimer. We further demonstrated a species-specific recognition of triacylated lipopeptides by boTLR2/1-fl in comparison to muTLR2/1 and huTLR2/1. The characterization of the structure-activity relationship and generation of a HEK293 cell line stably expressing boTLR2 and the full length TLR1 can hereafter improve our understanding of the action of species-specific pathogens and will represent a good tool to further explore and characterize the interaction of TLR with bacteria causing disease in cattle, e.g. mastitis.
ONLINE MATERIAL
Supplementary PDF file provided by the authors.
Acknowledgments
This work was supported in part by the Deutsche Forschungsgemeinschaft UL68/3-2. The excellent technical assistance by Mrs. Carola Schneider (Research Center Borstel), Marija Brcic and Kay-Sara Sauter (University of Bern) is highly appreciated.
Footnotes
Supplementary figures are available on line at www.vetres.org.
Werlin D., personal communication, Hatfield, UK.
References
- 1.Babu M.M., Priya M.L., Selvan A.T., Madera M., Gough J., Aravind L., Sankaran K., A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins, J. Bacteriol. (2006) 188:2761–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beutler B.A., TLRs and innate immunity, Blood (2009) 113:1399–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buwitt-Beckmann U., Heine H., Wiesmüller K.H., Jung G., Brock R., Akira S., Ulmer A.J., Toll-like receptor 6-independent signaling by diacylated lipopeptides, Eur. J. Immunol. (2005) 35:282–289 [DOI] [PubMed] [Google Scholar]
- 4.Buwitt-Beckmann U., Heine H., Wiesmüller K.H., Jung G., Brock R., Ulmer A.J., Lipopeptide structure determines TLR2 dependent cell activation level, FEBS J. (2005) 272:6354–6364 [DOI] [PubMed] [Google Scholar]
- 5.Buwitt-Beckmann U., Heine H., Wiesmüller K.H., Jung G., Brock R., Akira S., Ulmer A.J., TLR1- and TLR6-independent recognition of bacterial lipopeptides, J. Biol. Chem. (2006) 281:9049–9057 [DOI] [PubMed] [Google Scholar]
- 6.Diamantstein T., Ulmer A., Stimulation by cyclic GMP of lymphocytes mediated by soluble factor released from adherent cells, Nature (1975) 256:418–419 [DOI] [PubMed] [Google Scholar]
- 7.Dunne A., O’Neill L.A., The interleukin-1 receptor/toll-like receptor superfamily: signal transduction during inflammation and host defense, Sci. STKE (2003) 2003:re3. [DOI] [PubMed] [Google Scholar]
- 8.Farhat K., Riekenberg S., Heine H., Debarry J., Lang R., Mages J., et al., Heterodimerization of TLR2 with TLR1 or TLR6 expands the ligand spectrum but does not lead to differential signaling, J. Leukoc. Biol. (2008) 83:692–701 [DOI] [PubMed] [Google Scholar]
- 9.Farhat K., Sauter K.S., Brcic M., Frey J., Ulmer A.J., Jungi T.W., The response of HEK293 cells transfected with bovine TLR2 to established pathogen-associated molecular patterns and to bacteria causing mastitis in cattle, Vet. Immunol. Immunopathol. (2008) 125:326–336 [DOI] [PubMed] [Google Scholar]
- 10.Fujita M., Into T., Yasuda M., Okusawa T., Hamahira S., Kuroki Y., et al., Involvement of leucine residues at positions 107, 112, and 115 in a leucine-rich repeat motif of human toll-like receptor 2 in the recognition of diacylated lipoproteins and lipopeptides and Staphylococcus aureus peptidoglycans, J. Immunol. (2003) 171:3675–3683 [DOI] [PubMed] [Google Scholar]
- 11.Gautam J.K., Ashish, Comeau L.D., Krueger J.K., Smith M.F. Jr, Structural and functional evidence for the role of the TLR2 DD loop in TLR1/TLR2 heterodimerization and signaling, J. Biol. Chem. (2006) 281:30132–30142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gay N.J., Gangloff M., Structure of toll-like receptors, Handb. Exp. Pharmacol. (2008) 183:181–200 [DOI] [PubMed] [Google Scholar]
- 13.Grabiec A., Meng G., Fichte S., Bessler W., Wagner H., Kirschning C.J., Human but not murine toll-like receptor 2 discriminates between tri-palmitoylated and tri-lauroylated peptides, J. Biol. Chem. (2004) 279:48004–48012 [DOI] [PubMed] [Google Scholar]
- 14.Hajjar A.M., Ernst R.K., Tsai J.H., Wilson C.B., Miller S.I., Human toll-like receptor 4 recognizes host-specific LPS modifications, Nat. Immunol. (2002) 3:354–359 [DOI] [PubMed] [Google Scholar]
- 15.Hantke K., Braun V., Covalent binding of lipid to protein. Diglyceride and amide-linked fatty acid at the N-terminal end of the murein-lipoprotein of the Escherichia coli outer membrane, Eur. J. Biochem. (1973) 34:284–296 [DOI] [PubMed] [Google Scholar]
- 16.Janeway C.A. Jr, Approaching the asymptote? Evolution and revolution in immunology, Cold Spring Harb. Symp. Quant. Biol. (1989) 54:1–13 [DOI] [PubMed] [Google Scholar]
- 17.Jiang Z., Georgel P., Li C., Choe J., Crozat K., Rutschmann S., et al., Details of toll-like receptor:adapter interaction revealed by germ-line mutagenesis, Proc. Natl. Acad. Sci. USA (2006) 103:10961–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng G., Grabiec A., Vallon M., Ebe B., Hampel S., Bessler W., et al., Cellular recognition of tri-/di-palmitoylated peptides is independent from a domain encompassing the N-terminal seven leucine-rich repeat (LRR)/LRR-like motifs of TLR2, J. Biol. Chem. (2003) 278:39822–39829 [DOI] [PubMed] [Google Scholar]
- 19.Nakao Y., Funami K., Kikkawa S., Taniguchi M., Nishiguchi M., Fukumori Y., et al., Surface-expressed TLR6 participates in the recognition of diacylated lipopeptide and peptidoglycan in human cells, J. Immunol. (2005) 174:1566–1573 [DOI] [PubMed] [Google Scholar]
- 20.Nishiya T., Defranco A.L., Ligand-regulated chimeric receptor approach reveals distinctive subcellular localization and signaling properties of the toll-like receptors, J. Biol. Chem. (2004) 279:19008–19017 [DOI] [PubMed] [Google Scholar]
- 21.Omueti K.O., Beyer J.M., Johnson C.M., Lyle E.A., Tapping R.I., Domain exchange between human toll-like receptors 1 and 6 reveals a region required for lipopeptide discrimination, J. Biol. Chem. (2005) 280:36616–36625 [DOI] [PubMed] [Google Scholar]
- 22.Pandey S., Agrawal D.K., Immunobiology of toll-like receptors: emerging trends, Immunol. Cell Biol. (2006) 84:333–341 [DOI] [PubMed] [Google Scholar]
- 23.Reitermann A., Metzger J., Wiesmüller K.H., Jung G., Bessler W.G., Lipopeptide derivatives of bacterial lipoprotein constitute potent immune adjuvants combined with or covalently coupled to antigen or hapten, Biol. Chem. Hoppe Seyler (1989) 370:343–352 [DOI] [PubMed] [Google Scholar]
- 24.Roach J.C., Glusman G., Rowen L., Kaur A., Purcell M.K., Smith K.D., et al., The evolution of vertebrate toll-like receptors, Proc. Natl. Acad. Sci. USA (2005) 102:9577–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandor F., Latz E., Re F., Mandell L., Repik G., Golenbock D.T., et al., Importance of extra- and intracellular domains of TLR1 and TLR2 in NFkappa B signaling, J. Cell Biol. (2003) 162:1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauter K.S., Brcic M., Franchini M., Jungi T.W., Stable transduction of bovine TLR4 and bovine MD-2 into LPS-nonresponsive cells and soluble CD14 promote the ability to respond to LPS, Vet. Immunol. Immunopathol. (2007) 118:92–104 [DOI] [PubMed] [Google Scholar]
- 27.Takeda K., Takeuchi O., Akira S., Recognition of lipopeptides by toll-like receptors, J. Endotoxin. Res. (2002) 8:459–463 [DOI] [PubMed] [Google Scholar]
- 28.Takeuchi O., Kawai T., Mühlradt P.F., Morr M., Radolf J.D., Zychlinsky A., et al., Discrimination of bacterial lipoproteins by toll-like receptor 6, Int. Immunol. (2001) 13:933–940 [DOI] [PubMed] [Google Scholar]
- 29.Takeuchi O., Sato S., Horiuchi T., Hoshino K., Takeda K., Dong Z., et al., Cutting edge: role of toll-like receptor 1 in mediating immune response to microbial lipoproteins, J. Immunol. (2002) 169:10–14 [DOI] [PubMed] [Google Scholar]
- 30.Weber A.N., Morse M.A., Gay N.J., Four N-linked glycosylation sites in human toll-like receptor 2 cooperate to direct efficient biosynthesis and secretion, J. Biol. Chem. (2004) 279:34589–34594 [DOI] [PubMed] [Google Scholar]
- 31.Werling D., Jann O.C., Offord V., Glass E.J., Coffey T.J., Variation matters: TLR structure and species-specific pathogen recognition, Trends Immunol. (2009) 30:124–130 [DOI] [PubMed] [Google Scholar]
- 32.Wu H.C., Tokunaga M., Tokunaga H., Hayashi S., Giam C.Z., Posttranslational modification and processing of membrane lipoproteins in bacteria, J. Cell. Biochem. (1983) 22:161–171 [DOI] [PubMed] [Google Scholar]
- 33.Yang W., Zerbe H., Petzl W., Brunner R.M., Günther J., Draing C., et al., Bovine TLR2 and TLR4 properly transduce signals from Staphylococcus aureus and E. coli, but S. aureus fails to both activate NF-kappaB in mammary epithelial cells and to quickly induce TNFalpha and interleukin-8 (CXCL8) expression in the udder, Mol. Immunol. (2008) 45:1385–1397 [DOI] [PubMed] [Google Scholar]
- 34.Zähringer U., Lindner B., Inamura S., Heine H., Alexander C., TLR2 – promiscuous or specific?, A critical re-evaluation of a receptor expressing apparent broad specificity, Immunobiology (2008) 213:205–224 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary PDF file provided by the authors.