Abstract
In eukaryotes, sequence-specific DNA-binding proteins activate gene expression by recruiting the transcriptional apparatus and chromatin remodeling proteins to the promoter through protein-protein contacts. In many instances, the connection between DNA-binding proteins and the transcriptional apparatus is established through the intermediacy of adapter proteins known as coactivators. Here we describe synthetic molecules with low molecular weight that act as transcriptional coactivators. We demonstrate that a completely nonnatural activation domain in one such molecule is capable of stimulating transcription in vitro and in vivo. The present strategy provides a means of gaining external control over gene activation through intervention using small molecules.
Each of the roughly 100,000 genes encoded in the human genome is subject to individual dosage control. The systems that regulate gene expression respond to a wide variety of developmental and environmental stimuli, thus allowing each cell type to express a unique and characteristic subset of its genes, and to adjust the dosage of particular gene products as needed. The importance of dosage control is underscored by the fact that targeted disruption of key regulatory molecules in mice often results in drastic phenotypic abnormalities (1), just as inherited or acquired defects in the function of genetic regulatory mechanisms contribute broadly to human disease. These findings have fueled efforts aimed at understanding fundamental mechanisms of gene regulation, with a eye toward discovering means of overriding endogenous regulatory controls or of creating new signaling circuitry in cells (2–5). Of particular interest in this regard are synthetic molecules designed to modulate gene transcription in living cells (2–6). To date, attention has been focused mostly on the discovery of organic molecules that interact sequence-specifically with DNA and thereby antagonize transcriptional stimulation by activator proteins (6). Here we describe a strategy for conditional activation of gene expression using organic molecules that simultaneously target the transcriptional machinery and a DNA-binding protein.
The regulatory mechanisms controlling the transcription of protein-coding genes by RNA polymerase II have been extensively studied. In the current model, RNA polymerase II and its host of associated proteins are recruited to the core promoter through noncovalent contacts with sequence-specific DNA binding proteins (7, 8). An especially prevalent and important subset of such proteins, known as transactivators, typically bind DNA at sites outside the core promoter and activate transcription via through-space contacts with components of the transcriptional machinery, including chromatin remodeling proteins (7–10). The DNA-binding and activation functions of transactivators generally reside on separate domains whose operation is portable to heterologous fusion proteins (11). Though activation domains must be physically associated with a DNA-binding domain to attain proper function, the linkage between the two need not be covalent (2–4). In many instances, the activation domain does not appear to contact the transcriptional machinery directly, but rather through the intermediacy of adapter proteins known as coactivators (12, 13). Our objective in the present work was to design relatively small organic molecules (≈4 kDa) that would function as transcriptional coactivators in vitro and in vivo.
MATERIALS AND METHODS
Synthesis of the Nonnatural Coactivators L-1 and D-1.
The Boc-protected hexamethylenediamine carbamate derivative of FK506 was deprotected and N-bromoacetylated in situ by treatment with bromoacetic anhydride as described (14). The product was purified by flash chromatography and its structure was confirmed by fast atom bombardment-HRMS. The L and D activator peptides CGSDALDDFDLDMLGSDALDDFDLDMLGS-NH2 were synthesized by standard standard solid phase peptide synthesis (rink resin)/deprotection methods, purified by reversed-phase HPLC, and characterized by amino acid analysis and HRMS. The bromoacetylated FK506 derivative and activator peptide were coupled using a procedure reported for protein ligation (15). Briefly, the activator peptide (650 μg, 0.21 μmol) was combined with bromoacetylated-FK506 (250 μg, 0.23 μmol) in 300 μl of 95% dimethylformamide/5% 0.1 M sodium phosphate buffer (pH 7) and the reaction was allowed to proceed overnight at room temperature. The product was purified by anion-exchange chromatography on a Waters Gen-Pack FAX HPLC column (4.6 × 100 mm) using a gradient of 5–45% B over 40 min (eluent A: 25 mM Tris⋅HCl, pH 7.5/10% CH3CN; eluent B: eluent A + 1.0 M NaCl). After desalting on a C18 Waters Sep-Pack Cartridge, the product was eluted with 9:1 acetonitrile/water and lyophilized. L-1 and D-1 conjugates were quantified by amino acid analysis and characterized by electrospray-ionization mass spectroscopy (negative ion absorption mode).
In Vitro Transcription.
HeLa nuclear extracts were prepared as described (16). GAL4(1–147)-VP16(413–490) was overexpressed and purified as described (17). The expression vectors coding for GAL4(1–94), and GAL4(1–94)-FKBP12 were subcloned into pLM1 (18) and the resulting fusion proteins were overexpressed and purified to homogeneity essentially as described for GAL4(1–147)-VP16(413–490). In vitro transcription assays were performed as described (19). The mixtures contained 25 μl of HeLa nuclear extract (3.15 mg/ml) in Dignam D buffer (20 mM Hepes, pH 7.9/100 mM KCl/20% glycerol/0.2 mM EDTA/0.5 mM DTT/0.5 mM phenylmethylsulfonyl fluoride), 8 mM MgCl2, 10 mM ammonium sulfate, 1% PEG 8000, 0.1 mg/ml BSA, 8 units RNAguard (Pharmacia), 200 ng of pGEM3 as carrier, 30 ng of template pG5E4T and either no GAL4 protein or an amount sufficient to give >90% protein-DNA complex, as determined by independent gel-shift assays (data not shown). The optimal amount of compounds L-1 and D-1 was titrated by transcription in vitro (data not shown). GAL4-FKBP was preincubated for 10 min at room temperature with 10 molar equivalents of coactivator L-1 or D-1 followed by a 10 min incubation time with the reporter construct. After addition of the nuclear extract and further preincubation for 15 min at room temperature, the reaction was initiated by addition of 2 μl of 10 mM ribonucleotide triphosphates. After 1 h at 30°C, the reaction was terminated and the reaction products were purified and analyzed by primer extension as described (19). Each experiment was repeated a minimum of three times.
In Vivo Transcription.
Jurkat cells were maintained in RPMI 1640 media containing 10% (vol/vol) calf serum, l-glutamine and 1% (vol/vol) penicillin/streptomycin. Cells plated in a six-well tissue culture plate (2 × 106 cells per well) were transfected (6 μl DMRIE-C; GIBCO/BRL) with 2 μg each of G5IL2SX and GF3. After 24 h incubation, the medium was removed and the cells were resuspended in fresh Opti-Mem I reduced serum medium and aliquoted into a 96-well microtiter plate (2 × 106 cells per well). Various concentrations of L-1 and D-1 in DMRIE-C were added to the cells. After 24 h, aliquots were removed and assayed for secreted alkaline phosphatase (SEAP) activity as described (2). In competition experiments, 1 μM rapamycin was added at the same time as D-1.
RESULTS
Experimental Design.
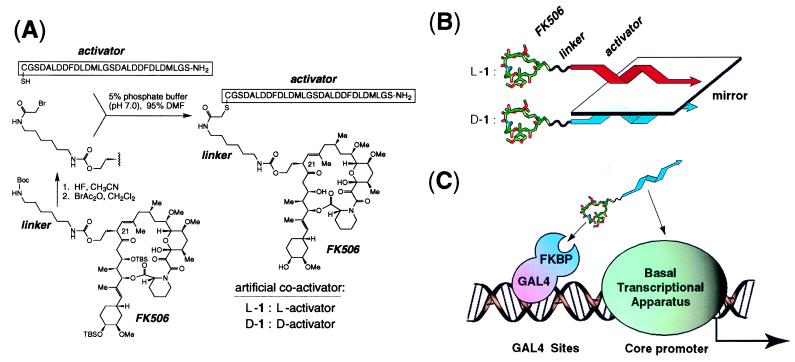
A molecule designed to serve as an intermediary between a DNA-binding protein and the transcriptional apparatus should incorporate binding elements for each of these two macromolecular targets. As the former, we chose the immunosuppressive drug FK506, which binds with high affinity (Kd = 0.4 nM) to the immunophilin FKBP12 (20). FK506 and certain of its derivatives can be targeted to the DNA binding domain of GAL4 by fusion of this domain to FKBP (GAL4-FKBP) (2). Modification of the calcineurin-binding surface of FK506 yields derivatives that lack immunosuppressive properties but retain the ability to bind FKBP with high affinity (2, 21). We equipped one such nonimmunosuppressive FK506 derivative, bearing a hydroxyethyl group at C-21, with an activator element through the addition of a linker, to which was attached a 29-amino acid L peptide containing a tandemly repeated undecamer sequence derived from the N-terminal portion of the VP16 activation domain (Fig. 1 A and B). This L peptide, when directly fused to the GAL4 DNA-binding domain, is a potent activator of transcription in vivo (22), most likely through binding directly to component(s) of the basal transcriptional apparatus (23). Thus, the FK506-peptide conjugate L-1 could in principle be capable of bridging GAL4-FKBP and the basal transcriptional apparatus (Fig. 1C).
Figure 1.
(A) Synthesis of the nonnatural coactivators L-1 and D-1. Abbreviations: TBS, tert-butyldimethylsilyl; Boc, t-butyloxycarbonyl. BrAc2O, bromoacetic acid anhydride; DMF, dimethylformamide; Me, methyl. (B) The structural relationship between L-1 and D-1. Both artificial activators contain an identical FK506 moiety (tubular structure) attached through an achiral linker (wavy line) to either of two enantiomeric 29-mer activator peptides (twisted arrow); L-1 (red) possesses the natural L stereochemical configuration, whereas D-1 (blue) possesses the nonnatural, mirror-image D configuration. (C) Schematic representation of the artificial coactivator serving as an intermediate between the DNA binding protein GAL4-FKBP and the basal transcriptional apparatus. The actual construct used in these experiments encodes GAL4 fused to three tandemly repeated FKBP domains (GAL4–FKBP3); only one FKBP domain is shown for graphical simplicity.
Synthetic Molecules That Coactivate Transcription of a Mammalian Gene in HeLa Nuclear Extracts.
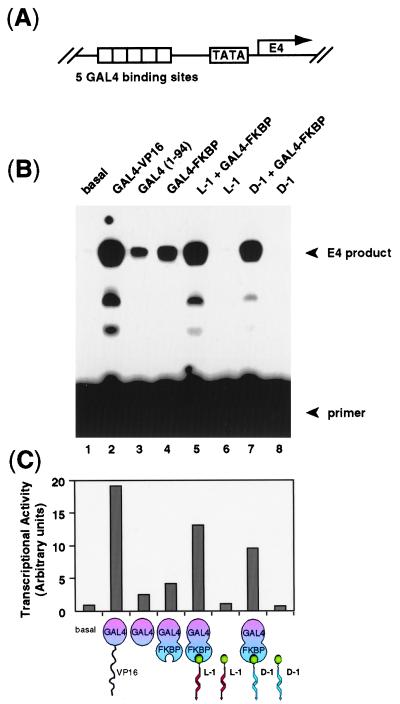
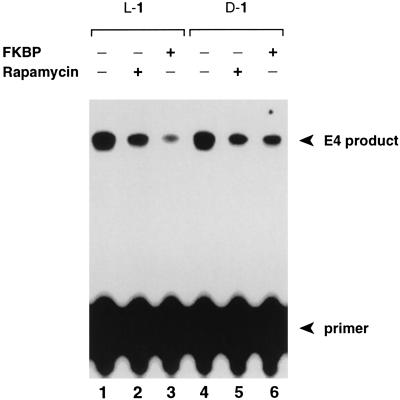
To assess the ability of L-1 to function as a transcriptional coactivator, we carried out in vitro transcription assays using HeLa nuclear extracts and a reporter construct pG5E4T containing 5 GAL4 sites upstream of an adenovirus E4 promoter (Fig. 2A). As shown in Fig. 2B (lane 5), L-1 stimulated transcription in the presence of GAL4-FKBP, but was unable to stimulate in the absence of GAL4-FKBP (lane 6). The activation potential of L-1 was significantly reduced in the presence of added rapamycin or FKBP, molecules that compete against the L-1/GAL4-FKBP interaction (Fig. 3, lanes 2 and 3) (24). These experiments demonstrate that L-1 functions as a coactivator with GAL4-FKBP in vitro.
Figure 2.
Transcriptional activation by the nonnatural coactivators L-1 and D-1. (A) The reporter construct pG5E4T contains five tandem 17-bp GAL4 binding sites (indicated by open boxes) positioned 23 bp upstream to the TATA box of a E4 promoter gene. (B) The template pG5E4T was transcribed in a crude HeLa cell nuclear extract in the presence of various added proteins and artificial coactivators (illustrated as in Fig. 1, except green sphere represents FK506). The primer and the E4 extension products are indicated by arrows. (C) Quantitation: The autoradiogram in B was scanned by PhosphorImager and the transactivator’s activity were plotted relative to the activity of the control nuclear extract.
Figure 3.
Competition Assay. The in vitro transcription was carried out as described (19), except GAL4-FKBP was preincubated 10 min at room temperature with 10 equivalents of compound L-1 or D-1 plus 100 equivalents of competitor, prior to the addition of the reporter construct and the HeLa nuclear extract. FKBP12 was expressed in Escherichia coli as a glutathione S-transferase fusion protein and purified on glutathione agarose beads.
Acyclic peptides having the natural L stereochemical configuration are highly susceptible to proteolysis in vivo, especially when they possess unmodified amide bonds (25). For this reason, it seemed unlikely that L-1 would function effectively to activate transcription in cells. On the other hand, peptides bearing the nonnatural D stereochemistry are often resistant to proteolysis, even in linear form (26). However, it is unknown whether a D configured peptide, or for that matter any nonnatural ligand, can function as a transcriptional activator. To test this issue directly, we synthesized D-1, an FK506 conjugate bearing the enantiomeric, D configured version of the VP16 activation peptide (Fig. 1 B and C). The results of in vitro transcription assays revealed that D-1 reproducibly stimulated transcription to a significant extent, though slightly less than L-1 (Fig. 2, lane 7). D-1 exhibited no activation in the absence of GAL4-FKBP (lane 8), and its activation potential was significantly reduced in the presence of added rapamycin or FKBP (Fig. 3, lanes 5 and 6). These results establish that the nonnatural molecule D-1 functions effectively as a transcriptional coactivator in vitro.
An Activation Domain Composed of All D Amino Acids Stimulates Transcription in Vivo.
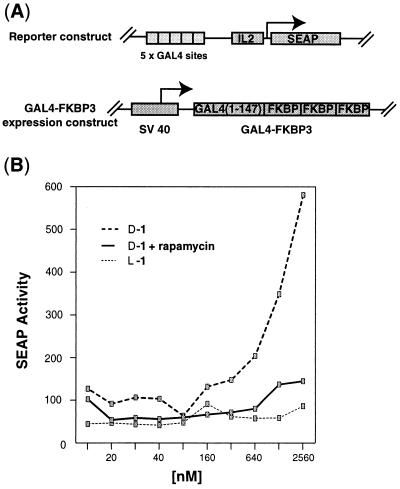
The in vitro results of D-1 prompted us to examine whether D-1 can function as a coactivator of transcription in living cells. To detect transcriptional activation signals, Jurkat cells were transiently cotransfected with (i) a reporter plasmid containing the SEAP cDNA and interleukin 2 promoter with five upstream GAL4 DNA binding sites, and (ii) a constitutive expression plasmid encoding the GAL4 DNA-binding domain fused to three tandemly repeated FKBP12s modules (GAL4-FKBP3, Fig. 4A). The transfected cells were subsequently treated with various concentrations of L-1 and D-1 incorporated into liposomes to enhance cell-permeability. When the cells were treated with D-1, expression of the SEAP reporter gene was stimulated in a dose-dependent manner (Fig. 4B). When the GAL4-FKBP3 expression plasmid was omitted during the transfection step, the cells were unresponsive to D-1 (data not shown), indicating that the activation stimulus was dependent upon a interaction between GAL4-FKBP3 and D-1. Consistent with this, rapamycin abolished the activation signal, presumably by competing with the FK506 portion of D-1 for the FKBP domain in GAL4-FKBP3. By contrast, L-1 showed no detectable ability to activate SEAP expression in transfected cells (Fig. 4B). As the L activation peptide is known to stimulate transcription in HeLa (22) and Jurkat cells (O.N. and G.L.V., unpublished results) when fused to the GAL4 DNA binding domain, and as FK506 derivatives are capable of saturating FKBP binding sites under the conditions of these experiments (2, 3), the failure of L-1 to activate transcription seems most likely to have resulted from intracellular proteolysis. The nuclear extracts used in vitro transcription assays almost certainly contain proteases as well; however, these are presumably rendered inactive by the proteases inhibitors included in the assays.
Figure 4.
Effect of artificial coactivators on reporter gene expression in vivo. (A) Schematic illustration of plasmids transfected into Jurkat cells in these experiments. The reporter construct (Top) G5IL2SX contains five tandem copies of the GAL4 response element upstream of the interleukin 2 minimal promoter and SEAP reporter (gift of G. R. Crabtree); the GAL4-FKBP3 (GF3) expression construct contains the GAL4 DNA-binding domain fused to three tandemly repeated FKBP domains (2) (gift of S.L.S.). (B) The artificial coactivator D-1, which contains a nonnatural D configured peptide, stimulates transcription in vivo; this effect is abolished by the addition of 1 μM rapamycin. The artificial coactivator containing the naturally configured L peptide fails to activate transcription. No activation was observed when L-1 or D-1 were administrated to cells in the absence of DMRIE-C. Highly acidic peptides such as the activator generally possess poor cell permeability.
DISCUSSION
Here we have shown that a ≈4-kDa synthetic molecule containing two linked binding elements—one that targets a DNA-binding protein and another that targets the transcriptional machinery—can coactivate transcription of a mammalian promoter. Specifically, we have shown that a designed coactivator containing a nonnatural completely D configured peptide stimulates transcription in vitro with only slightly less potency than the corresponding coactivator bearing the natural L configuration. Strikingly, the nonnatural molecule D-1 also stimulates transcription of a GAL4-driven promoter in vivo, when present in conjunction with GAL4-FKBP. The present experiments thus demonstrate both the feasibility of using small molecules to coactivate gene expression in vitro and in vivo, and the ability of completely nonnatural small molecules to serve this function.
The ability of a D configured peptide to serve as an activation domain raises a number of interesting mechanistic issues, such as whether the L and D peptides contact the same target. As the FK506 portion of either L-1 or D-1 almost certainly interacts much more strongly with its target than does the activator peptide portion, we envision that the synthetic coactivators first form a stable complex with the GAL4-FKBP fusion protein, and the resulting DNA-bound complex then recruits the transcriptional machinery to the promoter through direct peptide-protein contacts. The target of the L activator peptide is likely to be TFIIB, if indeed it contacts the same protein as the N-terminal portion of the VP16 activation domain, from which the 29-mer is derived (22, 23). Regardless whether the peptide is fused directly to a DNA-binding domain or bound noncovalently through the aegis of FK506-FKBP interactions, the target of the L peptide probably remains the same. No high-resolution structural information is available for any activation domain bound to its target. However, it has recently been demonstrated that an activation peptide derived from the C-terminal domain of VP16 folds into an α-helix upon interaction with its target, hTAFII31, with nonpolar contacts being made by hydrophobic amino acid side-chains that lie among one face of the helix, including one key contact made by a phenylalanine residue that apparently represents a common feature of several acidic activation domains (27). We note that the repeated sequence in the activation peptide not only contains a Phe residue that is known to serve an important functional role in the intact VP16 activation domain (28), but also contains additional hydrophobic residues at the i − 3 and i + 4 and i + 5 positions, which would all lie along one face of a putative α-helix. If indeed the L peptide contacts its transcriptional target using these residues, it is conceivable that the D peptide could make similar contacts, (though in reversed orientation with respect to the target), because the indicated nonpolar residues would all lie along one face of a D helix, and hydrophobic interactions can exhibit remarkable steric and geometric plasticity. Consistent with this notion, L and D configured calmodulin-binding peptides having the same amino acid sequence bind with similar strength to calmodulin (29). Of course, it remains a real possibility that the L-1 and D-1 target different components of the transcriptional apparatus.
The present demonstration that a nonnatural entity can activate transcription, together with prior findings that activators arise at a small but significant frequency in libraries of random fusion peptides (30), suggests it should also be possible to identify activation domains with low molecular weight from combinatorial libraries of organic molecules. A major limitation to any such screening effort has been the requirement that a prospective activator be physically associated with a DNA-binding domain. The system described here provides a means of overcoming this limitation by linking the activator to a cell-permeable organic ligand, FK506.
Acknowledgments
We thank Luc Gaudreau for providing the GAL4-VP16 construct (pJL2) and Brian D. Dynlacht for the HeLa cell line as well as for helpful discussion. Dusan Stanojevic prepared some of the reagents used in this investigation. This work was supported by a grant to G.L.V. from the National Science Foundation Presidential Young Investigator Program. O.N. was supported by fellowships from the Swiss National Foundation of Scientific Research and the Foundation Georgine-Claraz. M.U. was supported by fellowships from the Leukemia Society of America and Naito Foundation. D.J.A. was supported by a fellowship from the American Cancer Society.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: SEAP, secreted alkaline phosphatase.
A commentary on this article begins on page 13388.
References
- 1.Johnson R S, Spiegelman B M, Papaioannou V. Cell. 1992;71:577–586. doi: 10.1016/0092-8674(92)90592-z. [DOI] [PubMed] [Google Scholar]
- 2.Belshaw P J, Ho S N, Crabtree G R, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho S H, Biggar S R, Spencer D M, Schreiber S L, Crabtree G R. Nature (London) 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 4.Rivera V M, Clackson T, Natesan S, Pollock R, Amara J F, Keenan T, Magari S R, Phillips T, Courage N L, Cerasoli F, Jr, Holt D A, Gilman M. Nat Med. 1996;2:1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- 5.Spencer D M, Wandless T J, Schreiber S L, Crabtree G R. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 6.Gottesfeld J M, Neely L, Trauger J W, Baird E E, Dervan P B. Nature (London) 1997;387:202–205. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 7.Tjian R, Maniatis T. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 8.Stringer K F, Ingles C J, Greenblatt J. Nature (London) 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 9.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 10.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 11.Sadowski I, Ma J, Triezenberg S, Ptashne M. Nature (London) 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 12.Silverman N, Agapite J, Guarente L. Proc Natl Acad Sci USA. 1994;91:11665–11668. doi: 10.1073/pnas.91.24.11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. Nature (London) 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 14.Robey F A, Fields R L. Anal Biochem. 1989;177:373–377. doi: 10.1016/0003-2697(89)90068-7. [DOI] [PubMed] [Google Scholar]
- 15.Muir T W, Williams M J, Ginsberg M H, Kent S B H. Biochemistry. 1994;33:7701–7708. doi: 10.1021/bi00190a025. [DOI] [PubMed] [Google Scholar]
- 16.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chasman D I, Leatherwood J, Carey M, Ptashne M, Kornberg R D. Mol Cell Biol. 1989;9:4746–4749. doi: 10.1128/mcb.9.11.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sodeoka M, Larson C J, Chen L, LeClair K P, Verdine G L. Bioorganic Med Chem Lett. 1993;3:1089–1094. [Google Scholar]
- 19.Carey M, Leatherwood J, Ptashne M. Science. 1990;247:710–712. doi: 10.1126/science.2405489. [DOI] [PubMed] [Google Scholar]
- 20.Standaert R F, Galat A, Verdine G L, Schreiber S L. Nature (London) 1990;346:671–674. doi: 10.1038/346671a0. [DOI] [PubMed] [Google Scholar]
- 21.Bierer B E, Somers P K, Wandless T J, Burakoff S J, Schreiber S L. Science. 1990;250:556–559. doi: 10.1126/science.1700475. [DOI] [PubMed] [Google Scholar]
- 22.Seipel K, Georgiev O, Schaffner W. EMBO J. 1992;11:4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y-S, Ha I, Maldonado E, Reinberg D, Green M R. Nature (London) 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 24.Bierer B E, Mattila P S, Standaert R F, Herzenberg L A, Burakoff S J, Crabtree G, Schreiber S L. Proc Natl Acad Sci USA. 1990;87:9231–9235. doi: 10.1073/pnas.87.23.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saffran M, Kumar G S, Savariar C, Burnham J C, Williams F, Neckers D C. Science. 1986;233:1081–1084. doi: 10.1126/science.3526553. [DOI] [PubMed] [Google Scholar]
- 26.Wermuth J, Goodman S L, Jonczyk A, Kessler H. J Am Chem Soc. 1997;119:1328–1335. [Google Scholar]
- 27.Uesugi M, Nyanguile O, Lu H, Levine A J, Verdine G L. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 28.Cress W D, Triezenberg S J. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 29.Fisher P J, Prendergast F G, Ehrhardt M R, Urbauer J L, Wand A J, Sedarous S S, McCormick D J, Buckley P J. Nature (London) 1994;368:651–653. doi: 10.1038/368651a0. [DOI] [PubMed] [Google Scholar]
- 30.Ma J, Ptashne M. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]