Abstract
Osterix (Osx) is a zinc-finger-containing transcription factor that is expressed in osteoblasts of all endochondral and membranous bones. In Osx null mice osteoblast differentiation is impaired and bone formation is absent. In this study, we hypothesized that overexpression of Osx in murine bone marrow stromal cells (BMSC) would be able to enhance their osteoblastic differentiation and mineralization in vitro. Retroviral transduction of Osx in BMSC cultured in non-differentiating medium did not affect expression of Runx2/Cbfa1, another key transcription factor of osteoblast differentiation, but induced an increase in the expression of other markers associated with the osteoblastic lineage including alkaline phosphatase, bone sialoprotein, osteocalcin, and osteopontin. Retroviral transduction of Osx in BMSC also increased their proliferation, alkaline phosphatase activity, and ability to form bone nodules. These events occurred without significant changes in the expression of α1(II) procollagen or lipoprotein lipase, which are markers of chondrogenic and adipogenic differentiation, respectively.
Keywords: Osterix, BMSC, RCAS/TVA, Cbfa1/Runx2, Proliferation, Differentiation
Tissue engineering approaches are attempting to heal bone lesions above a critical size, by using resorbable scaffolds supplemented with regeneration-competent cells, such as embryonic stem (ES) cells derived from blastocyst stage embryos or adult bone marrow stromal cells (BMSC) [1–3]. Due to ethical controversy about the use of embryonic ES cells, research focus has shifted towards ex vivo expansion of adult BMSC for its use in bone regeneration studies. Following their isolation and expansion in tissue culture, BMSC are capable of repairing bone defects upon their transplantation into various animal models [4–6]. However, long-term ex vivo expansion of BMSC progressively leads to a decrease in their proliferation and loss of their osteogenic potential [5]. One approach that can be used to enhance and maintain a robust osteoblastic differentiation capacity in BMSC is through the use of ex vivo gene delivery [6] in which viral vectors are utilized to genetically modify osteoprogenitor cells to overexpress essential transcription factors for osteoblast differentiation and bone formation such as core binding factor α1 (Cbfa1)/runt-related gene (Runx)2 [7–9]. Runx2/Cbfa1 is expressed in an early multipotential mesenchymal cell population that can give rise to chondrogenic, osteogenic, and dentinogenic tissues as well as other lineages [10]. Runx2/Cbfa1 enhances osteoblast differentiation at an early stage and inhibits the late stage of osteoblast maturation [7–11]. Overexpression of Runx2/Cbfa1 in BMSC has been reported to enhance osteoblastic differentiation and mineralization in vitro [12,13] and promote bone formation in heterotopic (subcutaneous and intramuscular) and orthotopic (bone defect) sites [12]. Recently, a zinc-finger-containing transcription factor named osterix (Osx) has also been demonstrated to be critical in osteoblast differentiation and bone formation [14]. Osx acts downstream of Runx2/Cbfa1 to induce differentiation of preosteoblasts into fully functional osteoblasts [14] and overexpression of Osx has been shown to be sufficient to guide differentiation of ES towards the osteoblastic lineage in vitro [15]. The objectives of the current study were to examine the effects of Osx overexpression in BMSC proliferation and in their differentiation toward the osteoblastic, chondrogenic, and adipogenic lineages. Our results showed that Osx overexpression in BMSC can enhance their proliferation and osteogenic lineage commitment and hence holds a promising future in bone regeneration studies.
Materials and methods
Animals
Six- to eight-week-old B6D2F1 mice were maintained and used in accordance with recommendations in the Guide for the Care and Use of Laboratory Animals, prepared by the Institute on Laboratory Animal Resources, National Research Council (DHHS Publ. NIH 86-23, 1985). The animal protocol was approved by the Institutional Animal Use and Care Committee at the Tufts-New England Medical Center in Boston (MA).
Subcloning of viral constructs
To transduce BMSC we used the TVA-based retroviral system [16,17]. In this system, ectopic expression of the TVA receptor makes cells susceptible to infection with a replication-competent, subgroup-A avian leucosis viral (ASLV) vector, RCAS. The advantage of using this model is that cells expressing TVA can be repeatedly infected using RCAS vectors expressing different genes simultaneously or sequentially. To create the construct expressing TVA receptor, the tva fragment was cleaved out with EcoRI from pCR-tva-800 vector (a gift from Dr. Varmus’s laboratory, Memorial Sloan-Kettering Cancer Center, NY) and subsequently cloned it into the EcoRI site of pRc/CMV (Invitrogen, San Diego, CA). This new construct was named pRc/CMVtva. The empty viral vector RCAS (previously named by others RCASBP(A) [16–18]) was generously provided by Dr. Stephen Hughes (NCI/NIH). The 1.2 kb fragment of mouse Osx cDNA was kindly provided by Dr. Benoit de Crombrugghe (MD Anderson, Houston, TX). Osx cDNA was cloned into Cla I sites of RCAS construct through the Cla 12 Nco shuttle vector. This viral vector carrying the Osx cDNA was named RCAS-Osx. The orientation of the inserts was confirmed by restriction mapping and sequencing.
Production of high titer RCAS viral stocks
RCAS constructs were transfected into the established chicken fibroblast cell line, DF1 (CL-12203) to produce the viral stocks as previously described [18]. DF1 cells were purchased from the ATCC (Manassas, VA) and propagated in DMEM with 4 mM L-glutamine, 1.5 g/L sodium bicarbonate, and 4.5 g/L glucose (ATCC), 10% fetal bovine serum (Gibco-BRL), and 1% penicillin/streptomycin at 39 °C, and 5% CO2. Briefly, RCAS constructs were transfected into 80% confluent DF-1 cells using Superfect Lipofectin solution (Qiagen). Viral supernatant was harvested over 3 days and then concentrated about 30-fold by ultracentrifugation at 26,000 rpm at 4 °C. The viral pellet was re-suspended to a titer of 108 cfu/ml and stored at −80 °C.
BMSC culture conditions and viral transduction
BMSC were harvested from 6- to 8-week-old B6D2F1 mice. The femora and tibiae were excised immediately following euthanasia and the soft tissue was removed. The proximal end of the femur and the distal end of the tibiae were excised. α-MEM was used to flush gently marrow from the shafts with a 25-gauge needle. A single cell suspension was obtained by gently aspirating the cells sequentially through 20- and 23-gauge needles. The bone marrow cells were then seeded into T-75 flasks (Falcon Labware, Meylan Cedex, France) at a cell density of 4.0 × 105 nucleated cells/cm2 and cultured in non-differentiating maintenance media (α-MEM supplemented with 10% fetal bovine serum (FBS), 100 IU/mL penicillin, and 100 μg/mL streptomycin). Clonal BMSC cells were obtained essentially as previously described [19] and used in the experiments described in the manuscript. To control the specificity of the viral targeting we stably transfected clonal BMSC with pRc/CMVtva and then infected these cells with RCAS-vectors. The BMSC were transfected with pRc/CMVtva using the liposome reagent (Qiagen, Valencia, CA) as previously described [20]. Positive clones were then selected in G418-containing media for 14 days. Infection of BMSC was performed at 50% confluence. These BMSC clones stably expressing TVA were incubated with RCAS viral supernatant for 3 h at 37 °C, and then an equal volume of non-differentiating maintenance culture medium (α-MEM, 10% FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin) was added. Culture media were replaced after 24 h and then every 3 days. To induce osteogenic differentiation, confluent cells were cultured in osteogenic induction media (α-MEM supplemented with 10% FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin, 10−8 M dexamethasone, 10−2 M β-glycerophosphate, and 10−4 M L-ascorbic acid phosphate magnesium salt). To induce chondrogenesis, confluent BMSC were incubated in chondrogenic induction media (α-MEM supplemented with 10% FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin, 10−8 M dexamethasone, and 50 μg/mL ascorbic acid). To induce adipogenesis, BMSC cells were cultured in adipogenic induction media (α-MEM supplemented with 10% FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin, 10−8 M dexamethasone, and 6 ng/mL insulin).
Reverse transcriptase polymerase-chain reaction (RT-PCR) analysis
Total RNA was extracted by using an RNeasy MiniKit (Qiagen, Valencia, CA). Freshly isolated RNA was reverse transcribed with a SuperScript™ first-strand synthesis system (Invitrogen, Carlsbad, CA) following the manufacturer’s recommendations. The resulting cDNA was then amplified by PCR with the Platinum PCR supermix (Invitrogen, Carlsbad, CA). The sequences of the primers for amplification of mouse bone sialoprotein (BSP) were: 5′-GGAGGGGGCTTCACTGAT-3′ and 5′-AACAATCCGTGCCACCA-3′ (product size: 1048 bp); for mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were: 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′ (product size: 450 bp); for alkaline phosphatase (AP) were: 5′-GAAGACGTGGCGGTCTTTGC-3′ and 5′-GGGAATCTGTGCAGTCTGTG-3′ (product size: 457 bp); for osteopontin (OPN) were: 5′-CTCCCGGTGAAAGTGACTGA-3′ and 5′-GACCTCAGAAGATGAACTCT-3′ (product size: 831 bp); for osterix (Osx) were: 5′-ATTCTCCCATTCTCCCTCCCT-3′ and 5′-GGAAGGGTGGGTAGTCATTTGC-3′ (product size: 376 bp), for osteocalcin (OC) were: 5′-CTCCCGGTGAAAGTGACTGA-3′ and 5′-GACCTCAGAAGATGAACTCT-3′ (product size: 371 bp), for Runx2/Cbfa1 were: 5′-GAGGCCGCCGCACGACAACCG-3′ and 5′-CTCCGGCCCACAAATCTCAGA-3′ (product size: 294 bp), for α1(II) procollagen were 5′-CACACTGGTAAGTGGGGCAAGACCG-3′ and 5′-GGATTGTGTTGTTTCAGGGTTCGGG-3′ (product size: 172 bp), for lipoprotein lipase (LPL) were 5′-TCCAGAGTTTGACCGCCTTC-3′ and 5′-TTGGTCAGACTTCCTGCTACG-3′ (product size: 477 bp). Images of the amplified products in 1.2% agarose gels were captured with an UVP bioimaging system and processed by Adobe Photoshop 6.0 (Adobe Systems Incorporated, San Jose, CA) and Scion Image Beta 4.02 (Scion Image, Frederick, Maryland).
Proliferation assay
Cell proliferation was determined by the MTS assay, essentially as previously described [21,22]. Three thousand cells per well were seeded on 96-well plates and MTS readings at 405 nm were taken at different time points.
Alkaline phosphatase activity assay
An alkaline phosphatase (AP) assay was performed in cell layers by colorimetric assay of enzyme activity using an alkaline phosphatase kit from BioAssay system (Hayward, CA) as recommended by the manufacturer. To prepare the cell lysates, cell layers were washed three times with TBS buffer (50 mM Tris, pH 7.4, and 0.15 M NaCl) and then scraped into TBS/Triton buffer (0.1% Triton X-100) followed by sonication and centrifugation to remove cellular debris. One hundred microliter of lysate was then mixed with 100 μl of the freshly prepared colorimetric substrate para-nitrophenyl phosphate, and incubated at 37 °C for 30 min. The enzymatic reaction was stopped by adding 100 μl of 0.2 N NaOH solution. The optical density of the yellow product para-nitrophenol was determined spectrophotometrically at 405 nm. Protein concentration of the cell lysates was measured with a BCA Protein Assay Kit (Pierce, Rockford, IL), and AP activity was then expressed as para-nitrophenol produced in nmol/min/mg of protein.
In vitro differentiation of clonal BMSC
Accumulation of chondroid-appearing, alcianophilic matrix was used as evidence of BMSC chondrogenic potential and assessed by Alcian blue staining essentially as described [19]. To that end, cells were fixed with 4% phosphate-buffered formalin and stained with 0.1% Alcian blue (Sigma–Aldrich) in 0.1 N HCl for 2 h at room temperature and then washed three times with distilled water. Intracellular accumulation of neutral lipids, used as evidence of adipogenesis in vitro was demonstrated by Oil Red staining as described [23,24]. Formation of bone nodules, used as indicative of mineralization in vitro, was monitored by alizarin red staining. To that end, cells were fixed with 4% phosphate-buffered formalin and then rinsed three times with water. Cells were then treated with a 2% (weight/volume) solution of Alizarin red-S stain at pH 4.0 (Sigma, St. Louis, MO) for 10 min and then washed with 1× PBS for 15 min with gentle agitation. Digital images of the different stainings were taken at 10× magnification. Bone nodule number was counted in 6 different fields of each well and in three independent experiments.
Statistical analysis
All results are expressed as means ± SEM of 3 or more independent experiments. One-way ANOVA was used to test significance using the software package Statgraphic statistical graphics system (STSC, Rockville, MD). Linear and nonlinear fittings of time-course RT-PCR data were performed with Origin 6.1 (OriginLab, Northampton, MA). Regression coefficient closest to 1 was used to determine the best fit. Values of p lower than 0.05 were considered statistically significant.
Results and discussion
Clonal BMSC are pluripotent and exhibit different levels of expression of Osx and Cbfa1 depending on their differentiation commitment and cell culture conditions
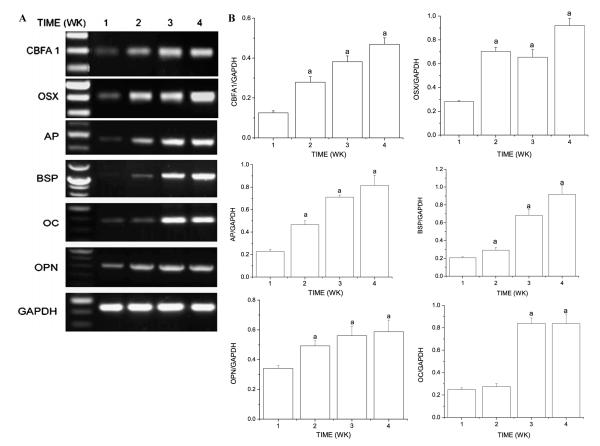
We first analyzed the time-dependent regulation of markers of osteoblastic (AP, BSP, OC, and OPN), chondrogenic (α1(II) procollagen), and adipogenic (LPL) differentiation as well as the levels of Cbfa1 and Osx in clonal BMSC cultured in differentiation media. When clonal BMSC were cultured under osteogenic conditions, expression levels of Cbfa1, Osx, BSP, OPN, and AP and OC increased in a time-dependent fashion (Figs. 1A and B). At 4 weeks, levels of Cbfa1/Runx2 and Osx were 5- and 3-fold higher, respectively, than those exhibited at the 1 week time point. Similarly, BSP, OPN, AP, and OC expression levels were 4.6-, 1.7-, 3.7-, and 3.4-fold higher at 4 weeks than at 1 week. When the same clonal BMSC were cultured under chondrogenic conditions, the expression levels of α1(II) procollagen, Cbfa1, and Osx were 1.5-, 1.5-, and 2-fold higher at 4 weeks than at 1 week (Figs. 2A and B). When the same clonal BMSC were cultured under adipogenic conditions, the expression levels of LPL and Osx expression increased for the first 3 weeks whereas Cbfa1 expression did not change significantly over a 4 week time period (Figs. 2C and D). Under these experimental conditions, levels of LPL and Osx were 2-fold higher at 3 weeks than at 1 week.
Fig. 1.
Clonal BMSC differentiate towards the osteoblastic lineage when cultured in osteogenic induction media. Clonal BMSC were cultured in osteogenic induction media for different times. (A) Representative image of semiquantitative RT-PCR analyses of extracellular matrix proteins (AP, BSP, OC, and OPN) characteristic of an osteoblastic phenotype and transcription factors (Cbfa1 and Osx) important for bone formation. (B) Levels of AP, BSP, OC, OPN, Cbfa1, and Osx were normalized with those of the loading control GAPDH in three independent experiments. ap < 0.05 vs. 1 week time point.
Fig. 2.
Clonal BMSC differentiate towards the chondrogenic and adipogenic lineages when cultured in chondrogenic and adipogenic induction media, respectively. Clonal BMSC were cultured in chondrogenic induction media (A,B) or in adipogenic induction media (C,D) for different times. (A) Representative image of semiquantitative RT-PCR analyses of α1(II) procollagen, a marker of chondrogenic differentiation and transcription factors (Cbfa1 and Osx) important for bone formation. (B) Levels of α1(II) procollagen, Cbfa1, and Osx were normalized with those of the loading control GAPDH in three independent experiments. ap < 0.05 vs. 1 week time point. (C) Representative image of semiquantitative RT-PCR analyses of LPL, a marker of adipogenic differentiation and transcription factors (Cbfa1 and Osx) important for bone formation. (D) Levels of LPL, Cbfa1, and Osx were normalized with those of the loading control GAPDH in three independent experiments. ap < 0.05 vs. 1 week time point.
These results demonstrated that the clonal BMSC used in our study were pluripotent, and that their differentiation towards the osteoblastic, chondrogenic, and adipogenic lineages induced by differentiating media was associated with different regulation in the expression of Cbfa1 and Osx. Whereas cells induced to differentiate into the osteoblastic or chondrogenic phenotypes exhibited an upregulation in the expression of both transcription factors, cells induced to differentiate into the adipogenic phenotype only experienced changes in Osx expression. Expression of Osx in BMSC committed towards the osteoblastic, chondrogenic, and adipogenic lineages is in agreement with previous reports suggesting that Osx may be expressed earlier than the preosteoblast stage by committed bipotential progenitors for bone/fat-forming cells and bone/cartilage-forming cells [25].
Expression pattern of Osx and RUNX2/Cbfa1 in BMSC transduced with RCAS/TVA retroviral infection system
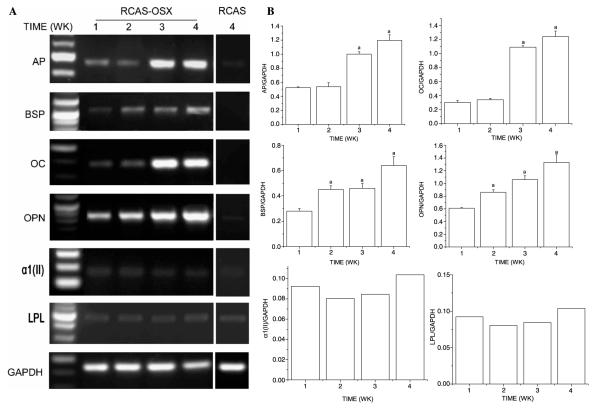
In order to overexpress Osx in BMSC, we used an RCAS/TVA retroviral system. Cells were then cultured in non-differentiating maintenance media that lack of dexamethasone and other factors in order to analyze their true differentiating ability in vitro. We then evaluated the expression pattern of Osx, Runx2/Cbfa1, and markers of osteogenic, chondrogenic, and adipogenic differentiation in BMSC transduced with RCAS-Osx or with the empty vector. As shown in Fig. 3A, RCAS-Osx transduced BMSC cultures showed a linear increase with time in their normalized levels of Osx. Thus, at 4 weeks levels of Osx were 5-fold higher than those exhibited at 1 week. In contrast, empty vector transduced cells did not express Osx at any of the time points examined, and the levels of Cbfa1 did not significantly change with time in either BMSC transduced with RCAS-Osx (Fig. 3A) or with the empty RCAS vector (Fig. 3B). As shown in Figs. 4A and B, levels of expression of the osteogenic markers AP, BSP, OPN or OC, as opposed to those of GAPDH, LPL, and α1(II) procollagen, increased with their time in culture in RCAS-Osx transduced BMSC. In RCAS-Osx transduced cells, BSP and OPN expression increased in a linear fashion, whereas AP and OC fitted a sigmoidal curve with time in culture. Thus, levels of BSP, OPN, AP, and OC were 2.3-, 2.1-, 2.3-, and 4.1-fold higher at 4 weeks than at 1 week. BSP and OC expression levels were undetectable at any of the time points examined in empty RCAS transduced BMSC (Fig. 4A and data not shown). However, some expression of OPN and AP was detectable in the latter cells but it did not increase with time in culture (Fig. 4A and data not shown). Thus, OPN and AP were expressed 17- and 28-fold higher in RCAS-Osx transduced BMSC than in those transduced with the empty vector at 4 weeks.
Fig. 3.
The RCAS/TVA retroviral system induces high levels of expression of Osx in BMSC cultured in maintenance media. RCAS-Osx transduced BMSC and empty vector transduced BMSC were cultured in maintenance media (non-differentiating) for different times. (A) Representative image of semiquantitative RT-PCR analyses of Osx expression. (B) Levels of Osx were normalized with those of the loading control GAPDH in three independent experiments. ap < 0.05 vs. 1 week time point. (C) Representative image of semiquantitative RT-PCR analyses of Cbfa1 expression. (D) Levels of Cbfa1 were normalized with those of the loading control GAPDH in three independent experiments.
Fig. 4.
RCAS-Osx transduced BMSC cultured in maintenance media exhibit increased expression of osteoblastic differentiation markers. RCAS-Osx transduced BMSC and empty vector transduced BMSC were cultured in non-differentiating maintenance media for different times. (A) Representative image of semiquantitative RT-PCR analyses of markers of osteoblastic differentiation (AP, BSP, OC, and OPN), chondrogenic differentiation (α1(II) procollagen), and adipogenic differentiation (LPL). (B) Levels of different differentiation markers were normalized with those of the loading control GAPDH in three independent experiments. ap < 0.05 vs. 1 week time point.
RCAS-Osx transduced BMSC exhibited significant increases with time in the expression of OC and AP mRNA, as previously described in overexpression studies with Osx in ES cells or C2C12 and C3H10T1/2 cell lines [14,15]. However, our study shows for the first time that overexpression of Osx in adult BMSC leads to the increased expression of osteogenic differentiation markers (OC, AP, BSP, and OPN) under experimental conditions that do not affect Runx2/Cbfa1 expression. The finding that RCAS-Osx transduced BMSC did not exhibit an upregulated expression of Runx2/Cbfa1 was not surprising, based on a previous report indicating that Osx functions downstream of Runx2/Cbfa1 in the osteoblastic differentiation pathway [14]. Authors of that study showed that Osx null mice did not express transcripts for BSP, OPN, but exhibited normal levels of Runx2/Cbfa1 [14]. In comparison, Runx2/Cbfa1 defective embryos, that are deficient in BSP and OPN expression [7,8], expressed normal levels of Osx [14]. However, overexpression of Osx in ES cells was unexpectedly found to upregulate Runx2/Cbfa1 expression [15] as a result of a possible feedback mechanism of Osx to Cbfa1. However, the overexpression of Osx in the latter study was performed in the presence of dexamethasone, which is a potent stimulator of osteogenic commitment and differentiation in vitro [26,27], and increases the expression of Runx2/Cbfa1 and Osx in differentiating fetal rat calvarial cells [28]. In our studies, osteogenic media that included 10−8 M dexamethasone also increased levels of Cbfa1 and Osx in murine BMSC. Therefore, the reported upregulation of Runx2/Cbfa1 as a consequence of Osx overexpression in ES cells [15] could have been the result of inclusion of dexamethasone in the culture media. It is also possible that the transcriptional requirements of ES cells to differentiate into the osteogenic lineage are different to those of BMSC. Indeed, control ES cells grown in osteogenic media exhibit a significant upregulation in Cbfa1 expression whereas levels of Osx do not change markedly with time [15]. In contrast, our RT-PCR results indicated that control BMSC grown in osteogenic media showed significant upregulation in the expression of both Cbfa1 and Osx.
Overexpression of Osx increases BMSC proliferation, AP activity, and mineralization in vitro
We then analyzed whether overexpression of Osx could affect BMSC proliferation within their first week of culture in maintenance medium. As shown in Fig. 5A, BMSC transduced with RCAS-Osx exhibited 2-fold higher proliferation rate than untransduced cells or those transduced with the empty RCAS vector. We then assessed whether even in the absence of osteogenic culture conditions, overexpression of Osx would be sufficient to increase AP activity at early stages of BMSC differentiation. As shown in Fig. 5B, BMSC transduced with RCAS-Osx, exhibited a linear increase in AP activity within the first 2 weeks in culture. In contrast, AP activities of non-transduced BMSC or those transduced with empty vector remained at background levels when cultured under the same experimental conditions (Fig. 5B). Thus, AP activities were 1.8- and 3.4-fold higher at 1 week and 2 weeks, respectively, in RCAS-Osx transduced cells compared to cells transduced with empty virus. Bone nodule formation in postconfluent cultures also increased in a linear fashion with time and reached the maximum level after 4 weeks in RCAS-Osx transduced cells cultured in regular maintenance media in a similar fashion than untransduced BMSC grown under osteogenic conditions (Fig. 5C). Importantly, untransduced BMSC or BMSC transduced with empty RCAS vector did not form any bone nodules when cultured in maintenance media for up to 4 weeks (data not shown).
Fig. 5.
RCAS-Osx transduced BMSC cultured in maintenance media exhibit increased proliferation, AP activity, and mineralization in vitro. RCAS-Osx transduced BMSC and empty vector transduced BMSC were cultured in non-differentiating maintenance media for different times. Untransduced BMSC were cultured in non-differentiating maintenance media (A,B) or osteogenic media (C) depending on the experiment. (A) Osx increases BMSC proliferation. MTS assays were performed in multiple 96-well plates containing 3000 cells per well at time 0 (100% proliferation) for up to 6 days. Experiments were performed in triplicate, and the mean values ± SE are shown. ap < 0.05 vs. empty vector transduced cells or untransduced BMSC. (B) Osx increases AP activity in a linear fashion. AP activity of transduced and transduced BMSC cultured in maintenance media was measured in triplicate and normalized by the protein levels. Mean values ± SE are shown. AP activity was significantly increased for RCAS-Osx transduced BMSC. ap < 0.05 vs. day 4. (C) Comparison of mineralization assays by RCAS-Osx transduced BMSC cultured in maintenance media and untransduced BMSC cultured in osteogenic media. Alizarin red S stainings were performed in three independent experiments with BMSC transduced with RCAS-Osx or with empty vector cultured in non-osteogenic media. The mineralization potential of non-transduced BMSC cultured in osteogenic media was also monitored for comparison purposes. Number of bone nodules was counted in pictures taken from three independent experiments at 10× magnification and expressed as number of nodules on each well (9.6 cm2 surface area). All values were significantly different (p < 0.05) vs. 1 week time point.
The ability of Osx to induce proliferation of BMSC in our study is in agreement with a recent report demonstrating that overexpression of Osx induces the proliferation of NIH3T3 fibroblasts [29]. In our study, AP activity, an early marker of osteoblast differentiation, was also upregulated in Osx-transduced BMSC cultured in non-osteogenic media for up to 2 weeks. More importantly, RCAS-Osx transduced BMSC were able to form bone nodules in vitro in the absence of any other osteogenic factors in the culture media. Similarly to our report, overexpression of Osx in ES cells has been shown to increase the formation of bone nodules [15]. In contrast, Osx did not promote osteoblast differentiation or mineralization in NIH3T3 fibroblasts [29]. Whether the inability of Osx to induce mineralization in cultures of NIH3T3 is due to suboptimal levels of expression of Osx requires further investigation.
Overexpression of Osx in BMSC does not favor chondrogenic or adipogenic lineage commitment
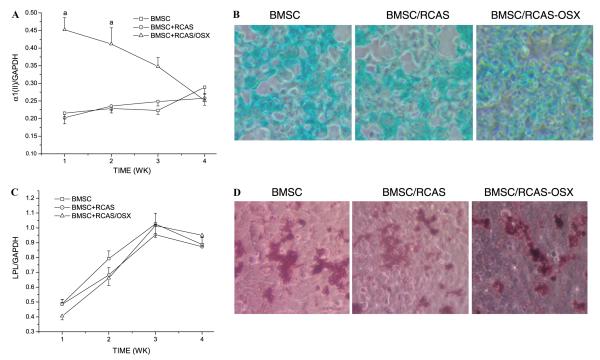
We then sought to investigate whether Osx overexpression would have any effect in the ability of BMSC to commit to the chondrogenic or adipogenic lineages. To that end, we analyzed the expression of LPL and α1(II) procollagen by semiquantitative RT-PCR in RCAS-Osx transduced cells versus untransduced cells or those infected with empty RCAS. As shown in Fig. 6A, α1(II) procollagen expression exhibited a 2-fold upregulation in RCAS-Osx transduced cells respect to untransduced cells or cells transduced with empty RCAS within the first 2 weeks. However, the levels of α1(II) procollagen expression were not significantly different among the three cell groups at 4 weeks (Fig. 6A). Consistent with these results and with the inability of Osx to induce chondrogenic differentiation, Alcian blue stainings of the three cell groups were not significantly different among each other at 4 weeks (Fig. 6B). On the other hand, LPL expression exhibited a similar pattern of time-dependent upregulation in the 3 cell groups (Fig. 6C). Consistent with these results and with the inability of Osx to induce adipogenic differentiation, Oil Red O stainings were not significantly different among the 3 cell groups at 4 weeks (Fig. 6D).
Fig. 6.
Overexpression of Osx in BMSC does not increase their chondrogenic or adipogenic potential. RCAS-Osx transduced BMSC and empty vector transduced BMSC and untransduced BMSC were cultured in chondrogenic induction media (A,B) or adipogenic induction media (C,D). (A) Levels of α1(II) procollagen, a chondrogenic differentiation marker were determined by RT-PCR and then normalized with those of the loading control GAPDH in three independent experiments. ap < 0.05 vs. untransduced BMSC or transduced with empty RCAS. Values at 4 weeks were significantly different (p < 0.05) vs. 1 week time point in the three groups. (B) Alcian blue stainings were performed to demonstrate chondrogenic differentiation at 4 weeks. (C) Levels of LPL, an adipogenic differentiation marker, were determined by RT-PCR and then normalized with those of the loading control GAPDH in three independent experiments. All values were significantly different (p < 0.05) vs. 1 week time point. (D) Oil Red O staining was performed to demonstrate adipogenic differentiation at 4 weeks.
Taken together, our report is the first one demonstrating that Osx can increase proliferation and promote osteoblast differentiation of adult BMSC. Therefore, use of ex vivo gene therapy to overexpress Osx in BMSC holds a great promise in bone regeneration studies aimed at healing bone defects above a critical size.
Acknowledgments
This work was supported by National Institutes of Health Grants DE11088 and DE14537 (to J.C.).
References
- [1].Petite H, Viateau V, Bensaid W, Meunier A, de Pollak C, Bourguignon M, Oudina K, Sedel L, Gillemin G. Tissue-engineered bone regeneration. Nat. Biotechnol. 2000;18:959–963. doi: 10.1038/79449. [DOI] [PubMed] [Google Scholar]
- [2].Heng BC, Cao T, Stanton LW, Robson P, Olsen BR. Strategies for directing the differentiation of stem cells into the osteogenic lineage in vitro. J. Bone Miner. Res. 2004;19:1379–1394. doi: 10.1359/JBMR.040714. [DOI] [PubMed] [Google Scholar]
- [3].Hutmacher DW, Garcia AJ. Scaffold-based bone engineering by using genetically modified cells. Gene. 2005;347:1–10. doi: 10.1016/j.gene.2004.12.040. [DOI] [PubMed] [Google Scholar]
- [4].Bianco P, Riminucci M, Gronthos S, Robey PG. Bone marrow stromal stem cells: nature, biology and potential applications. Stem Cells. 2001;19:180–192. doi: 10.1634/stemcells.19-3-180. [DOI] [PubMed] [Google Scholar]
- [5].Banfi A, Bianchi G, Notaro R, Luzzatto L, Cancedda R, Quarto R. Replicative aging and gene expression in long-term cultures of human bone marrow stromal cells. Tissue Eng. 2002;8:901–910. doi: 10.1089/107632702320934001. [DOI] [PubMed] [Google Scholar]
- [6].Derubeis AR, Cancedda R. Bone marrow stromal cells (BMSCs) in bone engineering: limitations and recent advances. Ann. Biomed. Eng. 2004;32:160–165. doi: 10.1023/b:abme.0000007800.89194.95. [DOI] [PubMed] [Google Scholar]
- [7].Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:677–680. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- [8].Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- [9].Otto F, Thornell AP, Cromptom T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblastic differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- [10].Bronckers AL, Sasaguri K, Cavender AC, D’Souza RN, Engelse MA. Expression of Runx2/Cbfa1/Pebp2alphaA during angiogenesis in postnatal rodent and fetal human orofacial tissues. J. Bone Miner. Res. 2005;10:428–437. doi: 10.1359/JBMR.041118. [DOI] [PubMed] [Google Scholar]
- [11].Liu W, Toyosawa S, Furuichi T, Kanatani N, Yoshida C, Liu Y, Himeno M, Narai S, Yamaguchi A, Komori T. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J. Cell Biol. 2001;155:157–166. doi: 10.1083/jcb.200105052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zheng H, Guo Z, Ma Q, Jia H, Dang G. Cbfa1/osf2 transduced bone marrow stromal cells facilitate bone formation in vitro and in vivo. Calcif. Tissue Int. 2004;74:194–203. doi: 10.1007/s00223-003-0004-x. [DOI] [PubMed] [Google Scholar]
- [13].Byers BA, Garcia AJ. Exogenous Runx2 expression enhances in vitro osteoblastic differentiation and mineralization in primary bone marrow stromal cells. Tissue Eng. 2004;10:1623–1632. doi: 10.1089/ten.2004.10.1623. [DOI] [PubMed] [Google Scholar]
- [14].Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- [15].Tai G, Polak JM, Bishop AE, Christodoulou I, Buttery LDK. Differentiation of osteoblasts from murine embryonic stem cells by overexpression of the transcription factor osterix. Tissue Eng. 2004;10:1456–1466. doi: 10.1089/ten.2004.10.1456. [DOI] [PubMed] [Google Scholar]
- [16].Federspiel MJ, Hughes SH. Retroviral gene delivery. Methods Cell Biol. 1997;52:179–214. [PubMed] [Google Scholar]
- [17].Holland EC, Varmus HE. Basic fibroblast growth factor induces cell migration and proliferation after glia-specific gene transfer in mice. Proc. Natl. Acad. Sci. USA. 1998;95:1218–1223. doi: 10.1073/pnas.95.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li L, Zhu J, Tu Q, Yamauchi M, Sodek J, Karsenty G, Tang J, Chen J. An in vivo model to study osteogenic gene regulation: targeting an avian retroviral receptor (TVA) to bone with the bone sialoprotein (BSP) promoter. J. Bone Miner. Res. 2005;20:1403–1413. doi: 10.1359/JBMR.050316. [DOI] [PubMed] [Google Scholar]
- [19].Satomura K, Krebsbach P, Bianco P, Robey PG. Osteogenic imprinting upstream of marrow stromal cell differentiation. J. Cell Biochem. 2000;78:391–403. [PubMed] [Google Scholar]
- [20].Tu Q, Yamauchi M, Pageau SC, Chen JJ. Autoregulation of bone sialoprotein gene in pre-osteoblastic and non-osteoblastic cells. Biochem. Biophys. Res. Commun. 2004;316:461–467. doi: 10.1016/j.bbrc.2004.02.068. [DOI] [PubMed] [Google Scholar]
- [21].Valverde P, Obin MS, Taylor A. Role of Gas6/Axl signaling in lens epithelial cell proliferation and survival. Exp. Eye Res. 2004;78:27–37. doi: 10.1016/j.exer.2003.10.002. [DOI] [PubMed] [Google Scholar]
- [22].Valverde P, Tu Q, Chen J. BSP and RANKL induce osteoclastogenesis and bone resorption synergistically. J. Bone Miner. Res. 2005;20:1669–1679. doi: 10.1359/JBMR.050511. [DOI] [PubMed] [Google Scholar]
- [23].Banfi A, Muraglia A, Dozin B, Mastrogiacomo M, Cancedda R, Quarto R. Proliferation, kinetics and differentiation potential of ex vivo expanded human bone marrow stromal cells: Implications for their use in cell therapy. Exp. Hematol. 2000;28:707–715. doi: 10.1016/s0301-472x(00)00160-0. [DOI] [PubMed] [Google Scholar]
- [24].Bosch P, Pratt SL, Stice SL. Isolation, characterization, gene modification, and nuclear reprogramming of porcine mesenchymal stem cells. Biol. Reprod. 2006;74:46–57. doi: 10.1095/biolreprod.105.045138. [DOI] [PubMed] [Google Scholar]
- [25].Tai G, Christodoulou I, Bishop AE, Polak JM. Use of green fluorescent fusion protein to track activation of transcription of the transcription factor osterix during early osteoblast differentiation. Biochem. Biophys. Res. Commun. 2005;333:1116–1122. doi: 10.1016/j.bbrc.2005.05.195. [DOI] [PubMed] [Google Scholar]
- [26].Buttery LD, Bourne S, Xynos JD, Wood H, Hughes FJ, Hughes SP, Episkopou V, Polak JM. Differentiation of osteoblasts and in vitro bone formation from murine embryonic stem cells. Tissue Eng. 2001;7:89–99. doi: 10.1089/107632700300003323. [DOI] [PubMed] [Google Scholar]
- [27].Beresford JN, Joyner CJ, Devlin C, Triffitt JT. The effects of dexamethasone and 1,25-dihydroxyvitamin D3 on osteogenic differentiation of human marrow stromal cells in vitro. Arch. Oral Biol. 1994;39:941–947. doi: 10.1016/0003-9969(94)90077-9. [DOI] [PubMed] [Google Scholar]
- [28].Igarashi M, Kamiya N, Hasegawa M, Kasuya T, Takahashi T, Takagi M. Inductive effects of dexamethasone on the gene expression of Cbfa1, Osterix and bone matrix proteins during differentiation of cultured primary rat osteoblasts. J. Mol. Histol. 2004;35:3–10. doi: 10.1023/b:hijo.0000020883.33256.fe. [DOI] [PubMed] [Google Scholar]
- [29].Kim YJ, Kim HN, Park EK, Lee BH, Ryoo HM, Kim SY, Kim IS, Stein JL, Lian JB, Stein GS, van Wijnen AJ, Choi JY. The bone-related Zn finger transcription factor Osterix promotes proliferation of mesenchymal cells. Gene. 2005;25 doi: 10.1016/j.gene.2005.08.021. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]