Abstract
As a case-study of biosimulation model integration, we describe our experiences applying the SemSim methodology to integrate independently-developed, multiscale models of cardiac circulation. In particular, we have integrated the CircAdapt model (written by T. Arts for MATLAB) of an adapting vascular segment with a cardiovascular system model (written by M. Neal for JSim). We report on three results from the model integration experience. First, models should be explicit about simulations that occur on different time scales. Second, data structures and naming conventions used to represent model variables may not translate across simulation languages. Finally, identifying the dependencies among model variables is a non-trivial task. We claim that these challenges will appear whenever researchers attempt to integrate models from others, especially when those models are written in a procedural style (using MATLAB, Fortran, etc.) rather than a declarative format (as supported by languages like SBML, CellML or JSim's MML).
1. Integrating multiscale biosimulation models
A goal of many biosimulation researchers is to develop libraries of re-usable simulation modules that can be combined in a plug-n-play manner. Ideally, biosimulation researchers should be able to retrieve a module from such a library and easily integrate it with their own models. Although this vision has been well-described[1, 2], to our knowledge there have been no published processes or methodologies by which model integration might occur.
In this paper, we* report on our experiences integrating two independently developed models for cardiovascular circulation. As we describe in greater detail in Section 2, our first goal was to make the shape of the aortic pressure waveform in our cardiovascular simulation model (CV) more canonical by raising the pulse pressure. Our second goal was to test and extend the semantic simulation (SemSim) framework[3] for biosimulation model reuse with a challenging case of cross-platform integration: integrating MATLAB code with code for the JSim environment. Our most important results and findings have to do with three critical challenges to understanding and translating procedural biosimulation code, such as MATLAB, into a declarative environment, such as JSim[4].
1.1 SemSim modeling
We used the declarative SemSim modeling framework for our model integration task. SemSim models (called “AMO models” in [3]) are light-weight ontologies that leverage knowledge in larger reference ontologies to match concepts across biosimulation models. The SemSim framework captures the biological meaning of model components along with the mathematical/logical dependencies necessary for model simulation. We have designed this framework to facilitate model sharing and reuse by automating the processes of merging and translating legacy biosimulation models. Presently these tasks are accomplished through hand-coding—a very costly and error-prone process.
Figure 1 shows the representational schema by which a SemSim model serves as a middle layer between biosimulation code (on the right) and biomedical reference ontologies (on the left). SemSim model variables and equations are represented by SemSim Data structure and Computation classes in a Computational model and then mapped (via encodes relations) to corresponding Physical properties and Property dependencies classes in a Physical model. The Physical properties in turn are unambiguously referenced (has_property relation) to a Physical entity that references a specific class in the Foundational Model of Anatomy (FMA)[5]. Physical property, on the other hand, is referenced to a Physical property class of the Ontology of Physics for Biology (OPB [6]). For example, the physical property Volume of blood in systemic arteries is defined using the class Blood in systemic arterial tree from the FMA and the class Fluid volume in the OPB. Such annotations uniquely encode and identify the biological meaning of model variables in a computable form that allows concepts to be mapped across SemSim models.
Figure 1.
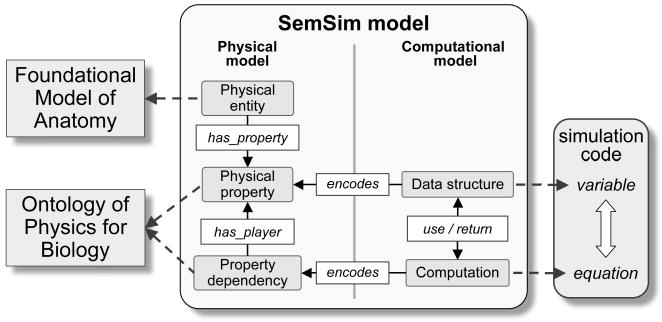
A diagram of the structure of a SemSim ontology. These ontologies can improve the reusability of biosimulation code (on right) by connecting variables and equations with knowledge stored in reference ontologies (on left).
1.2 Procedural versus declarative code
Procedural programming languages, such as MATLAB and Fortran, specify the steps computers must take to reach a desired outcome. including calls to subroutines and ODE solvers. In contrast, declarative languages simply specify the problem without specifying a procedure for solution. SBML, CellML and JSim's mathematical modeling language (MML, not to be confused with MathML) are declarative languages. There are strengths and weaknesses to both styles of programming. However, our SemSim architecture for reuse is declarative. Thus, we needed to express the models in a declarative manner.
In Section 3, we identify three important challenges to integrating procedural code into the declarative SemSim architecture for reuse:
First, the overall structure of the procedural model must be understood. Most critically, multiscale models may solve equations and update variables at more than one time scale, and these different scales must be clearly articulated and differentiated.
Second, different languages, whether procedural or declarative, employ a variety of data structures for storing and manipulating variables. Languages such as Fortran and MATLAB can use matrices, arrays, lists, etc., whereas SBML, CellML[7] and JSim's MML have a much smaller range of variable representational choices.
Third, the dependencies among biosimulation variables much be understood. In procedural code, this can be challenging, as the relationships among variables can be scattered throughout the code, and variable names can change as they are passed into and out of computational functions.
We believe these challenges are general—they will exist whenever researchers integrate externally developed procedural code into their models, whether or not they use the SemSim approach. Sections 3 and 4 below describe in detail how we overcame these challenges for our particular case study. We also discuss how some extensions to SemSim (section 4) will help address these challenges. However, these general problems may indicate that no model integration system can be fully automatic. Researchers should be forewarned and prepared for these complexities when integrating multiscale models.
2. Motivation and modeling results
We encountered the CircAdapt modeling of tissue and organ level cardiovascular dynamics at PSB08[8] and recognized an opportunity to improve our own CV model and to stress-test the representational capabilities of our SemSim modeling framework. Our modeling motivation was to improve the realism of our aortic pressure curve by integrating the CircAdapt systemic arterial component which included fluid inertial effects of pulsatile blood flow.
Figure 2 is a roadmap of our model merging process as described in this paper. We needed to: (1) translate the relevant portions of the CircAdapt MATLAB code into JSim MML code, (2) create a SemSim model of the CircAdapt systemic arterial system, (3) merge the SemSim version of the CircAdapt systemic arterial component with the SemSim version of our CV model and (4) re-encode the merged SemSim model into MML for execution in JSim.
Figure 2.
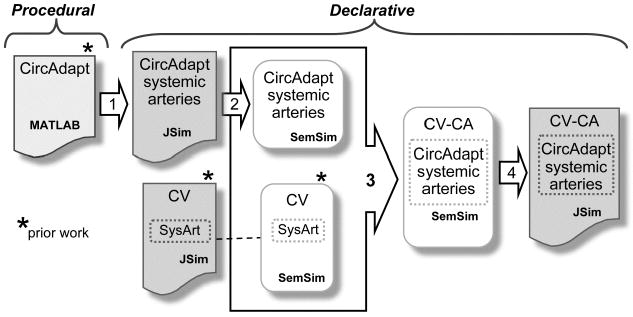
Overview of the 4-step process by which the CircAdapt MATLAB systemic arterial components were extracted and merged with the UW CV model. 1: Extract relevant procedural CircAdapt code and encode in functionally equivalent JSim MML code (results shown in Figure 3). 2: Derive a SemSim model from JSim version of CircAdapt Systemic arteries model. 3: Use Protégé Prompt plug-in to merge SemSim models and replace original CV systemic arterial (SysArt) components with CircAdapt systemic arteries. 4: Use SemSim Coder software to automatically generate executable JSim MML code from the merged model (results shown in Figure 4).
We validated our MATLAB-to-JSim integration in two ways. First, we compared the response of the native MATLAB systemic arterial pulse pressure profiles with those generated by the JSim version by driving the JSim version with aortic flow data produced by CircAdapt. Figure 3 shows that the numerical results were virtually identical and any discrepancies were probably attributable to differences in the ODE solver routines. Second, we found that incorporating the more sophisticated modeling of systemic arterial dynamics of the CircAdapt model did indeed provide a much more canonical aortic pressure waveform than our original CV model (Figure 4).
Figure 3.
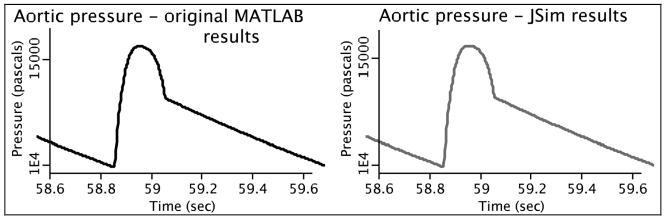
We reproduced the CircAdapt MATLAB model's steady state aortic pressure (left) in our JSim version of the systemic arterial component (right).
Figure 4.
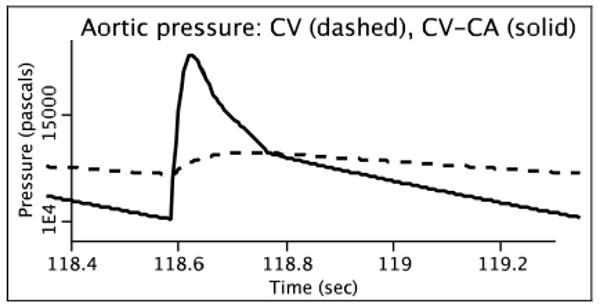
Steady state aortic pressure results from the CV-CA merged model (solid) showing improved pulse pressure over the original CV model results (dashed).
3. Translating from procedural to declarative models
We encountered three main challenges when translating the procedural code of CircAdapt's systemic arterial model to JSim's declarative MML code: 1) simulating multiple time scales, 2) resolving differences in the simulation environments' allowed data structures, and 3) translating the mathematical dependencies from the original model completely and faithfully.
3.1 Assessing computational architecture and identifying time scales
To reproduce and reuse biosimulation models, researchers must rely on several sources. Some source code may be publicly available in model repositories but in other cases, the primary source is a publication (e.g., the publication we used:[9]) which generally contains only incomplete details about model code implementation. Thus, direct contact with model authors must often be established to obtain source code and crucial clarifications about modeling choices. In this case, we had no prior collaboration with the T. Arts group, and given the geography, communication was limited to a handful of emails. We believe this will be common for many reuse scenarios, and adds to the challenge of understanding and integrating code developed by others.
We first assessed the CircAdapt MATLAB model's computational structure which consists of 14 separate MATLAB files and a README documentation file. Model execution is under the control of the CircAdaptMain.m file which calls a MATLAB ODE solver (ode23) to calculate circulatory volume and flow state variables for a single heart beat. At the end of each heart beat, CircAdapt discretely updates several model variables to simulate the slower process of genetically-controlled blood vessel adaptation to hemodynamic variables. The adapted variables are then used to restart the ODE solver for the next heart beat repeatedly until a user-specified end time. Therefore, the CircAdapt model operates on two different time scales: a faster one for simulating within-beat fluid dynamics and a slower one for genetic adaptations of blood vessel geometry. This aspect of the model was not immediately apparent until we assessed the procedural code in detail. In section 3.3, we detail the workarounds in our JSim code to handle integrations on dual time scales.
Given our experiences, we believe that it is important for model annotations to explicitly describe the time scales upon which models operate. This will be especially salient when models are translated from a procedural to a declarative language, since procedural languages offer more control over how the model is solved. We recognize, however, that these annotations are only a first step in solving the more general and more difficult computational problem of efficient and accurate numerical integration across multiple time-scales. Our current focus is on identifying temporal semantics, a first and necessary step before devising efficient mathematical methods for simulating dynamics on multiple time-scales.
3.2 Identifying data structures and physiological variables
Our modeling goal was to include only the dynamics of the systemic arterial tree of the CircAdapt model. Thus, we needed to identify and extract only those variables relevant to that portion of the CircAdapt model. CircAdapt uses a single, globally-accessible MATLAB tree-like data structure called Par that represents physiological variables (blood pressures, flows, volumes, etc.) as leaves. This structure allows the developer to group related variables together as sub-trees, so that they can be passed together into various functions. For example the Par.TubeLArt subtree includes variables for the systemic arterial vessels whereas the Par.TubeLArt.Adapt subtree contains variables for the adaptive calculations. Thus, for example, the variable representing volume of blood in the systemic arteries is accessed via Par.TubeLArt.V, and the corresponding pressure via Par.TubeLArt.p. Some information about this data structure was supplied by T. Arts in a README file.
JSim's MML does not support the complex data type used for Par. In general, whenever translating across simulation platforms and languages, there will be some level of mismatch among supported data types. Because we were using only a single component of the CircAdapt model, we had little use for the grouping advantages this data structure provides. Thus, we simply flattened the list of variables and parameters while retaining CircAdapt's naming conventions: variables like Par.TubeLArt.V became Par_TubeLArt_V
In general, it may be that some information will be lost during translation from complex data structures to simpler ones. Although we could flatten the CircAdapt Par tree structure, multi-dimensional matrices cannot be so easily accommodated. Model authors should be explicit about their data structure choices, and researchers that wish to reuse model components should be aware of potential problems translating such structures.
3.3 Assessing variable dependencies
Assessing variable dependencies is critical to understanding a model: which variables depend on time vs. which variables are fixed in the model, which variables are inputs vs. those that are outputs, and, critically, how variables within the model depend upon one another. Once we understood the overall CircAdapt architecture, data structures and dependencies we created a corresponding list of parameter and variable declarations in the MML model that included the role players in CircAdapt relevant for reproducing the systemic arterial dynamics (such as Par.TubeLArt.V). The next task was to determine the mathematical dependencies among these variables.
As described above, some variables, like systemic arterial volume, are solved continuously within the MATLAB ode23 call, but other variables depend on processing the results from the previous heartbeat and are updated discretely following each heart cycle. Reproducing the mathematical dependencies present in the CircAdapt ode23 call was straightforward within JSim. However, to update the slower adaptation variables at the end of each heart cycle we used MML “realState” variables, which can be updated discretely. Additionally, some of these discrete variables in CircAdapt were updated using mean values of pressure and flow data from the previous heartbeat. Because MATLAB allows more procedural control over how model data is produced, we could not directly replicate these computations in MML. We instead wrote a new set of equations conducive to the JSim simulation engine to compute these mean values.
Some time-continuous variables are computed piecewise throughout the original CircAdapt code execution, and these separated dependencies had to be merged together in the MML version. In general, dependencies among variables in procedural code can be scattered and possibly buried deep within nested function calls. Tracking such nested dependencies was difficult, time-consuming and depended on direct communication with the CircAdapt author (T. Arts).
Communication with T. Arts was important throughout the translation process. These communications helped the team understand the structure, inherent assumptions and computational implementation of the CircAdapt model much more readily. Specifically, Dr. Arts: 1) identified those model codewords that are global constants versus those that update discretely versus those that update continuously, 2) identified the units used for the model constants and variables, 3) clarified that the model was designed to conserve blood volume, and 4) identified appropriate CircAdapt output data for validating the JSim results.
4. SemSim model-merging and code generation
In this section we describe our use of the SemSim architecture to merge the CircAdapt systemic arterial component into our CV model, and then to generate code for the combined system (steps 2-4 in Figure 2).
4.1 Creating a SemSim model of the CircAdapt systemic arteries
The first step in our merging process was to create a SemSim model of the JSim code that implements the CircAdapt systemic arterial model. Using Protégé [10] and the SemSim framework (see Figure 1), we began by creating a Physical property class for each variable in the CircAdapt JSim model. Thus, the JSim variable Par_TubeLArt_V (Volume of blood in the systemic arteries) was represented by a Physical property subclass Fluid volume, which references the Fluid volume class in the Ontology of Physics for Biology. We linked this Property class to the Physical entity class Blood in the systemic arterial tree, which references the corresponding class in the Foundational Model of Anatomy.
Next, we created classes describing the quantitative Physical dependencies (see Figure 1) between Physical properties. For example, the Systemic arterial blood volume dependency class has a positive flow rate role player Blood flow through aortic valve and a negative rate player Blood flow through systemic arterial tree. This Physical dependency describes how Systemic arterial blood volume is solved as the temporal integral of the fluid analog of Kirchoff's current law (change in vessel fluid volume equals vessel inflow minus outflow).
In addition to these Physical model components of the CircAdapt SemSim module, we completed the SemSim model by building appropriate Datum and Computation classes under the SemSim Computational model component class. We have developed a prototype SemSim Coder program that uses the Protégé API to access a SemSim file and automatically generate simulation code from its Computational model component contents. In the future, this program will use platform-independent mathematical pseudo-code, but currently, it is restricted to generating MML code for JSim.
We tested the completeness of the SemSim version of the CircAdapt systemic arteries by running it through the SemSim Coder program and checking the numerical simulation results against the MML version. This process revealed several manual data entry errors, but these were easily corrected. For relatively small biosimulation models such as the CircAdapt systemic arteries, creating a SemSim model is straightforward, and it will become easier as we improve our tools.
4.2 Merging the SemSim models with Prompt
We next used the Protégé Prompt plug-in [11] to merge models and create a new circulatory system that retains most components of the CV model but replaces its original systemic arterial component with the CircAdapt systemic arteries component. Prompt can be custom-tailored; however, we simply used the off-the-shelf capabilities of this ontology merging tool.
First, we used the Prompt plug-in to copy all the classes of the previously built SemSim CV model and the new SemSim CircAdapt model into one file. Next, we had to identify and resolve the points of overlap between the CircAdapt module and the CV model. From our own knowledge we knew a priori which physiological properties had to be resolved: those properties shared by the two models that we as the modelers considered equivalent. Prompt automatically recognized three of these shared properties: blood flow through the aortic valve, pressure in the aorta, and heart rate. However, we also wished to replace the property Volume of blood in systemic arteries in capillaries in the CV model with Volume of blood in the systemic arteries from the CircAdapt module. For Prompt to identify this kind of resolution, it must recognize that there is a structural relationship between the participating entities in these two properties. This information was already encoded in the SemSim CV model, where Blood in systemic arteries and capillaries → has_part → Blood in systemic arteries. In the future we plan to implement a Prompt plug-in or standalone program as part of our SemGen package that analyzes the structural relationships between anatomical entities shared by both models before it suggests points of resolution.
Next, we hand-resolved these four points of overlap between the models. For each of the shared Physical Properties between the models, we picked the one we wanted to use in the merged system. For example, because we wanted to compute aortic pressure using the CircAdapt equations, we used the Pressure in aorta property class from the CircAdapt module, and not the one from the CV model. For each matching Property pair the values in the Input_Role_Player_In slot from the unused property were copied into the used property in order to preserve the dependencies specified in the unused property. Next we identified all those Datum classes which had unused property classes in their Computational_Component_For slots. All values in the Input_Data_For slots of these unused Datum classes were copied into the Datum classes that referred to the corresponding used property. This step preserved dependency information from the unused Datum classes. We then deleted all the Datum classes that referred to Properties that were not going to be used in the merged system (like Paorta, which referred to the unused Pressure in aorta property from the CV model). Then we deleted all the unused property classes, all the computation classes that had no Output_Data slot entries, and all the dependencies that had no Has_Output_Player slot entries.
To resolve the shared Physical entity classes of the two models, we arbitrarily chose to keep those entities from the CircAdapt model, and delete their twin entities in the CV model but only after copying all the slot values from the CV classes into their twin CircAdapt classes. This step preserved all the information from the CV Physical entity classes in the new merged model. To remove irrelevant Physical entities from the resulting merged system we deleted any Physical entity classes in which the Has_Physical_Property, Part_of, Adjacent_to, and Continuous_with slots were empty.
Based on our work merging these two models and others in the past, we plan to generalize this process of rectifying resolution points and create a tool within our SemGen program that better automates the steps described above.
4.3 Generating MML code from the merged SemSim model
Finally, we generated MML code from the merged CV-CA SemSim model using our SemSim Coder program. After entering the parameter values and initial conditions from the original CV and CircAdapt models, we attempted to run the merged model. However, the initial run failed because our model merging had introduced a circular dependency among variables. In particular, the equation for aortic pressure depended on the change in systemic arterial volume which depended on aortic flow which in turn depended on aortic pressure. This circular dependency was not present in either of the individual models, but rather was a feature of the new merged system. In general, this type of integration problem may be common when merging models. We argue that our SemSim approach, with its declarative specification of variables and dependencies, will make it easier to detect and resolve such circularities. In our case, we changed the equation for arterial volume to an ODE so that this variable could be set with an initial start value, and thus break the circular dependency.
After we corrected the circularity, the model was too stiff to solve numerically. Checking over our parameter values, we noticed that the value for heart rate in the merged model was incorrect, given its units. This error appeared because both the CV and CircAdapt models used the same codeword to represent heart rate (HR) but used different units and thus, different numerical values. In their respective parameter sets, which do not include unit designations, the numbers for HR are an order of magnitude apart, and when the parameters for the individual models were loaded into the merged system, HR was reparameterized with a value from the wrong model. To fix the problem, we simply corrected the value of the parameter in the MML code. Aside from HR, no changes were made to the parameters of the merged model. As presented in section 2, Figure 4 shows that aortic pressure in the CV-CA merged model has a more canonical waveform than the original CV model.
5. Discussion and Conclusions
A pervasive problem in reusing and merging biosimulation code is their inconsistent and incomplete annotations and explanations. There are ongoing efforts to standardize model annotation (e.g., [1]) in terms of units, nomenclature, etc. We contend that one of the most important aspects of every biosimulation model is its biological meaning—what physical aspects of which biological structures are being modeled and to what purpose. Thus we are developing the SemSim approach to provide a machine-readable, and thus searchable, annotation for models that are formally referenced to ontologies and constrained vocabularies of biological structure and biophysics.
Whereas we believe a declarative approach for model representation will be important for model reuse and integration, we recognize that procedural programming is widely used among modelers and offers important advantages in efficiency and customization. In our case study we illuminated three important challenges for representing procedural models in a declarative framework:
Models may include procedures that occur on multiple time scales
Data structures and naming conventions used to represent model variables may not translate across simulation platforms
Dependencies among model variables may be obscured due to piecewise computation and/or variable renaming within functions
We claim that these challenges will appear whenever researchers attempt to integrate models from others, especially when those models are written in a procedural style (using MATLAB, Fortran, etc.) rather than a declarative format (as supported by languages like SBML, CellML or JSim's MML). We were able to solve these challenges in our particular case study, and our solutions will inform the further development of our SemGen software, and more broadly, our SemSim architecture. Our goals is to provide modelers with useful tools to annotate, create, and merge multiscale biosimulation models.
Given the inherent complexity of biosimulation modeling and the variety of independent modeling approaches for solving specific problems, it is no wonder that these challenges can be major impediments to model reuse and integration. A major premise of our SemSim approach is borrowed from the Java mantra: “write once, run anywhere.” Thus, we propose that the hard work of translating native biosimulation code into a SemSim model can be performed once, and using more automated code re-writing methods, new models can be generated to run anywhere.
Acknowledgments
This work was partially funded by the NIH: for M. Neal: #T15 LM007442-06, and for J. Gennari & D. Cook: #R01HL087706-01.
Footnotes
This research involved two independent research groups. For narrative simplicity, this paper was written from the point of view of the University of Washington authors. Contributions from the University of Maastricht author are indicated explicitly in the text.
References
- 1.Le Novere N, Finney A, Hucka M, Bhalla US, Campagne F, Collado-Vides J, et al. Minimum information requested in the annotation of biochemical models (MIRIAM) Nat Biotechnol. 2005;23(12):1509–15. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- 2.Hunter PJ, Borg TK. Integration from proteins to organs: the Physiome Project. Nat Rev Mol Cell Biol. 2003;4(3):237–43. doi: 10.1038/nrm1054. [DOI] [PubMed] [Google Scholar]
- 3.Gennari JH, Neal ML, Carlson BE, Cook DL. Integration of multi-scale biosimulation models via light-weight semantics. Pac Symp Biocomput. 2008;13:414–425. [PMC free article] [PubMed] [Google Scholar]
- 4.JSim. The JSim Home Page at NSR. http://physiome.org/jsim/index.html.
- 5.Rosse C, Mejino JLV. A Reference Ontology for Bioinformatics: The Foundational Model of Anatomy. Journal of Biomedical Informatics. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Cook DL, Mejino JLV, Neal ML, Gennari JH. Bridging Biological Ontologies and Biosimulation: The Ontology of Physics for Biology. AMIA. 2008 in press. [PMC free article] [PubMed] [Google Scholar]
- 7.CellML. CellML. http://www.cellml.org.
- 8.Lumens J, Delhaas T, Kirn B, Arts T. Modeling ventricular interaction: a multiscale approach from sarcomere mechanics to cardiovascular system hemodynamics. Pac Symp Biocomput. 2008:378–89. [PubMed] [Google Scholar]
- 9.Arts T, Delhaas T, Bovendeerd P, Verbeek X, Prinzen FW. Adaptation to mechanical load determines shape and properties of heart and circulation: the CircAdapt model. Am J Physiol Heart Circ Physiol. 2005;288(4):H1943–54. doi: 10.1152/ajpheart.00444.2004. [DOI] [PubMed] [Google Scholar]
- 10.Gennari JH, Musen MA, Ferguson RW, Grosso WE, Crubezy M, Eriksson H, et al. The evolution of Protege: an environment for knowledge-based systems development. Int J Human–Computer Studies. 2003;58:89–123. [Google Scholar]
- 11.Noy NF, Musen MA. The PROMPT suite: Interactive tools for ontology merging and mapping. Int J of Human-Computer Studies. 2003;59(6):983–1024. [Google Scholar]


