Abstract
MicroRNAs, processed by the RNAase III enzyme Dicer, are ~22 nucleotide endogenous noncoding small RNAs. The function of Dicer in the mouse central nervous system (CNS) development is not well understood. Here we show that specifically deleting Dicer expression in the CNS and in the cerebral cortex using two Cre lines results in reduced progenitor numbers, abnormal neuronal differentiation and thinner cortical wall. Incomplete Dicer deletion during early embryonic stages contributes to normal development of early-born neurons in the cortex and motor neurons in the spinal cord. However, at late embryonic stages when Dicer is completely ablated in the CNS, the migration of late-born neurons in the cortex and oligodendrocyte precursor expansion and differentiation in the spinal cord are greatly affected. Our studies of different timings of Dicer deletion demonstrate the importance of the Dicer-mediated microRNA pathway in regulating distinct phases of neurogenesis and gliogenesis during the CNS development.
Keywords: Dicer, microRNAs, mouse central nervous system, neurogenesis, gliogenesis
Introduction
The development of the central nervous system (CNS) initiates from the induction of the neural tissue in the ectoderm, the formation of the neural plate and afterward the folding and closure of the neural tube (Tanabe and Jessell, 1996). Subsequently, the neural tube is divided into several constrictions that correspond to the presumptive regions of the CNS: the forebrain, midbrain, hindbrain and spinal cord, along the anterior and posterior neural tube.
The forebrain consists of the cerebral cortex and the subcortical regions, such as the striatum. In the embryonic mouse cerebral cortex, radial glial cells in the ventricular zone (VZ) represent the majority of neural progenitor cells and they normally undergo asymmetric division to generate one radial glial cell and one neuron (Anthony et al., 2004; Kriegstein, 2005). Intermediate (or basal) progenitors, which reside in the subventricular zone (SVZ), divide symmetrically to generate postmitotic neurons (Noctor et al., 2004; Hevner et al., 2006; Sessa et al., 2008). Radial glial cells and intermediate progenitors give rise to projection neurons that migrate away from the VZ/SVZ to the cortical plate (CP). The CP is organized into an inside-to-outside layer structure due to the migration of late-born neurons passing through the early-born neurons. In the striatum, progenitors produce interneurons that tangentially migrate to the cerebral cortex (Corbin et al., 2001; Wonders and Anderson, 2006). Moreover, gliogenesis of astrocytes and oligodendrocytes occurs mostly in postnatal stages in developing cortices (Richardson et al., 2006). However, the molecular control that is critical for proper cortical neurogenesis and gliogenesis is not well understood.
In the mouse spinal cord, progenitors in the ventricular zone give rise to motor neurons in the ventral region and distinct interneurons in the dorsal region by early embryonic day 10.5 (E10.5). The production of motor neurons and interneurons is controlled by cross-interactions of multiple transcription factors (Jessell, 2000; Lee and Pfaff, 2001). Oligodendrocyte progenitors and astrocytes are also derived from the ventral neural tube by complex gene expression regulators at a later stage by E12.5 (Rowitch, 2004; Richardson et al., 2006). The molecular program that regulates development of distinct cell types in the spinal cord remains an exciting and unclear question.
The recent discoveries of microRNAs have revealed a new layer of gene expression regulation during development. MicroRNAs (miRNAs) are ~22 nucleotide (nt) endogenous noncoding small RNAs (Lee et al., 1993; Wightman et al., 1993). MiRNA precursors are processed into mature miRNAs by the RNAase III enzyme Dicer (Kim, 2004; Hammond, 2005). Mature miRNAs regulate gene expression by recognizing the 3′-untranslated region (3′-UTR) of target genes and silencing protein translation. Dicer plays an important role during development. For example, Dicer null mice die at E7.5 and lack multi-potent stem cells (Bernstein et al., 2003). In maternal–zygotic dicer mutant zebrafish, the early development of the nervous system is severely disrupted (Giraldez et al., 2005). However, little is known about the role of miRNA-mediated post-transcriptional gene regulation during the development of mammalian central nervous system (Kosik, 2006).
Here we show that the RNAase III enzyme Dicer plays an important role in mouse CNS development. Using the Cre-loxp system, we conditionally deleted Dicer expression in the CNS and in the cortex using two Cre lines, Emx1-Cre and Nestin-Cre, and found that neural progenitors undergo cell death and abnormal differentiation in the cortex and striatum of Dicer knockout mice. The different timings of complete Dicer deletion affect early-born and late-born cortical neurons in the Emx1-Cre and Nestin-Cre generated knockout lines, respectively. In the spinal cord, the development of motor neurons appears normal, which is likely caused by incomplete Dicer deletion at early embryonic stages, due to the weak activity of the Nestin-Cre line. However, in late embryonic stages, Dicer deletion causes reduced oligodendrocyte precursors and decreased differentiation of oligodendrocytes in the spinal cord. Our results demonstrate that Dicer function is required for the proper development of neurons and glia in the mouse central nervous system during early and late embryonic stages.
Results
Cortical defects in CNS-specific and cortical-specific Dicer conditional knockout mice
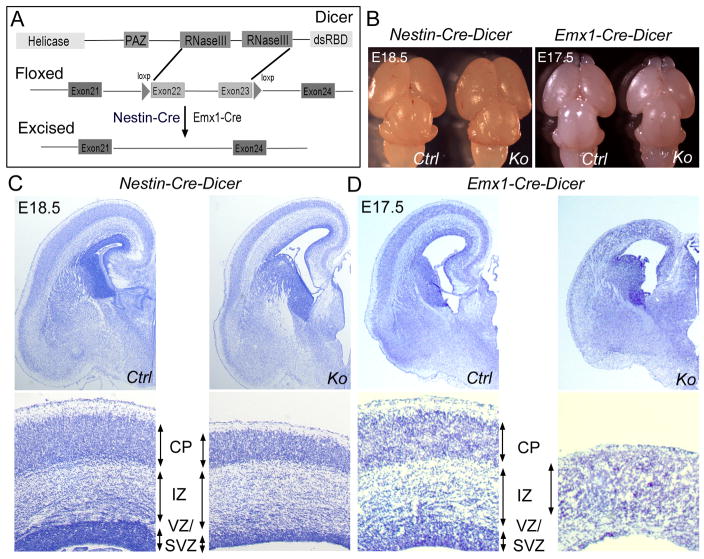
To examine whether Dicer function is required for the development of mouse central nervous system (CNS), we deleted Dicer expression in the CNS using a Cre-loxp system. Floxed Dicer mice (Dicerloxp/loxp) with two loxP sites flanking exon 22 and exon 23 for Dicer, which encode the RNAase III domains, were bred with a Nestin-Cre line and an Emx1-Cre line (Fig. 1A). The Nestin-Cre line will delete Dicer expression in the entire CNS, and the Emx1-Cre line will ablate Dicer only in the cerebral cortex (Tronche et al., 1999; Gorski et al., 2002). The breeding strategy and genotyping results are shown in Figure S1.
Figure 1.
Dicer function is required for normal brain development. (A) The targeting strategy for Dicer conditional deletion in the central nervous system. The N-terminal RNA helicase domain, PAZ domain, two Ribonuclease III domains and a double-stranded RNA-binding domain (RBD) are labeled. Nestin-Cre and Emx1-Cre were used to exercise two loxp sites in Dicer. (B) In E18.5 Nestin-Cre-Dicer conditional knockout (Ko) mice, the brain size was similar to that of controls (Ctrl). In E17.5 Emx1-Cre-Dicer mice, the cerebral cortex was significantly smaller than controls. (C) In coronal sections of E18.5 Nestin-Cre-Dicer cortices with Nissl staining, the ventricular zone (VZ), subventricular zone (SVZ) and the cortical plate (CP) were thinner than controls, while the intermediate zone (IZ) was thicker. (D) In coronal sections of E17.5 Emx1-Cre-Dicer cortices, the CP was slightly enlarged, but the VZ/SVZ and the IZ were greatly reduced. The hippocampus was not detectable in Emx1-Cre-Dicer cortices.
While the Nestin-Cre line produced very few Dicer conditional knockout embryos (Nestin-Cre-Dicer or Ko) (6.8% at embryonic day 18.5), Dicer knockout embryos from the Emx1-Cre line (Emx1-Cre-Dicer) were collected at Mendelian ratios (24.8%) (Table 1 and Table S1). Additionally, no surviving newborns of Nestin-Cre-Dicer mice were detected. On the other hand, Emx1-Cre-Dicer mice could survive until postnatal day 30 (P30).
Table 1.
Genotyping results of Nestin-Cre-Dicer (Nestin-Cre:Dicerflox/flox ) knockout mice.
| Stages | Nestin-Cre: Dicerflox/flox | Nestin-Cre: Dicerflox/+ | Dicerflox/flox, Dicerflox/+ | Total | |
|---|---|---|---|---|---|
| E9.5–E13.5 | Total number | 7 | 31 | 51 | 89 |
| Percentage (%) | 7.9 | 34.8 | 57.3 | 100 | |
| E15.5 | Total number | 6 | 46 | 57 | 109 |
| Percentage (%) | 5.5 | 42.2 | 52.3 | 100 | |
| E18.5 | Total number | 10 | 58 | 79 | 147 |
| Percentage (%) | 6.8 | 39.5 | 53.7 | 100 |
A very low percentage of Nestin-Cre-Dicer (Nestin-Cre:Dicerflox/flox) knockout mice was detected. No surviving newborns of Nestin-Cre-Dicer mice were recovered.
At embryonic day 18.5 (E18.5), the brain size of Nestin-Cre-Dicer mice was compatible with that of controls (wild type or heterozygous mice) (Fig. 1B). However, in coronal sections of E18.5 cortices with Nissl staining, the dorsal and lateral cortical wall in the Nestin-Cre-Dicer mice was thinner and the ventricles were larger than controls (Fig. 1C). In the Nestin-Cre-Dicer cortex, the ventricular zone (VZ) and the subventricular zone (SVZ), residing dividing cells, were reduced. The cortical plate (CP) that contains postmitotic neurons was thinner in the Ko than controls, but the intermediate zone (IZ) was expanded in E18.5 Nestin-Cre-Dicer mice (Fig. 1C).
Emx1-Cre-Dicer mice had noticeably smaller cerebral cortices even at E17.5 (Fig. 1B). The cortical wall in the Emx1-Cre-Dicer brain was significantly thinner than the control (Fig. 1D). The VZ and IZ were greatly reduced and the CP was slightly enlarged in the Ko cortices. In addition, the hippocampus was not detectable in the Emx1-Cre-Dicer brain, suggesting a dramatic defect of neurogenesis.
Our results indicate that blocking miRNA biogenesis by deleting Dicer expression in developing brains causes severe defects in neuronal production and results in a great reduction of the cortical wall and an increase of the ventricles.
Defects of early-born neurons in Emx1-Cre-Dicer cortices
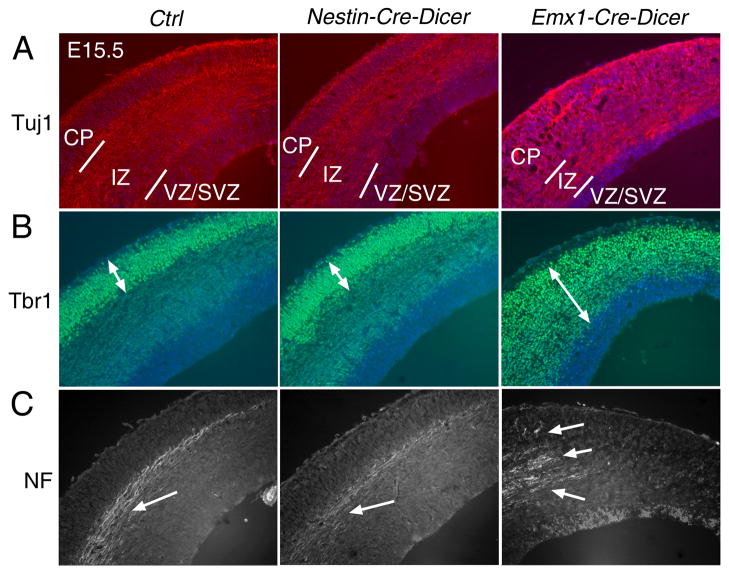
To reveal the underlying mechanisms that cause reduced cortical wall in Dicer Ko mice, we examined cortical neurogenesis. In E15.5 control cortices, Tuj1 was highly expressed in the IZ, Tbr1 was detected in early-born neurons in the CP, and the expression of Neurofilament (NF) was observed in axonal projections in the IZ (Fig. 2). Similar expression patterns of Tuj1, Tbr1 and NF were detected in E15.5 Nestin-Cre-Dicer cortices, suggesting that early-born neuron development appears normal (Fig. 2). In contrast, E15.5 Emx1-Cre-Dicer cortices had high Tuj1 and Tbr1 expression in both the IZ and CP. While, normal Tbr1+ cells were detected in E13.5 Emx1-Cre-Dicer cortices as in controls (Figs. S2A and B), by E15.5 the total number and percentage of Tbr1+ cells were significantly increased in Emx1-Cre-Dicer cortices without significant changes in cell density (Figs. S2C–E). Our data suggest an early differentiation of neural progenitors and abnormal neuronal migration in Emx1-Cre-Dicer cortices. Finally, instead of being expressed as a bundle of axons, NF was expressed as split axonal projections in Emx1-Cre-Dicer cortices (Fig. 2C). Our results indicate that early neurogenesis is not affected in Nestin-Cre-Dicer but is significantly altered in Emx1-Cre-Dicer cortices.
Figure 2.
Defects of early-born neurons in Emx1-Cre-Dicer but not in Nestin-Cre-Dicer cortices at E15.5. (A) Tuj1 expression was detected in the intermediate zone (IZ) in the control (Ctrl) and Nestin-Cre-Dicer cortices but in the IZ and cortical plate (CP) in Emx1-Cre-Dicer cortices. The ventricular zone (VZ) and the subventricular zone (SVZ) are labeled. (B) Early-born neurons, labeled with Tbr1, were detected in the CP in the control and Nestin-Cre-Dicer cortices but in the IZ and CP in Emx1-Cre-Dicer cortices. (C) Neurofilament (NF) expression appeared in a bundle of axons (arrow) in the IZ in the control and Nestin-Cre-Dicer cortices but in split axons (arrows) in Emx1-Cre-Dicer cortices.
Defects of late-born neurons in Nestin-Cre-Dicer cortices
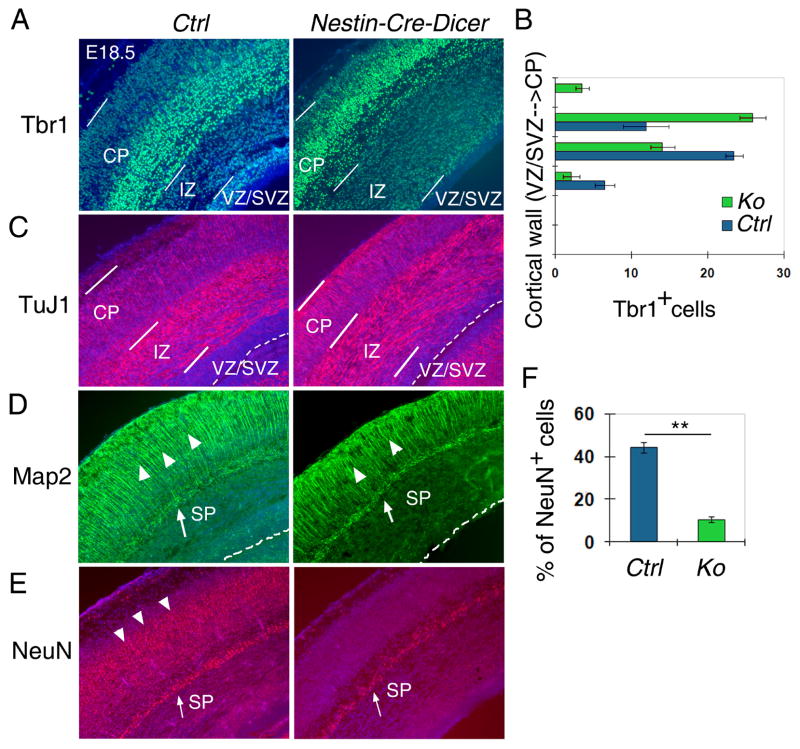
In Nestin-Cre-Dicer cortices, while the production of early-born neurons was not affected, late-born neurons displayed defects. At E18.5, while Tbr1+ cells in control cortices were concentrated in the deeper layers of the CP, Tbr1+ cells in Nestin-Cre-Dicer cortices were expressed throughout the entire CP (Figs. 3A and B). More Tuj1+ cells were detected in the IZ in Nestin-Cre-Dicer cortices than controls, indicating a migration defect of postmitotic neurons (Fig. 3C). Additionally Map2 expressing neurons in the CP were greatly reduced in Nestin-Cre-Dicer cortices (Fig. 3D). These observations are consistent with the thinner CP but thicker IZ in Nestin-Cre-Dicer cortices, as detected by Nissl staining and Tbr1 expression (Fig. 1C and 3A).
Figure 3.
Defects of late-born neurons in Nestin-Cre-Dicer cortices at E18.5. (A) While most Tbr1 positive cells were detected in the deeper layers in the cortical plate (CP) in controls (Ctrl), Tbr1 positive cells were observed in the entire CP in Nestin-Cre-Dicer (Ko) cortices. While the ventricular zone (VZ) and the subventricular zone (SVZ) were reduced, the intermediate zone (IZ) was enlarged in Nestin-Cre-Dicer cortices. (B) Quantification of the distribution of Tbr1+ cells from the VZ/SVZ to the CP in the cortical wall. Fields: n=6. (C) More Tuj1+ cells were detected in the IZ in Nestin-Cre-Dicer cortices. The CP in Dicer Ko cortices was thinner than controls. (D) Fewer Map2+ neurons were detected in the CP in Nestin-Cre-Dicer cortices than controls (arrowheads). Map2 expression in the subplate (SP) (arrow) was not changed. (E and F) Fewer mature neurons, labeled with NeuN, were detected in the CP in Nestin-Cre-Dicer cortices than controls (arrowheads). NeuN expression in the SP (arrow) was not changed. Fields: n=6, **: p<0.0002. At least 3 Nestin-Cre-Dicer and 3 control animals were used for all statistical analyses.
In E18.5 control cortices, the mature neuron marker NeuN was highly expressed in the subplate (SP) and the cortical plate. And while NeuN+ cells were detected in the SP of Nestin-Cre-Dicer cortices, there was a significantly decreased expression of NeuN+ cells in the CP (Figs. 3E and F). The decrease of NeuN+ cells is likely caused by delayed neurogenesis but not cell death of postmitotic neurons, since no significant apoptosis was detected in the CP of E15.5 and E18.5 Nestin-Cre-Dicer cortices (Figs. S3A and S4A). A great reduction of NeuN+ cells was also observed in P0 Emx1-Cre-Dicer cortices (Fig. S2F). The decrease of NeuN+ cells is likely caused by defects in differentiation and progressive cell death of postmitotic neurons, as detected by TUNEL assays in the CP of Emx1-Cre-Dicer cortices (Fig. S5). Our data suggests that Dicer deletion using the Nestin-Cre line causes abnormal development of late-born neurons, and Dicer deletion using two Cre lines affects maturation of postmitotic neurons.
In summary, our results indicate that Dicer cortical deletion in the two Cre lines causes distinct defects: early differentiation of neural progenitors and ectopic early-born neurons are detected in Emx1-Cre-Dicer cortices, and a reduced number of late-born neurons and abnormal migration are observed in Nestin-Cre-Dicer cortices.
Dicer cortical deletion results in abnormal neuronal migration
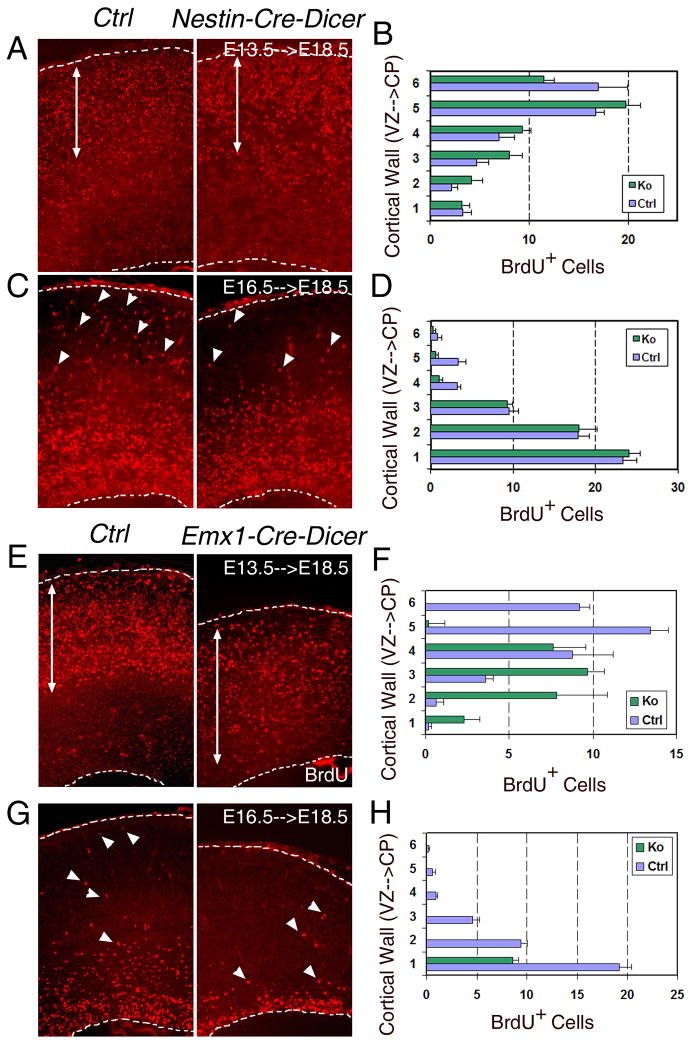
To monitor the migration of cortical cells in the CP, we conducted a Bromodeoxyuridine (BrdU) birthdating assay by injecting BrdU at E13.5 or E16.5 and collecting brains at E18.5. The BrdU injection at E13.5 labels early born neurons that migrate to the deeper layers of the CP by E18.5. In Nestin-Cre-Dicer cortices, BrdU+ cells were detected only in the CP with similar numbers as in controls, suggesting a normal production and migration of early-born neurons (Figs. 4A and B). However, in Emx1-Cre-Dicer cortices, BrdU+ cells were distributed throughout the entire cortical wall, suggesting abnormal migration of early-born neurons (Figs. 4E and F).
Figure 4.
Dicer deletion affects cortical neuronal migration. (A and B) In E18.5 Nestin-Cre-Dicer (Ko) cortices, a majority of cells labeled by BrdU injected at E13.5 migrated to the CP as in controls (Ctrl). (C and D) While many cells (arrowheads) labeled by BrdU injected at E16.5 migrated to the CP in controls, fewer BrdU+ cells (arrowheads) migrated into the upper layers in E18.5 Nestin-Cre-Dicer cortices. (E and F) While a majority of cells labeled by BrdU injected at E13.5 migrated to the CP in controls, BrdU+ cells were detected in the entire cortical wall in E18.5 Emx1-Cre-Dicer (Ko) cortices. (G and H) While many cells (arrowheads) labeled by BrdU injected at E16.5 migrated to the CP in the controls, only very few BrdU+ cells (arrowheads) migrated away from the ventricular zone (VZ) and the subventricular zone (SVZ) in E18.5 Emx1-Cre-Dicer cortices.
The BrdU injection at E16.5 labels late-born neurons that migrate toward the upper layers of the CP by E18.5. While the control cortices showed BrdU+ cells migrating to the upper layers of the CP, only a few scattered BrdU+ cells migrated to the upper layers of the CP in Nestin-Cre-Dicer cortices (Figs. 4C and D). Similarly, in Emx1-Cre-Dicer cortices, only a few BrdU+ cells migrated away from the VZ/SVZ (Figs. 4G and H). In the Nestin-Cre line, the numbers of BrdU+ cells were not significantly reduced in the Ko cortices compared to the control cortices. In contrast, the number of BrdU+ cells in Emx1-Cre-Dicer cortices was significantly lower than the control cortices. This suggests that Dicer deletion in the Nestin-Cre line primarily affects migration of late-born neurons whereas Dicer deletion in the Emx1-Cre line causes both reduced neuronal production and abnormal migration by E18.5.
Delayed defects of cortical progenitor development in Nestin-Cre-Dicer cortices
To further understand the cause of altered neuronal production and delayed migration in Dicer Ko mice, we examined the development of cortical progenitors. Our studies and others found that at E14.5, Emx1-Cre-Dicer cortices displayed severe cell death and reduced progenitors (De Pietri Tonelli et al., 2008) (data not shown). Although no obvious defects in neural progenitors, which is demonstrated by a TUNEL assay and with progenitor markers for Pax6 and Tbr2, were observed in Nestin-Cre-Dicer cortices at E15.5 (Fig. S3), by E18.5 a large amount of cell death was detected, particularly in the VZ and SVZ (Figs. S4A and B).
To further examine the cell cycle progression of Dicer deficient neural progenitors in Nestin-Cre-Dicer mice, we calculated the progenitor labeling index (LI) with a 30-minute pulse of BrdU. While BrdU labels cells in the S-phase, Ki-67 labels cells in the G1, S, G2 and M-phase. The LI is measured by calculating the proportion of BrdU+/Ki-67+ versus Ki-67+ progenitors, and a higher LI suggests a shorter cell cycle length (Chenn and Walsh, 2002; Kee et al., 2002). As shown, the cell cycle length of progenitors as detected by the LI was not affected in Nestin-Cre-Dicer cortices (Figs. S3C and D). Transcription factor Tbr2, which is highly expressed in intermediate progenitors in the SVZ (Englund et al., 2005; Sessa et al., 2008), was also labeled in Nestin-Cre-Dicer cortices, and the number of Tbr2- and BrdU-positive progenitors was not significantly reduced (Figs. S4E and F).
Therefore, while Emx1-Cre-Dicer cortices had severe defects in progenitor development as early as E14.5 (De Pietri Tonelli et al., 2008), Nestin-Cre-Dicer cortices displayed normal progenitor development at E15.5 and later showed significant progenitor cell death at E18.5.
Mild defects of interneuron development in Nestin-Cre-Dicer brains
Because interneurons are generated in the striatum of the subcortical region and then migrate to the cerebral cortex (Corbin et al., 2001; Wonders and Anderson, 2006), we examined whether the thinner cortices in Nestin-Cre-Dicer mice were associated with abnormal interneuron production. At E15.5, no cell death and progenitor defects were detected in Dicer Ko striatum (data not shown). But by E18.5, many dead cells were observed in the VZ in the striatum of Nestin-Cre-Dicer as opposed to none in the controls (Fig. S6A). There were more progenitors, labeled with Ki-67, BrdU and PH3, migrated away from the VZ in Dicer Ko than control striatum (arrows in Fig. S6A). Our results indicate that there is a late defect of survival and migration of interneuron progenitors in Nestin-Cre-Dicer striatum.
Transcription factors Lhx6 and Lhx7 are first expressed in interneuron progenitors and later in migrating interneurons. Lhx6- and Lhx7-expressing cells were readily detected in E15.5 and E18.5 Nestin-Cre-Dicer cortices (Fig. S6B and data not shown). Moreover, migrating interneurons, labeled by Gad67 and Lhx6, were also detected in Dicer Ko cortices at E18.5 (Fig. S6B). Thus, despite the higher incidence of cell death in the striatum, interneuron production and migration are not significantly affected by Dicer deletion in the embryonic brain using the Nestin-Cre line at E18.5.
Delayed Dicer deletion in Nestin-Cre-Dicer brains
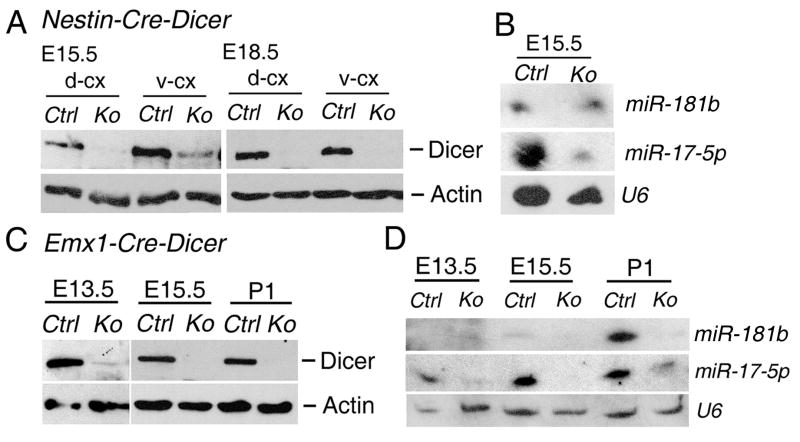
To test whether the distinct cortical defects are due to the activity of the two different Cre lines (Emx1-Cre and Nestin-Cre), we examined expression levels of Dicer proteins using the Western blotting assay. At E13.5 and E15.5, weak Dicer expression was observed in the dorsal cortical region (cerebral cortex) and low Dicer proteins still remained in the ventral cortical region (striatum) in Nestin-Cre-Dicer brains (Fig. 5A and data not shown). At E18.5 when defects became obvious in Nestin-Cre-Dicer cortices, Dicer proteins were completely abolished in the dorsal and ventral cortical regions (Fig. 5A). On the contrary, in Emx1-Cre-Dicer mice, very low Dicer expression was detected in the dorsal cortical region at E13.5, and Dicer proteins were completely abolished in the cerebral cortices at E15.5 and P1 (Fig. 5C). Thus, the weaker Cre activity in Nestin-Cre-Dicer mice, compared to Emx1-Cre-Dicer mice, allows a delayed ablation of Dicer proteins in the cortex.
Figure 5.
Incomplete Dicer deletion in developing brains of Nestin-Cre-Dicer mice. (A) At E15.5, very low Dicer proteins were detectable in the dorsal cortical region (d-cx) in Nestin-Cre-Dicer knockout (Ko) mice compared to controls (Ctrl), as detected by Western blotting analyses. Actin served as a loading control. Dicer proteins were present in the ventral cortical region (v-cx) in Dicer Ko mice. At E18.5, Dicer expression was abolished in the dorsal and ventral cortical regions in Nestin-Cre-Dicer mice. (B) At E15.5, miRNA biogenesis was not completely blocked in Nestin-Cre-Dicer cortices, as detected by miR-181b and miR-17-5p expression using Northern blotting analyses. The small nuclear RNA (snRNA) U6 which is ubiquitously expressed was used as a loading control. (C) In Emx1-Cre-Dicer (Ko) mice, very low Dicer protein was detectable in the cortex at E13.5 and Dicer proteins were absent in cortices at E15.5 and P1. (D) MiRNA expression, such as miR-17-5p and miR-181b, was undetectable in cortices of E13.5, E15.5 and P1 Emx1-Cre-Dicer mice.
As a consequence, mature miRNAs, such as miR-181b and miR-17-5p, were still detected in E15.5 cortex in Nestin-Cre-Dicer mice whereas miRNA biogenesis was greatly blocked in cortices of Emx1-Cre-Dicer mice from E13.5 to P1 (Figs. 5B and D). Thus, the remaining miRNAs produced by incomplete Dicer ablation in the cortex of Nestin-Cre-Dicer mice may have contributed to the normal development of early-born neurons and delayed defects of neural progenitors, compared to Emx1-Cre-Dicer mice.
Normal development of motor neurons in the spinal cord of Nestin-Cre-Dicer mice
A few surviving embryos of Nestin-Cre-Dicer mice were recovered, but no Nestin-Cre-Dicer newborns survived (Table 1). This may have been caused by impaired development of motor neurons, which is essential for movement control. Thus, we examined the development of the spinal cord in Nestin-Cre-Dicer mice. Motor neuron progenitors in the ventral spinal cord are generated by E10.5 and specified into distinct motor neuron subtypes later on (Jessell, 2000; Lee and Pfaff, 2001).
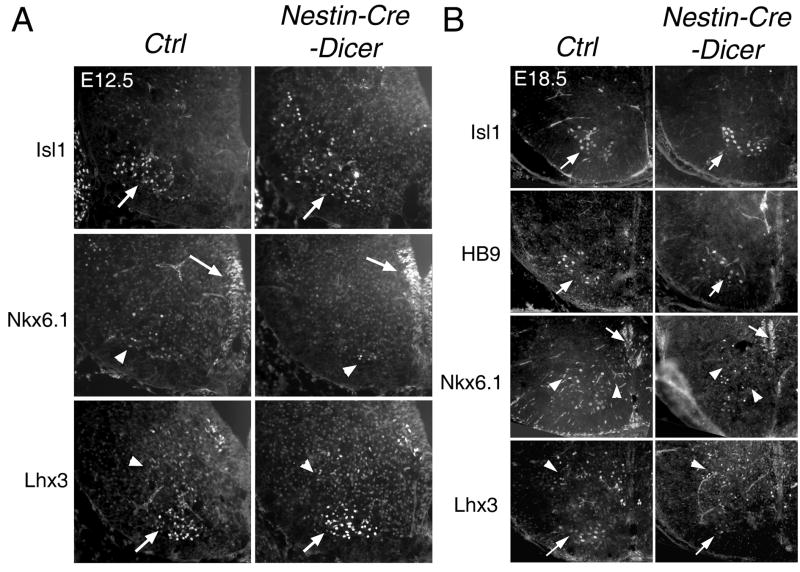
Motor neuron development appeared normal in E12.5 and E15.5 Nestin-Cre-Dicer spinal cord, as detected by motor neuron markers (Fig. 6A and data not shown). Notably, the distribution of Isl1+ cells was more scattered, and the number of Nkx6.1+ motor neurons was slightly reduced in E12.5 Nestin-Cre-Dicer spinal cords than controls. At E18.5, the expression patterns of motor neuron markers, Isl1, HB9 and Nkx6.1, were indistinguishable between the Nestin-Cre-Dicer and control spinal cords (Fig. 6B). And the expression of Lhx3, which labels a population of interneurons and motor neurons, did not show an obvious difference in Dicer Ko and control spinal cords. Therefore, the overall development of motor neurons appears unaffected in the Nestin-Cre-Dicer spinal cord.
Figure 6.
Motor neuron development is not significantly affected in by Dicer deletion in the Nestin-Cre-Dicer spinal cord. (A, B) The numbers and expression pattern of motor neurons (arrows), labeled with Isl1 or HB9, did not show obvious difference in E12.5 and E18.5 Nestin-Cre-Dicer spinal cord. Nkx6.1-expressing motor neurons (arrowheads) and progenitors (arrows), and Lhx3-exressing motor neurons (arrows) and interneurons (arrowheads) appeared normal in Nestin-Cre-Dicer spinal cord.
Reduced oligodendrocytes in the Nestin-Cre-Dicer spinal cord
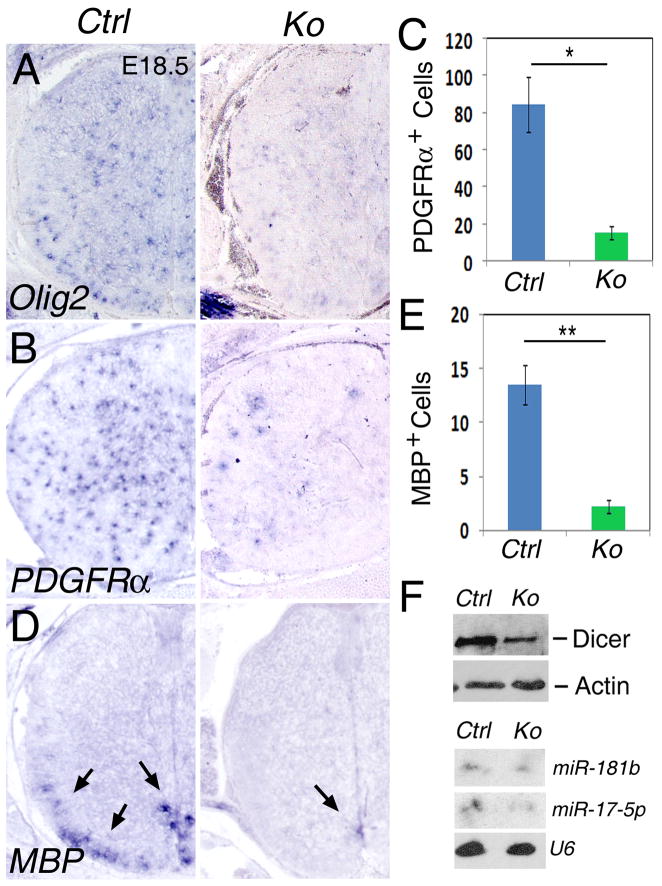
Oligodendrocyte precursors are derived from the ventral spinal cord by E12.5, later than motor neuron progenitors (Richardson et al., 2000). We examined oligodendrocyte development using markers for precursors and mature oligodendrocytes in Nestin-Cre-Dicer mice. As shown, fewer Olig2-expressing oligodendrocyte precursors were detected in E18.5 Dicer Ko spinal cord than controls (Fig. 7A). Platelet derived growth factor and its receptor alpha (PDGFRα) play a critical role in oligodendrocyte development (Fruttiger et al., 1999). PDGFRα+ cells were greatly reduced and mainly concentrated in the ventral region of the Nestin-Cre-Dicer spinal cord, compared to scattered PDGFRα+ cells throughout the entire spinal cord in controls (Figs. 7B and C). No significant dead cells were detected in the Nestin-Cre-Dicer spinal cord, indicating that the reduction of oligodendrocyte precursors is not caused by cell death (data not shown). Myelin basic protein (MBP), which is expressed in differentiated oligodendrocytes, was greatly reduced in the ventral spinal cord of Nestin-Cre-Dicer mice, suggesting a defect in oligodendrocyte differentiation caused by Dicer deletion (Figs. 7D and E).
Figure 7.
Dicer function is required for the proper development of oligodendrocytes. (A and B) Fewer oligodendrocyte precursors, labeled by Olig2 and PDGFRα, were detected in E18.5 Nestin-Cre-Dicer knockout (Ko) spinal cords compared to controls (Ctrl). The expression levels of Olig2 and PDGFRα were also reduced in Dicer Ko spinal cord. (C) Quantification of PDGFRα-expressing oligodendrocyte precursors. n=5, *: p<0.05. (D and E) Differentiated oligodendrocytes, labeled with myelin basic protein (MBP), were significantly decreased in Nestin-Cre-Dicer spinal cords. n=7, **: p<0.001. (F) At E15.5, lower levels of Dicer proteins were detectable in the spinal cord of Dicer Ko mice compared to controls, as detected by Western blotting analyses. Actin served as a loading control. MiRNA expression, such as miR-181b and miR-17-5p, was not completely blocked in Dicer Ko spinal cord, as detected using Northern blotting analyses. The small nuclear RNA (snRNA) U6 which is ubiquitously expressed was used as a loading control. At least 3 Nestin-Cre-Dicer Ko and 3 control animals were used for all statistical analyses.
Consistent with Dicer deletion in the cortex, Dicer proteins were detectable in E15.5 Nestin-Cre-Dicer spinal cord with reduced levels compared to controls (Fig. 7F). Reduced mature miRNAs, such as miR-181b, were still detected in E15.5 spinal cord in Nestin-Cre-Dicer mice (Fig. 7F). As a result, while the delayed Dicer ablation in the Nestin-Cre-Dicer spinal cord may have contributed to normal motor neuron development, the reduced levels of Dicer and mature miRNAs have affected normal oligodendrocyte development later on.
Discussion
The proper development of the central nervous system requires precise gene regulation. The RNAase III enzyme Dicer mediated miRNA pathway represents a novel machinery of post-transcriptional gene regulation during development (Kloosterman and Plasterk, 2006; Stefani and Slack, 2008). To test the importance of Dicer in the development of the CNS, we conditionally deleted Dicer expression, in turn blocking miRNA biogenesis, using both the Nestin-Cre line (CNS-specific) and Emx1-Cre line (cortical specific). Here we show that the lack of miRNA function affects survival and differentiation of cortical neural progenitors, and results in thinner cortical walls and larger ventricles. The progressive ablation of Dicer by Nestin-Cre restores the development of early-born neurons but not late-born neurons in the cortex. In Nestin-Cre-Dicer mice, although the development of interneurons in the cortex, and motor neurons in the spinal cord was not severely affected during the early neurogenesis stage, oligodendrocytes in the spinal cord were greatly reduced during the late gliogenesis stage. In conclusion, our results identify the essential role of Dicer in the proper development of the central nervous system.
Distinct cortical defects in Nestin-Cre-Dicer and Emx1-Cre-Dicer mice
Previous work has shown that Dicer null mice die at E7.5 (Bernstein et al., 2003) and in maternal–zygotic dicer mutant zebrafish, the development of the nervous system was severely disrupted (Giraldez et al., 2005). In our studies, Emx1-Cre-Dicer embryos and newborns display normal numbers as expected Mendelian ratios (Table S1), and Emx1-Cre-Dicer mice die after P30 due to severe cortical defects. Low numbers of surviving embryos and no surviving newborns of Nestin-Cre-Dicer mice (Table 1) are caused by abnormal development in the CNS, as demonstrated by the late defects in the cortex and spinal cord. In addition, although the Nestin-Cre is mainly active in the CNS, the early lethality of Nestin-Cre-Dicer mice may have also been caused in part by unexpected Cre excision from non-CNS regions, leading to weak activity in scattered cells in the kidney and heart (Tronche et al., 1999).
Dicer expression is deleted in the cerebral cortex by the Nestin-Cre and Emx1-Cre lines, but Nestin-Cre-Dicer and Emx1-Cre-Dicer mice display distinct cortical defects. In the Emx1-Cre-Dicer cortices, we and others found that neural progenitors display cell death even at E12.5 (De Pietri Tonelli et al., 2008). The neural progenitors are greatly reduced due to cell death. We also found over-production of early-born neurons such as Tbr1 and Tuj1, which is likely caused by early differentiation, suggesting that the Dicer-mediated miRNA pathway is essential for proper neuronal differentiation (Fig. 2). In Nestin-Cre-Dicer cortices, neural progenitors only display cell death after E17.5 and E18.5 (Figs. S3 and S4). While early-born neuron production is normal, the development of late-born neurons and their migration are affected (Figs. 3 and 4). The late defects of neural progenitor and postmitotic neuron development in Nestin-Cre-Dicer cortices further support the importance of Dicer function in maintaining cortical development.
Furthermore, the maturation defects of cortical neurons in the cortical plate, as detected by reduced NeuN+ cells, are observed in both Nestin-Cre-Dicer and Emx1-Cre-Dicer cortices. While differentiation defects contributed to decreased NeuN+ cells in both mouse lines, apoptosis in postmitotic neurons of Emx1-Cre-Dicer cortices leads to more severe cortical defects, such as a noticeably thinner cortical wall (Fig. S5). Our results are consistent with the severe cell death in differentiated neurons and smaller cortex of the Dicer knockout mice generated by a CaMKII-Cre line, which is active in postmitotic neurons in the cortex and hippocampus (Davis et al., 2008).
Proper Dicer expression maintains the neural progenitor pool
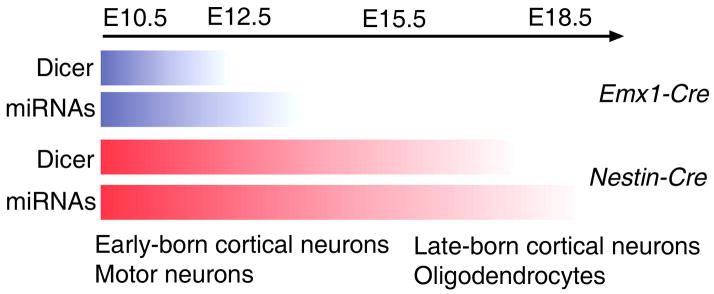
What may cause the distinct cortical phenotypes in Dicer Ko mice generated using two Cre lines? The activity of both Cre lines commences at a similar time by E10.5-E11, suggesting that the timing of Cre activity initiation does not affect distinct cortical defects (Tronche et al., 1999; Gorski et al., 2002). We found that while Dicer proteins are abolished in the dorsal cortical region at E15.5 in Emx1-Cre-Dicer mice, weak Dicer proteins are still detectable in E13.5 and E15.5 Nestin-Cre-Dicer cortices (Fig. 5). The delayed Dicer deletion may allow a low level of Dicer proteins to continue to process miRNAs (Harfe et al., 2005) and the processed miRNAs will continue to function in cortical progenitors and postmitotic cells for some time before being completely degraded. These remaining miRNAs likely maintain normal early CNS development in Nestin-Cre-Dicer cortices until E18.5 when miRNAs are completely abolished (Fig. 8). Therefore, the different activities of the two Cre lines in ablating Dicer expression contribute to distinct defects in Nestin-Cre-Dicer and Emx1-Cre-Dicer cortices.
Figure 8.
A model of the timing effect of Dicer deletion on neurogenesis and gliogenesis in the developing mouse central nervous system. Early Dicer deletion (by E12.5) using the Emx1-Cre line causes ectopic production of early-born neurons in the cortex. Delayed Dicer deletion (by E15.5) using the Nestin-Cre line affects development of late-born cortical neurons and oligodendrocytes in the spinal cord but not early-born cortical neurons and motor neurons.
Our results are largely consistent with previously reported work on Dicer deletion using the Emx1-Cre line (De Pietri Tonelli et al., 2008), but in addition, we have made two distinct discoveries. First, we found an increase of early born neurons, marked by Tbr1+ and Tuj1+ cells, as early as E15.5 (Fig. 2). The increase of early-born neurons is not caused by changes of cell density in the cortical wall, suggesting a role of Dicer in cortical neuronal differentiation (Fig. S2). Second, while we similarly found that the development of neural progenitors appears normal in E13.5 Emx1-Cre-Dicer cortices despite cell death, our causal interpretation is different. De Pietri Tonelli et al concluded that normal early development of neural progenitors in E13.5 Emx1-Cre-Dicer cortices indicates that miRNAs are not essential for progenitor expansion (De Pietri Tonelli et al., 2008). However, in our view, normal progenitor development is likely carried out by miRNAs processed prior to Dicer ablation (earlier than E10.5). Moreover, we found that Dicer-deficient neural stem cells collected from E12.5 Emx1-Cre-Dicer cortices form very few neurospheres compared to control neural stem cells, suggesting a critical function of Dicer-mediated miRNA pathway in expansion of neural stem cells/progenitors (Kawase-Koga and Sun, unpublished data). In addition, the incomplete Dicer deletion and normal development of early neural progenitors in Nestin-Cre-Dicer cortices further support the importance of Dicer in maintaining the neural progenitor pool (Fig. S3 and Fig. 5). Thus, our studies indicate that Dicer functions are required for both early and late neural progenitor development.
Cell death of progenitors and postmitotic neurons is observed in several mouse models in which Dicer is deleted by various Cre lines (Schaefer et al., 2007; Cuellar et al., 2008; Davis et al., 2008; De Pietri Tonelli et al., 2008). Our preliminary studies using proteomic approaches have revealed that Dicer deletion causes increased activities of pro-apoptosis pathways (Kawase-Koga and Sun, unpublished data). Thus, continuous Dicer activity and miRNA biogenesis may play a central role in the survival of multiple cell types by protecting cells from apoptosis and subsequently promoting the proper development of progenitors and postmitotic neurons (Schaefer et al., 2007; Cuellar et al., 2008; Davis et al., 2008; De Pietri Tonelli et al., 2008).
Dicer function is essential for neurogenesis and gliogenesis in the CNS
In E18.5 Nestin-Cre-Dicer cortices, interneuron progenitors also display cell death and abnormal migration, suggesting Dicer’s role in interneuron progenitor development (Fig. S6A). We found normal distribution of migratory interneurons in Nestin-Cre-Dicer embryos, which is likely a result of delayed Dicer protein ablation in interneuron progenitors in the ventral cortex (Fig. 5). Additionally we did not find obvious defects in cerebellum development in Nestin-Cre-Dicer mice during embryonic stages (data not shown). The postnatal lethality of Nestin-Cre-Dicer mice has limited our further examination of Dicer function in interneuron migration and cerebellum development. However, recent reports have shown that deleting Dicer and miRNA biogenesis in postmitotic neurons in the postnatal cerebral cortex and cerebellum causes cell death and neurodegeneration (Schaefer et al., 2007; Davis et al., 2008). Therefore, Dicer function is essential for the development of the cerebellum and cerebral cortex at postnatal stages.
The initiation of motor neuron development in the spinal cord and sensory neuron formation in the hindbrain commences very early during development (by E10.5) (Jessell, 2000; Qian et al., 2001) and the earliest activity of Nestin-Cre is detected by E11 (Tronche et al., 1999). Thus, at E11, some miRNAs may have already been processed in neural stem cells or progenitors in the CNS before Dicer deletion is activated. Moreover, the delayed Dicer protein deletion also allows continuous miRNA processing and function, which may serve as an explanation for the normal sensory neuron formation in the hindbrain (data not shown) and the development of motor neurons in the spinal cord in Nestin-Cre-Dicer mice (Fig. 6). Thus, the normal formation of sensory neurons and motor neurons in Nestin-Cre-Dicer mice does not exclude a role of Dicer and miRNAs in the proper development of the hindbrain and spinal cord (Fig. 8).
Oligodendrocyte development in the spinal cord begins after motor neuron formation and the number of oligodendrocyte precursors expands during late embryonic stages (Richardson et al., 2000). At E18.5, when Dicer proteins are greatly reduced by Nestin-Cre excisions, we detected reduced oligodendrocyte precursors in the spinal cord of Nestin-Cre-Dicer mice (Fig. 7). And since no obvious dead cells were detected in the Nestin-Cre-Dicer spinal cord, oligodendrocyte precursor defect is likely caused by a decreased expansion of the oligodendrocyte precursor pool. Moreover, the differentiation of mature oligodendrocytes is also decreased in Nestin-Cre-Dicer mice. Our results further demonstrate that the different timings of Dicer deletion affect both neurogenesis and gliogenesis (Fig. 8). Recent work has identified more than 43 miRNAs that are essential for oligodendrocyte differentiation (Lau et al., 2008). Thus, our studies and others demonstrate a critical role of Dicer and miRNAs in glial cell development in the spinal cord.
MiRNAs are essential for normal development (Hobert, 2005; Kosik, 2006). Our results and other previous work indicate that the proper Dicer expression and miRNA biogenesis are required for survival of neural progenitors and as well for differentiation and maturation of postmitotic neurons in the developing cortices. We further show that Dicer plays an important role in the development of neurons and glia in the striatum and the spinal cord. Our research demonstrates the global effects of blocking miRNAs biogenesis on CNS development but can’t address which specific miRNAs are involved in neurogenesis and/or gliogenesis. Revealing functions of individual miRNAs critical for the development of neuronal and glial progenitors and differentiation of distinct cell types will help us better understand the complex gene regulation during the central nervous system development in future work.
Experimental Procedures
Generation of Dicer conditional knockout mice
The floxed Dicer transgenic mice (Dicerflox/flox) (C57/BL6 × 129 background) (kindly provided by the Greg Hannon’s lab at the Cold Spring Harbor Laboratory) (Murchison et al., 2005) were bred with Nestin-Cre (Stock Number: 003771, C57/BL6 background) and Emx1-Cre mice (Stock Number: 005628, C57/BL6 background) (The Jackson Laboratory) to generate Nestin-Cre-Dicer (Nestin-Cre:Dicerflox/flox) and Emx1-Cre-Dicer (Emx1-Cre:Dicerflox/flox) animals (Fig. S1). The activities of the Nestin-Cre and Emx1-Cre were tested by breeding Nestin-Cre and Emx1-Cre mice with the Rosa26-LacZ reporter line (The Jackson Laboratory).
For staging of embryos, midday ofthe day of vaginal plug formation is considered embryonicday 0.5 (E0.5), and the first 24 hours after birth is defined as postnatal day 0 (P0). Animal use was overseen by the Animal Facility at the Weill Cornell Medical College.
Genotyping of Dicer conditional knockout mice
Mouse tail tip biopsies were used for genotyping by PCR reactions using the following primer pairs: for Cre, 5′– TAAAGATATCTCACGTACTGACGGTG-3′ and 5′-TCTCTGACCAGAGTCATCCTTAGC-3′ (product size: 350 bp); for Dicer, 5′ ATTGTTACCAGCGCTTAGAATTCC-3′ and 5′- GTACGTCTACAATTGTCTATG- 3′ (product sizes: 767 bp from Dicerflox allele and 560 bp from the wild type Dicer gene).
Tissue preparation and immunohistochemistry
Mouse tissues from the central nervous system were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) over night, incubated in 25–30% Sucrose in PBS, embedded in OCT and stored at −80°C until use. Cortices and the spinal cords were sectioned(10 μm) using a cryostat. For antigen recovery, sections were incubated in heated (95–100°C ) antigen recovery solution (1 mM EDTA, 5 mM Tris, pH 8.0) for 15–20 minutes, and cooled down for 20–30 minutes. Before applying antibodies, sections were blocked in 10% normal goat serum (NGS) in PBS with 0.1% Tween-20 (PBT) for 1 hour. Sections were incubated with primary antibodies at 4°C overnight and visualized using goat anti-rabbit IgG Alexa 488 and/or goat anti-mouse IgG Alexa 594 (1:350, Molecular Probes) for 1 hour at room temperature. Images were captured usinga Leica digital camera under a fluorescent microscope (Leica DMI6000B).
Primary antibodies against the following antigens were used: Phospho-Histone H3 (PH3) (1:1,000, Upstate), Bromodeoxyuridine (BrdU) (1:100, Molecular Probes), Ki-67 (1:500, Abcam), Pax6 (1:30, DSHB), Tbr1 (1:2,500) and Tbr2 (1:2,000, kindly provided by Dr. R. Hevner, University of Washington, Washington), β-tubulin III (TuJ1) (1:600, Chemicon), Neurofilament (NF) (1:20, DSHB), NeuN (1:300, Chemicon), Map2 (1:750, Chemicon), Isl-1 (1:20, DSHB), HB9 (1:20, DSHB), Nkx6.1 (1:20, DSHB) and Lhx3 (1:20, DSHB).
TUNEL Assay
To identify apoptotic cells in the cortex, we performed a TUNEL assay using an Apop Tag Fluorescein in situ Apoptosis detection kit (Chemicon) on 10 μm frozen sections. This assay was performed according to the manufacturer’s instructions.
Cell cycle labeling index (LI)
Timed-pregnant female mice were injected intraperitoneally (i.p.) with a single pulse 5-bromodeoxyuridine (BrdU, 50 μg/g body weight) 30 minutes before collecting tissues at E13.5, E15.5 and E18.5, and followed with BrdU and Ki-67 antibody staining. Briefly, sections (10 μm) were incubated with 6N HCl for 15 minutes at room temperature to unmask the antigen, followed by three washes with PBS, incubated with blocking solution (10% NGS in PBT) for 1 hour at room temperature and subsequently incubated with anti-BrdU and anti-Ki-67 antibodies in the blocking solution at 4°C overnight. The LI was calculated as ratios of BrdU+ and Ki-67+ cells versus total Ki-67+ cells per field in the control and Dicer knockout (Ko) cortices.
BrdU birthdating assay
Time-pregnant females were injected intraperitoneally with a single pulse BrdU (50 μg/g body weight) at E13.5 and E16.5. Brain tissues were collected at E18.5 and followed with BrdU antibody staining.
Nissl Staining
Sections (10 μm) were processed through incubation in the following solutions in order: ethanol/chloroform (1:1, overnight), 100% ethanol (30 sec), 95% ethanol (30 sec), distilled water (30 sec, twice), cresyl violet (3–5 min), distilled water (2 min, three times), 50% ethanol (2 min), 95% ethanol (5–30 min), 100% ethanol (5 min, twice), xylene (3 min, twice), and then mounted with a cover slip.
In situ Hybridization
Digoxygenin (DIG)-labeled sense and antisense mRNA probes were produced by in vitro transcription. The in situ hybridization on sections was performed as described (Sun et al., 2000). Briefly, the sections were hybridized at 65°C overnight and washed. After blocking for 2 hrs, sections were labeled with anti-DIG antibody (1:1,500, Roche) at 4°C overnight and washed, stained with BM purple (Roche) at room temperature until ideal intensity. The images of in situ hybridization were collected using a Leica digital camera under a dissection scope (Leica, MZ16F).
Cell counting in the cortical wall and the spinal cord
Coronal sections were collected in the medial cortical region (at levels between the anterior commissure and the anterior hippocampus). At least four sections from each brain and three brains from different litters were chosen for antibody labeling and TUNEL assay. Positive cells were counted in each bin in a field (totally 6 bins from the ventricular surface to the pial surface) using the method described previously (Takahashi et al., 1993). Positive cells and percentage of positive cells are presented as in a field in the cortical wall.
For the spinal cord, cross sections from the thoracic region were selected. PDGFRα+ and MBP+ cells were counted in half of the spinal cord.
Western blotting analysis
15 μg protein extracts from dorsal and ventral cortices and spinal cord of E13.5, E15.5, E18.5 and P1 mice were fractionated by SDS-PAGE, immunoblotted and probed with a rabbit anti-Dicer antibody (ab1416, 1:1,000; kindly provided by Drs. D. Livingston and C. Kanellopoulou, Dana-Farber Cancer Institute, Boston) (Kanellopoulou et al., 2005), and a rabbit anti-Actin antibody (1:200, Sigma), subsequently, with peroxidase-conjugated secondary antibodies (1:20,000; Sigma). The signals were detected with Supersignal West Pico Chemiluminescent Substrate (Pierce).
Northern blotting analysis
Total RNA was isolated from the dorsal and ventral cortical regions and the spinal cord of E13.5, E15.5 and P1 mice using the Trizol reagent (Invitrogen) according to manufacturer’s instructions. 5–10 μg total RNA was loaded onto 13% denatured polyacrylamide gels and separated at 65 mA for 1.5 h at room temperature, and transferred into nitrocellulose membrane overnight using a semi-dry transfer system. After cross-linking for 4 hours at 80°C, membranes were hybridized using locked nucleic acid (LNA) antisense probes specific for miR-181b, miR-17-5p and U6 (Exiqon). The probes were 3′-end labeled with digoxigenin (DIG)-ddUTP with terminal transferase using the DIG-3′-end labeling kit (Roche). After hybridization at 55°C overnight, membranes were washed 10 min in 2x SSC with 0.1% SDS for six times. The miRNA and U6 signals were detected using the CDP-star chemiluminescent substrate (Roche).
Statistic analysis
At least 3 Dicer knockout (Ko) and 3 control (Ctrl) animals were used for all statistical analyses. Data were shown as mean±SEM. Statistical comparison was made by analysis of variance (unpaired t test or ANOVAs).
Supplementary Material
Acknowledgments
We thank C. Lois, J. Meschter and J. Hong for critical reading of this manuscript. We thank G. Hannon for floxed Dicer mice, R. Hevner for anti-Tbr1 and Tbr2 antibodies, D. Livingston and C. Kanellopoulou for anti-Dicer antibodies and L. Selleri for Rosa26-LacZ mice. We are grateful for technique support from L. Shi, Z. Huang and S. Glickstein. This work was supportedby the Whitehall Foundation (T. S.), the Ellison Medical Foundation (T. S.), the Alice Bohmfalk Charitable Trust (T. S.) and a grant R01MH083680 from the NIH/NIMH (T. S.).
This work was supportedby the Whitehall Foundation (T. S.), the Ellison Medical Foundation (T. S.), the Alice Bohmfalk Charitable Trust (T. S.) and a grant R01MH083680 from the NIH/NIMH (T. S.).
References
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Corbin JG, Nery S, Fishell G. Telencephalic cells take a tangent: non-radial migration in the mammalian forebrain. Nat Neurosci. 2001;4(Suppl):1177–1182. doi: 10.1038/nn749. [DOI] [PubMed] [Google Scholar]
- Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, McManus MT. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci U S A. 2008;105:5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruttiger M, Karlsson L, Hall AC, Abramsson A, Calver AR, Bostrom H, Willetts K, Bertold CH, Heath JK, Betsholtz C, Richardson WD. Defective oligodendrocyte development and severe hypomyelination in PDGF-A knockout mice. Development. 1999;126:457–467. doi: 10.1242/dev.126.3.457. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Hodge RD, Daza RA, Englund C. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci Res. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Hobert O. MicroRNAs: all gone and then what? Curr Biol. 2005;15:R387–389. doi: 10.1016/j.cub.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee N, Sivalingam S, Boonstra R, Wojtowicz JM. The utility of Ki-67 and BrdU as proliferative markers of adult neurogenesis. J Neurosci Methods. 2002;115:97–105. doi: 10.1016/s0165-0270(02)00007-9. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14:156–159. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Kriegstein AR. Constructing circuits: neurogenesis and migration in the developing neocortex. Epilepsia. 2005;46(Suppl 7):15–21. doi: 10.1111/j.1528-1167.2005.00304.x. [DOI] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28:11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci. 2001;4(Suppl):1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Qian Y, Fritzsch B, Shirasawa S, Chen CL, Choi Y, Ma Q. Formation of brainstem (nor)adrenergic centers and first-order relay visceral sensory neurons is dependent on homeodomain protein Rnx/Tlx3. Genes Dev. 2001;15:2533–2545. doi: 10.1101/gad.921501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7:11–18. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson WD, Smith HK, Sun T, Pringle NP, Hall A, Woodruff R. Oligodendrocyte lineage and the motor neuron connection. Glia. 2000;29:136–142. doi: 10.1002/(sici)1098-1136(20000115)29:2<136::aid-glia6>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Rowitch DH. Glial specification in the vertebrate neural tube. Nat Rev Neurosci. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- Schaefer A, O’Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Sun T, Jayatilake D, Afink GB, Ataliotis P, Nister M, Richardson WD, Smith HK. A human YAC transgene rescues craniofacial and neural tube development in PDGFRalpha knockout mice and uncovers a role for PDGFRalpha in prenatal lung growth. Development. 2000;127:4519–4529. doi: 10.1242/dev.127.21.4519. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Nowakowski RS, Caviness VS., Jr Cell cycle parameters and patterns of nuclear movement in the neocortical proliferative zone of the fetal mouse. J Neurosci. 1993;13:820–833. doi: 10.1523/JNEUROSCI.13-02-00820.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe Y, Jessell TM. Diversity and pattern in the developing spinal cord. Science. 1996;274:1115–1123. doi: 10.1126/science.274.5290.1115. [DOI] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.