Abstract
The toolbox of rat genetics currently lacks the ability to introduce site-directed, heritable mutations into the genome to create knockout animals. Using engineered zinc-finger nucleases (ZFNs) designed to target an integrated reporter and two endogenous rat genes, Immunoglobulin M (IgM) and Rab38, we demonstrate that a single injection of DNA or mRNA encoding ZFNs into the one-cell rat embryo leads to a high frequency of animals carrying 25–100% disruption at the target locus. These mutations are faithfully and efficiently transmitted through the germline. Our data demonstrate the feasibility of targeted gene disruption in multiple rat strains within four months time, paving the way to a humanized monoclonal antibody platform and additional human disease models.
The laboratory rat is a well-established model for the genetic dissection of human disease-related traits (1) despite the fact that targeted modification of its genome is largely intractable. We investigated the application of engineered zinc-finger nucleases (ZFNs;(2)) for the elimination of specific rat gene function and generation of “knockout” rats. ZFNs induce site-specific, double-strand DNA breaks that can be repaired by the error-prone non-homologous end joining DNA repair pathway to result in a targeted mutation (Fig. 1A). In the fruit fly and zebrafish, direct embryo injection of ZFN-encoding mRNA has been used to generate heritable knockout mutations at specific loci (2).
Fig. 1.
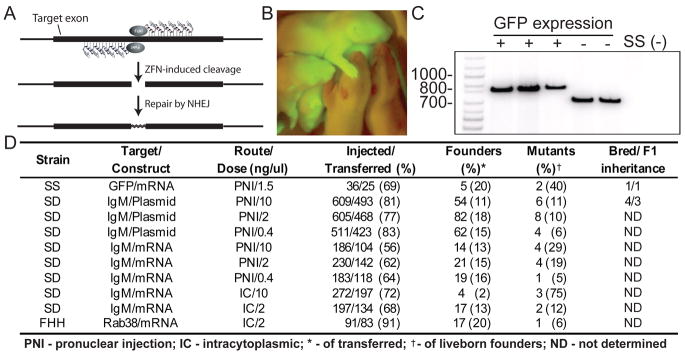
ZFN-mediated gene disruption in rat embryos. (A) ZFNs containing 5–6 fingers were designed to target coding sequences of interest (gray lines) for site-specific cleavage. (B) Two of five pups born after microinjection of GFP-targeted ZFNs were devoid of GFP expression. (C) Polymerase chain reaction using GFP-specific primers revealed truncated, but no wild-type sequence in each of the GFP negative pups compared to positive littermates. SS- Dahl S control DNA; NT indicates no template. (D) Table of injection data revealing successful mutagenesis of the three gene targets after multiple delivery methods and doses in three rat strains.
The design and validation of ZFN reagents to target a single-copy Green Fluorescent Protein (GFP) transgene inserted in a rat chromosome and two endogenous rat genes, IgM and Rab38, was performed as described (3) and is detailed in the supplemental materials. To take advantage of the potential for greater specificity-of-action afforded by longer (and therefore rarer) targets, we used 5- and 6- finger ZFNs.
We delivered these ZFNs to 36 hemizygous GFP-transgenic (4) inbred SS (Dahl S), 91 inbred FHH (Fawn-hooded hypertensive), and 2793 outbred SD (Sprague Dawley) embryos by pronuclear or intracytoplasmic injection of ZFN-encoding DNA or mRNA at different concentrations (Fig. 1D). Screening 295 founder animals yielded 35 (12%) that harbored targeted mutations.
Full knockout of the GFP transgene was achieved, as mutant animals lacked both GFP expression and wild-type GFP sequence (Fig. 1, B and C). Thirty-two IgM mutants and the single Rab38 mutant carried 25–100% disrupted target chromosomes (Fig. S1). Sequence analysis of 18 founders revealed deletion alleles ranging from 3–187 base pairs; of note, one animal carried biallelic mutations in IgM (Table S1). Furthermore, ZFN-mediated gene disruption demonstrated high fidelity for each target sequence since no ZFN-induced mutations were detected in target-gene-disrupted animals at any of 20 predicted ZFN off-target sites (Figs. S2, S3). After breeding to wild-type animals, 1 out of 1 GFP and 3 out of 4 IgM mutations were transmitted through the germline, one of which was subsequently bred to homozygosity (Figs. 1D, S4).
The high percentage of disrupted chromosomes demonstrates that ZFNs are active in early rat embryos from three strains, leading to both mono- and biallelic gene disruption. Although we observed no cleavage at predicted off-target sites, such events could be segregated away from the desired mutation by backcrossing to the parental strain. ZFN-driven gene disruption and germline transmission can be accomplished in 4 months’ time, and ZFNs can be engineered against a broad range of sequences (5, 6). Hence this strategy adds a valuable tool to an increasingly powerful rat genetic tool box, opening up a range of new experiments and models of human disease.
Supplementary Material
Acknowledgments
We thank R. Jaenisch, R. Hammer, P. Sullivan, and three anonymous referees for helpful suggestions; D. Smoller and E. Lanphier for support; E. Eastlund for the Rab38 ZFN mRNA; R. DeKelver and R. Amora for technical assistance, and Caliper Life Sciences, Inc. for excellent service. Supported by NIH grants 5U01HL066579-08 and 5P01HL082798-03, a sponsored research agreement between the Medical College of Wisconsin and Sigma-Aldrich, and the American Physiological Society Fellowship in Physiological Genomics to A.M.G. The authors are filing patents based on the results reported in this paper.
Contributor Information
Howard J. Jacob, Email: jacob@mcw.edu.
Roland Buelow, Email: rbuelow@omtinc.net.
References
- 1.Aitman TJ, et al. Nat Genet. 2008 May;40:516. doi: 10.1038/ng.147. [DOI] [PubMed] [Google Scholar]
- 2.Carroll D. Gene Ther. 2008 Nov;15:1463. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyon Y, et al. Nat Biotech. 2008 Jun;26:702. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michalkiewicz M, et al. Am J Physiol Heart Circ Physiol. 2007 Jul;293:H881. doi: 10.1152/ajpheart.00060.2007. [DOI] [PubMed] [Google Scholar]
- 5.Pabo CO, Peisach E, Grant RA. Annu Rev Biochem. 2001;70:313. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 6.Klug A. Proc Jpn Acad. 2005;81:87. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.