Abstract
Estrogen receptor α (ERα) plays a key role in mammary gland development and is implicated in breast cancer through the transcriptional regulation of genes linked to proliferation and apoptosis. We previously reported that hexamethylene bisacetamide inducible protein 1 (HEXIM1) inhibits the activity of ligand-bound ERα and bridges a functional interaction between ERα and positive transcription elongation factor b (P-TEFb). To examine the consequences of a functional HEXIM1-ERα-P-TEFb interaction in vivo, we generated MMTV/HEXIM1 mice that exhibit mammary epithelial-specific and doxycycline-inducible expression of HEXIM1. Increased HEXIM1 expression in the mammary gland decreased estrogen-driven ductal morphogenesis and inhibited the expression of cyclin D1 and serine 2 phosphorylated RNA polymerase II (S2P RNAP II). In addition, increased HEXIM1 expression in MCF-7 cells led to a decrease in estrogen-induced cyclin D1 expression, while downregulation of HEXIM1 expression led to an enhancement of estrogen-induced cyclin D1 expression. Studies on the mechanism of HEXIM1 regulation on estrogen action indicated a decrease in estrogen-stimulated recruitment of ERα, P-TEFb and S2P RNAP II to promoter and coding regions of ERα-responsive genes, pS2 and CCND1, with increased HEXIM1 expression in MCF-7 cells. Notably, increased HEXIM1 expression decreased only estrogen-induced P-TEFb activity. While there have been previous reports on HEXIM1 inhibition of P-TEFb activity, our studies add a new dimension by showing that E2/ER is an important regulator of the HEXIM1/P-TEFb functional unit in breast cells. Together, these studies provide novel insight into the role of HEXIM1 and ERα in mammary epithelial cell function.
Introduction
Mammary gland morphogenesis and development requires input from several genetic and epigenetic pathways regulated by hormones and growth factors including estrogens (1, 2). Estrogens mediate their actions through estrogen receptors (ERs), ERα, and ERβ, nuclear steroid receptors that classically regulate transcription either by directly binding to estrogen-response elements (EREs) of target genes (3-5) or indirectly via protein-protein interactions with other transcription factors like SP1 or AP-1 (6). In both cases, coregulatory proteins are also recruited to the promoter, and together ERs and these factors elicit changes in mRNA and protein levels of ER target genes, and ultimately, a physiological response (4-6). Since estrogen signaling controls the balance of growth and apoptosis in normal breast epithelial cells, a disruption of this balance contributes to abnormal cell growth and can lead to tumorigenesis (4, 7). Therefore, it is important to identify and elucidate the mechanism of action of ERs and their coregulators that give better insight into ER-mediated transcriptional regulation (8).
In eukaryotic transcription, the elongation stage is highly regulated and important for the generation of full-length mRNA transcripts (9-11). One of the positive regulators, positive transcription elongation factor b (P-TEFb) has an essential role in RNA polymerase II (RNAP II) transcription elongation (9, 10). In many human cell types, the predominant form of P-TEFb consists of cyclin dependent kinase 9 (CDK9) and its regulatory partner, cyclin T1 (11). It phosphorylates and thereby inhibits the activity of negative elongation factors, NELF and DSIF (DRB-sensitivity inducing factor) (9). It also phosphorylates the carboxy-terminal repeat domain (CTD) of the largest subunit of RNAP II (9, 10). The RNAP II CTD consists of multiple repeats of the heptapeptide sequence, YSPTSPS, phosphorylated at serine 5 by general transcription factor TFIIH during initiation and at serine 2 by P-TEFb during elongation (9, 10). These phosphorylation events are crucial for effective transition from an abortive to a productive phase of elongation (11, 12). P-TEFb is essential for productive HIV-1 transcriptional elongation and several studies have shown that various transcription factors bind to and recruit P-TEFb to specific promoters stimulating elongation (12, 13).
In previous studies, we identified an ERα-interacting protein, estrogen down-regulated gene-1 (EDG1) (also known as hexamethylene inducible protein 1 (HEXIM1)), and found that it is an inhibitor of ERα transcription and breast cell growth (14). Additionally, we demonstrated that HEXIM1 expression was lower in human breast tumors when compared to adjacent normal tissue, suggesting a role for HEXIM1 in breast tumorigenesis (14). Concurrent studies identified HEXIM1 as a P-TEFb-interacting factor that also inhibits P-TEFb activity (15, 16). Studies have also shown that HEXIM2, a paralog of HEXIM1, has the same inhibitory effect on P-TEFb (17, 18). We demonstrated that HEXIM1 modulates a functional interaction between ERα and cyclin T1 in breast epithelial cells, and inhibits the recruitment of ligand-bound ERα (E2/ERα) to the pS2 gene promoter (19). We also found that cyclin T1 appeared to be necessary for E2-induction of cyclin D1 protein expression (19). Cyclin D1 (CCND1) is a D-type cyclin that regulates G1-S cell cycle progression during cell proliferation (20). CCND1 is also E2/ERα responsive and is thought to play major roles in mammary gland development and breast cancer (1, 21). However, the contribution of E2/ERα to HEXIM1 action in breast cells is not well defined. In addition, the precise mechanism by which P-TEFb regulates CCND1 transcription and how this can be linked to mammary gland development and tumorigenesis needs to be further defined.
To address these questions, we developed a transgenic mouse model in our laboratory that is doxycycline-inducible and selectively expresses HEXIM1 in the mammary gland under the control of the mouse mammary tumor virus long terminal repeat (MMTV-LTR) promoter previously described (22). Using this model, we demonstrate that increased HEXIM1 expression decreased E2-induced CCND1 and serine 2 phosphorylated (S2P) RNAP II expression in the mouse mammary gland. In addition, increasing HEXIM1 expression inhibits E2-induced recruitment of ERα, P-TEFb and S2P RNAP II to ER-responsive genes, pS2 and CCND1 in MCF-7 cells. Finally, we observed that E2 stimulates P-TEFb activity and that HEXIM1 inhibits only E2-induced, and not basal, P-TEFb activity. These results elucidate the functional consequences of modulating HEXIM1 expression on E2/ERα-driven transcription in the mammary gland and its implications for estrogen-dependent breast cancer.
Materials and Methods
Materials and Antibodies
17β-Estradiol (E2) and CDK9 inhibitor, 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) were obtained from Sigma Chemical Co. (St. Louis, MO). Commercially available antibodies used for immunoprecipitation and Western blot analysis are described in the Supplementary section.
Transgenic mice (MMTV/HEXIM1) generation
MMTV-rtTA mice were generated as described by Gunther et al (22). To generate pTET-HEXIM1 mice, a plasmid construct was made by subcloning the coding sequence of human HEXIM1 gene downstream of the tetracycline-dependent minimal promoter in the pTet-splice transgene construct. After purification, the resulting plasmid was used for pronuclear injection into FVB oocytes (Case Western Reserve University Transgenic and Targeting Facility). To achieve mammary gland-specific expression of HEXIM1 in a doxycycline-dependent manner, pTET-HEXIM1 mice were crossed with the MTB line, which expressed the reverse tetracycline-dependent transcriptional activator (rtTA) under the control of the mouse mammary tumor virus long terminal repeat (MMTV-LTR). From these matings the bigenic mice, MMTV/HEXIM1, were created. Transgene expression was induced by adding 2 mg/ml doxycycline to the drinking water. Bigenic mice were identified by screening genomic DNA from tail biopsies for the presence of the transgenes using PCR and verified by Western blot analysis. See Supplemental Table 1 for list and sequences of primers used.
Whole-mount histology & Immunodetection
Mice were induced with doxycycline at 9 weeks of age, ovariectomized and treated with E2 at 12 weeks. E2 was administered as daily subcutaneous injections of sesame oil solution containing 1μg of E2. Mammary glands from the MMTV/HEXIM1 mice were collected 25 days after start of E2 treatment for whole-mount staining via the Carmine-alum technique and Western blot analyses. Immunohistochemistry using sections from mammary glands are described in the Supplementary section.
Reverse Transcription (RT) PCR Analyses
Human breast epithelial cells, MCF-7, were maintained as described (19) and were transiently transfected with pCMV-TAG2B or pCMV-TAG2B-HEXIM1 using FuGENE HD reagent (Roche, IN) according to the manufacturer's instructions. Forty-eight hours later, cells were treated with ethanol vehicle or 100 nM E2 for 3 hours. Total RNA was extracted using the TRIzol reagent (Invitrogen, CA) and subjected to RT-PCR analyses as described in the Supplementary section.
Western Analyses
Western blot analyses using extracts from mammary glands and MCF-7 cells are described in the Supplementary section.
RNA interference
A Pol II promoter-driven miRNA expression vector system (Invitrogen, CA) was used. To make pcDNA-HEXIM1 miR, miRNA oligos (see Supplemental Table 3 for list and sequences) were annealed and cloned into the pcDNA 6.2 GW/EmGFP vector (Invitrogen) according to the manufacturer's instructions. MCF-7 cells were transfected with pcDNA 6.2-GW/EmGFP-miR expression vectors containing either the HEXIM1 miRNA insert or a control LacZ miRNA insert. Following blasticidin selection, cells expressing the highest level of GFP were flow-sorted and expanded. During experiments, cells were treated with ethanol vehicle or 100 nM E2 for 3 hours and harvested as described above for Western blot analyses.
Chromatin immunoprecipitation (ChIP) assays
MCF-7 cells were plated on 150 mm plates and transfected as described with pCMV-TAG2B or pCMV-TAG2B-HEXIM1. Forty-eight hours later, cells were treated with ethanol vehicle or 100 nM E2 for 45 and 90 minutes. ChIP assays were carried out as previously described (19). See Supplemental section for further details of immunoprecipitation and analyses.
CTD kinase assays
Kinase assays were performed according to previously described protocols with some modifications (23, 24). See Supplemental section for complete description of assay.
Data analyses
Statistical significance was determined using Student's t test comparison for unpaired data and was indicated as follows: *, P < 0.05; **, P < 0.005.
Results
Increase in HEXIM1 expression inhibits E2-driven mammary gland development by decreasing cell proliferation and increasing apoptosis
To examine the functional consequences of an interaction between HEXIM1, ERα and P-TEFb in the mammary gland, we generated a double transgenic mouse model, MMTV/HEXIM1, which inducibly overexpress HEXIM1 in the mammary gland when the mice are treated with doxycycline (+DOX). We first examined the effects of elevated levels of HEXIM1 on E2-driven mammary gland development by having the MMTV/HEXIM1 mice ovariectomized and treated with E2 as described in “Materials and Methods”. In comparing whole mounts of mammary glands from -DOX and +DOX animal groups, we observed decreased ductal branching in the mammary glands of +DOX mice when compared to -DOX mice, as well as compared to single transgenics (MMTV alone) (Figure 1A, −/+DOX panel insets). We quantified the level of increase of HEXIM1 expression in the mammary gland as approximately 24% over endogenous levels (data not shown, see Figure 2A for immunoblot), so we do not foresee that an overwhelming amount of HEXIM1 is needed to dictate these physiological changes. Since ductal elongation and branching in the mammary gland have been shown to be E2/ERα-dependent (1, 25), these data suggest that HEXIM1 inhibits E2/ERα-driven mammary gland morphogenesis.
Figure 1. Increased HEXIM1 expression inhibits estrogen-regulated mammary gland morphogenesis due to changes in proliferation and apoptosis.
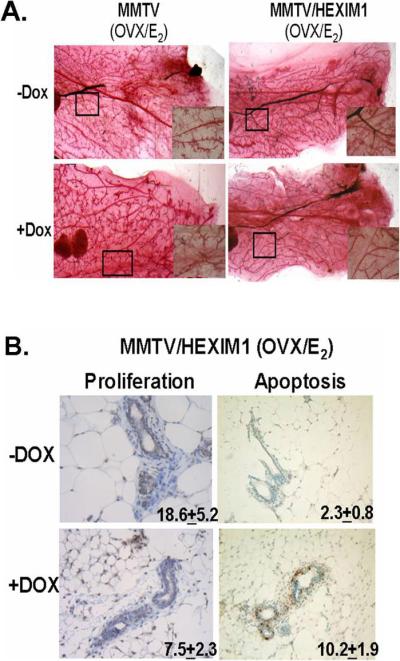
A. MMTV/HEXIM1 mice were treated as described in Materials and Methods. Representative whole mounts were obtained from MMTV and MMTV/HEXIM1 mice with Carmine alum stain. Original magnification is X40 for all panels.
B. BrdU-labeled nuclei were detected by immunostaining and apoptotic nuclei were stained by TUNEL. Quantitation of positively-labeled MMEC nuclei from at least 1000 nuclei each from 5 mice per group (−/+DOX) is shown. Original magnification is X40 for all panels.
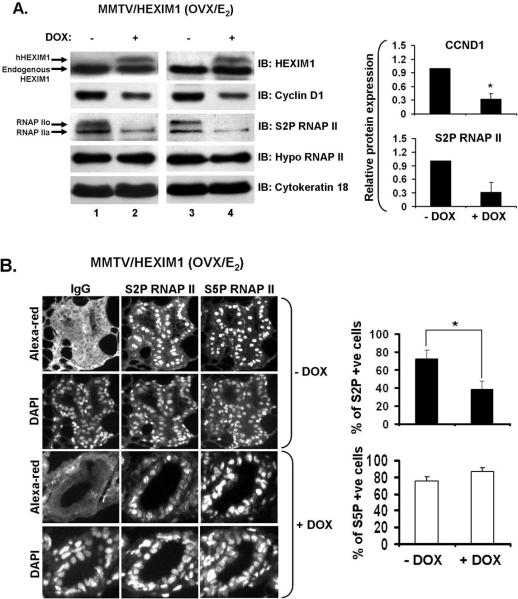
Figure 2. Increased HEXIM1 expression inhibits E2-induced cyclin D1 expression and serine 2 phosphorylation of RNAP II in mouse mammary gland.
A. MMTV/HEXIM1 mice were treated as described in Materials and Methods. Mammary gland tissue extracts from MMTV/HEXIM1 mice were subjected to Western blot using indicated antibodies for immunoblotting. Anti-cytokeratin 18 was used as an epithelial cell marker and a loading control. RNAP IIa and IIo indicate the “hypo” and “hyper” phosphorylated forms of RNAP II respectively. Samples were collected from 3-5 mice per group (−/+Dox) and columns represent quantitation of Western analyses for cyclin D1 and S2P RNAP II; bars, SE; *, P< 0.01.
B. Immunofluorescent detection of S2P and S5P RNAP II. Representative images of mammary gland sections from MMTV/HEXIM1 mice (−/+ Dox) stained as described in Materials and Methods with S2P and S5P RNAP II-specific antibodies. Nuclei were counterstained with DAPI as indicated. Original magnification is X20 for all panels. Quantitation of S2P or S5P positively stained (S2P+ or S5P+) luminal epithelial cells was performed as described in Materials and Methods. Columns represent percentage (mean ± SEM) of total S2P/S5P+ luminal epithelial cells with 3-4 mice per group (−/+ Dox) and a minimum of 1000 cells per animal analyzed; bars, SE; *, P<0.05
Previously, we found that HEXIM1 inhibits ERα transcription (14, 19), so this physiological effect could be due to a dysregulation of ER-responsive genes involved in proliferation and apoptosis. To investigate this, MMTV/HEXIM1 mice were injected with BrdU two hours prior to being sacrificed. BrdU-labeled nuclei in the mammary gland were detected by immunostaining and apoptotic nuclei were stained by TUNEL. Quantitation of positively-labeled mouse mammary epithelial cell nuclei revealed both a decrease in epithelial cell proliferation and an increase in apoptosis (Figure 1B). Taken together, our data suggest a critical role for HEXIM1 in E2/ERα-driven processes in the mammary gland.
Increased HEXIM1 expression regulates cyclin D1 protein expression levels and Serine 2 phosphorylation of RNA polymerase II in vivo
Our findings that overexpression of HEXIM1 decreased ductal branching and cell proliferation in the mammary gland prompted us to investigate the effect of HEXIM1 overexpression on E2/ERα signaling. As an output, we examined changes in cyclin D1 (CCND1) and c-Myc because both genes are E2/ERα responsive and involved in proliferation integral to mammary gland development and breast cancer (26, 27). MMTV/HEXIM1 mice were ovariectomized and treated with E2 as described and we found that increased HEXIM1 expression (+DOX) resulted in decreased CCND1 protein expression levels (~67%) in mouse mammary gland cell extracts (Figure 2A). However, c-Myc expression levels did not significantly change regardless of HEXIM1 expression (Supplemental Figure 1A), suggesting a difference in sensitivity of ERα target genes to HEXIM1.
In addition, since HEXIM1 inhibits P-TEFb activity (12), we examined the effect of increased HEXIM1 expression on P-TEFb activity in the mouse mammary gland by investigating the expression levels of the serine 2 phosphorylated form of RNAP II (S2P RNAP II). In +DOX mice, we observed a decrease in S2P RNAP II levels (~68%) (Figure 2A). It is important to note here that the antibody used to detect S2P RNAP II recognizes both phosphorylated (RNAP IIo) and unphosphorylated (RNAP IIa) forms of RNAP II. We also blotted with another antibody that detects only the RNAP IIa form (Hypo RNAP II) and found that there was no observable change between −/+DOX mice groups (Figure 2A). Also, HEXIM2, a paralog of HEXIM1, inhibits P-TEFb activity (12), but in the mammary gland, HEXIM1 protein expression levels are significantly higher (Supplemental Figure 1B), suggesting a dominant role for HEXIM1 in the mammary gland.
We also examined the expression levels of the serine 5 phosphorylated form of RNAP II (S5P RNAP II), associated with initiation, using immunofluorescent labeling of epithelial nuclei in mammary gland lumen from MMTV/HEXIM1 mice. We did not observe any changes in the percentage of S5P RNAP II positively-stained nuclei (S5P +ve cells) when we compared −/+DOX animal groups (Figure 2B). However, in the +DOX animal group, the percentage of S2P RNAP II positively-stained nuclei (S2P +ve cells) within the mammary gland lumen was significantly decreased by 50% (P < 0.05) when compared to the −DOX group (Figure 2B), verifying our results in the Western blot (Figure 2A). Given that E2/ERα regulates CCND1 expression (21, 27) and HEXIM1 inhibits P-TEFb activity and associates with ERα (12, 19), the effect of increased HEXIM1 expression levels on CCND1 and S2P RNAP II expression suggest that these changes reflect a complex interaction of multiple pathways that converge at the level of E2/ERα-mediated transcription.
HEXIM1 regulates cyclin D1 and pS2 expression levels in vitro
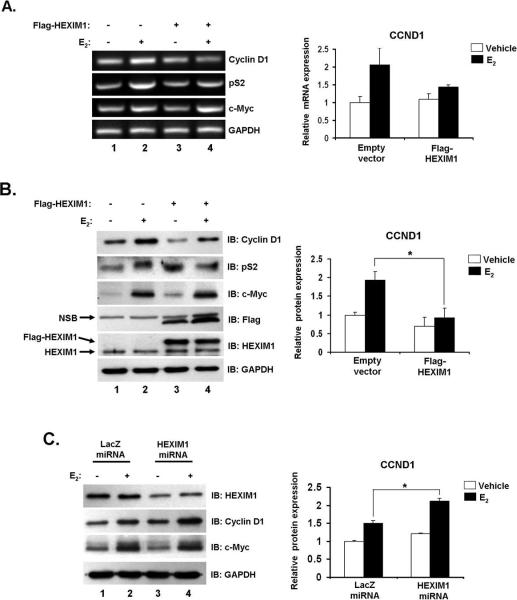
The effects of increased HEXIM1 expression on CCND1 and S2P RNAP II expression levels could also be the sum result of the disruption of E2/ERα activity in multiple cell types in the mammary gland. To verify that our observations can be attributed to a more localized event in epithelial cells, we investigated the effects of increased HEXIM1 expression in mammary epithelial MCF-7 cells. MCF-7 cells were transfected with pCMV-Tag2B-HEXIM1 or control vector and treated with ethanol vehicle or E2 for 3 hours. In control cells, we observed an average 1.5 to 2-fold E2 induction of both CCND1 mRNA and protein levels (Figures 3A and B); consistent with what has been demonstrated in other studies (8, 21). As expected, increased HEXIM1 expression diminished E2-induction of CCND1 mRNA and protein expression (Figures 3A and B, compare lanes 2 and 4). For CCND1 protein expression levels, this was quantified as a 52% decrease in expression by HEXIM1 when normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression (Figure 3B).
Figure 3. HEXIM1 regulates E2-induced cyclin D1 expression in breast epithelial cells.
A. MCF-7 cells were transfected with pCMV-Tag2B-HEXIM1 or empty vector, treated with 100 nM E2 or ethanol vehicle for 3 hours. RNA was extracted and subjected to RT-PCR as described in “Materials and Methods”. RT-PCR results quantitated for changes in cyclin D1 mRNA using GAPDH as a control. Columns, mean of four independent replicates; bars, SE; *, P<0.05
B. Representative Western blot using indicated antibodies for immunoblotting. Changes in cyclin D1 protein levels were quantitated and normalized to GAPDH. Columns, mean of three independent replicates; bars, SE; *, P<0.05
C. Representative Western blot indicating miRNA-mediated silencing of HEXIM1 in MCF-7 cells. E2-induced changes in cyclin D1 between LacZ and HEXIM1 miRNA stable cells were quantitated and normalized to GAPDH. Columns, mean of three independent replicates; bars, SE; *, P<0.01
We also investigated the effect of increased HEXIM1 expression on other ER-responsive genes and found increased HEXIM1 levels decreased E2-induced increases in pS2 mRNA and protein levels (Figures 3A and B, Supplemental Figure 2A). The E2-induced mRNA levels of other genes including c-Myc, Progesterone receptor (PR) and Cathepsin D remained unchanged with increased HEXIM1 expression (Figure 3A, Supplemental Figure 2A). In addition, E2-induction of c-Myc protein levels was also unchanged with increased HEXIM1 expression (Figure 3B, with quantitation in Supplemental Figure 2B). These data also suggest that increased HEXIM1 expression may not have a similar effect on all ERα-target genes.
Having observed that increased HEXIM1 expression decreased E2-induced CCND1 and pS2 expression, it stood to reason that HEXIM1 silencing would result in an increase in the induction of both genes by E2 when compared to control cells. HEXIM1 gene expression knockdown (~50%) using miRNA-mediated RNAi resulted in a statistically significant (P < 0.01) enhancement of E2-induced CCND1 protein expression levels (Figure 3C). We did not observe a similar effect with pS2 (data not shown) and E2-induced c-Myc protein expression levels remained unchanged (Figure 3C). Because multiple regulatory proteins are involved in CCND1 regulation (28), we cannot rule out possible actions of other factors in vivo. Also, we cannot rule out the possibility of HEXIM1 regulation of other key E2 responsive genes involved in cell proliferation and mammary carcinogenesis not studied here. Nonetheless, taken together our data support a novel physiological role for HEXIM1 in E2-driven mammary gland development via regulation of CCND1 levels and serine 2 phosphorylation of RNAP II.
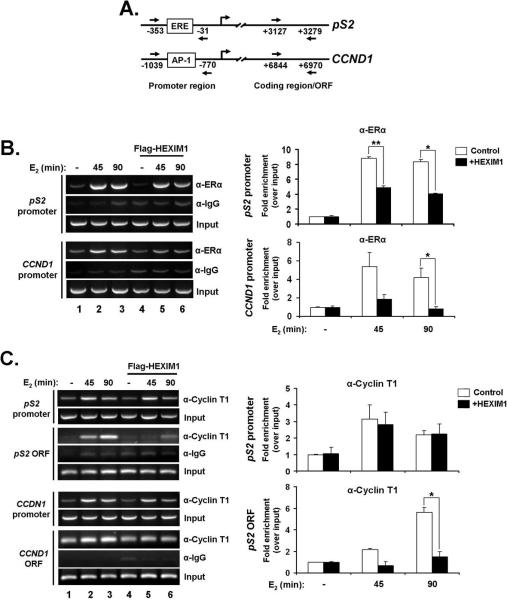
HEXIM1 inhibits E2-induced recruitment of ERα and cyclin T1 (P-TEFb) to ERα-responsive genes
Given our observations on the effect of HEXIM1 expression on CCND1 and pS2, it is critical to characterize the molecular events involved in HEXIM1 transcriptional regulation of these ER-responsive genes. We previously reported that HEXIM1 associates with ERα after 90 minutes of E2 stimulation in MCF-7 cells and increased HEXIM1 expression led to a decrease in E2/ERα recruitment to the pS2 promoter (19). Other reports have shown that P-TEFb associates with the elongating form of RNAP II, S2P RNAP II, which is thought to predominate in the coding regions of genes with increased occupancy towards the 3’ end of genes (29-31). Therefore, we hypothesized that increased HEXIM1 levels would (1.) inhibit the recruitment of ERα to the promoter region of ER target genes and (2.) inhibit the recruitment of P-TEFb to regions downstream from the promoter at ER-responsive genes, and this would in turn verify the potential regulation of transcriptional elongation of these genes. To investigate this, we carried out chromatin immunoprecipitation (ChIP) assays in MCF-7 cells and examined the effect of E2 on ERα and P-TEFb occupancy at two ERα-responsive genes, pS2 and CCND1.
We found that an increase in HEXIM1 expression in MCF-7 cells correlated with a 2-fold increase in HEXIM1 occupancy at E2-responsive regions within the pS2 and CCND1 (32) promoters (Supplemental Figure 3A) that was not significantly affected by E2 treatment. We also found that increased HEXIM1 expression inhibited the recruitment of E2/ERα to the promoter regions of pS2 and CCND1 (Figure 4B). Now, it is well documented that E2/ERα cycles on and off the promoter of ER-responsive genes and ERα binding is typically diminished after 90 minutes of E2 stimulation (33, 34). Other studies have also shown that the absolute timing of ERα cycling differs and there can still be significant E2/ERα enrichment at 90 minutes (35, 36). In our studies, it is clear that E2/ERα binding is less at 90 minutes when compared to 45 minutes (Figure 4B, compare lanes 2 and 3), and we see a significant decrease at 3 hours (data not shown) indicating E2/ERα is cycling on DNA in our experiments. Regardless of the time point of E2 stimulation, increased HEXIM1 inhibits E2/ERα recruitment to DNA.
Figure 4. Effect of increased HEXIM1 expression on E2-dependent recruitment of ERα and P-TEFb (cyclin T1) to ER-responsive genes.
A. Primers used in ChIP assays are directed at regions indicated for pS2 and CCND1 genes.
B. Increased HEXIM1 expression leads to decrease in E2/ERα recruitment to promoter regions of ER target genes, pS2 and CCND1. Samples were prepared from MCF-7 cells transiently transfected with pCMV-Tag2B-HEXIM1 or empty vector, treated with ethanol vehicle or 100 nM E2 for 45 and 90 minutes. Chromatin immunoprecipitation was done with antibodies against ERα and rabbit IgG (as an IP control). Panels on the left, DNA fragments were analyzed by PCR with primers specific for the promoter-proximal region of pS2 and CCND1 as indicated in (A). Panels on the right, Quantitations of ERα IP enrichment at pS2 and CCND1 promoters. Columns, mean of two independent replicates; bars, SE; *, P<0.05; **, P<0.005.
C. HEXIM1 inhibits E2-dependent recruitment of cyclin T1 to coding regions of pS2 and CCND1. MCF-7 cells were treated as described in (B). ChIP analysis of cyclin T1 recruitment to E2-responsive region of pS2 and CCND1 promoter and coding regions was carried out using primers specific for the regions indicated in (A). Panels on the left, DNA fragments were analyzed by PCR with primers specific for the regions indicated. Panels on the right, Quantitations of cyclin T1 IP enrichment at pS2 gene. Columns, mean of 2-3 independent replicates; bars, SE; *, P<0.05
For P-TEFb (via cyclin T1), we observed an E2-dependent recruitment pattern that differed slightly at the promoter and coding regions of both pS2 and CCND1 genes. At the promoters of both pS2 and CCND1, cyclin T1 follows a similar trajectory as ERα in terms of recruitment in control cells, but there is no significant effect on cyclin T1 recruitment with increased HEXIM1 expression (Figure 4C). However, at the coding regions of both genes, we observed a gradual increase in cyclin T1 recruitment with over time with E2 stimulation and, increased HEXIM1 expression inhibited the recruitment of cyclin T1 (Figure 4C). As a further control, we looked at cyclin T1 recruitment to the GAPDH ORF at a region that has been shown to have significant RNAP II enrichment during GAPDH transcription (37). We did not observe an E2-dependent recruitment pattern and there were no significant changes in cyclin T1 recruitment with increased HEXIM1 expression (Supplemental Figure 3B).
These data suggest that HEXIM1 inhibits ERα transcriptional elongation by inhibiting PTEFb recruitment, via cyclin T1, to the coding region of some ER-responsive genes. This result is consistent with other studies showing that the recruitment of P-TEFb to promoter-proximal and coding regions stimulates transcriptional elongation (30, 38). Also, in previous studies, we observed a decrease in the co-recruitment of cyclin T1 with ERα at the pS2 promoter with increased HEXIM1 expression (19), but it appears that the total occupancy of cyclin T1 at the promoter region does not change with increased HEXIM1 expression (Figure 4C). In this context, our data suggests a dual role for P-TEFb in ERα transcription. On one hand, it acts as a coactivator for ERα-driven transcription by directly associating with E2/ERα (19), but it also serves its general purpose as a transcription elongation factor, with E2 acting as an enhancer of P-TEFb occupancy in the context of ER-responsive genes.
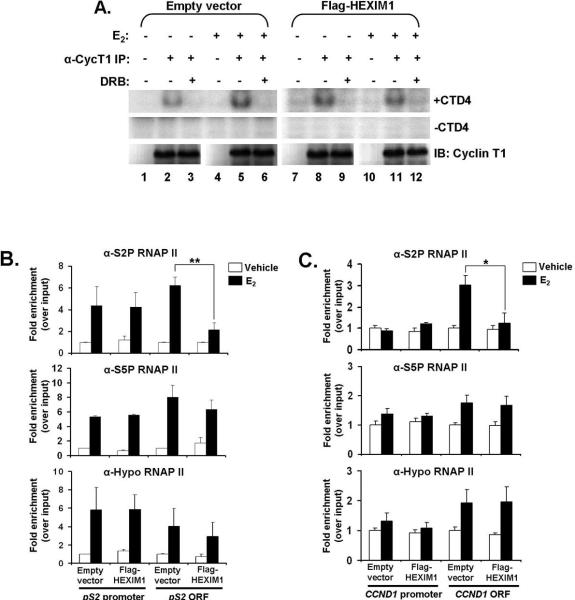
HEXIM1 inhibits E2-induced P-TEFb activity and recruitment of serine 2 phosphorylated RNA polymerase II to coding regions of ER-responsive genes
Since E2 enhanced P-TEFb recruitment to pS2 and CCND1 genes, we investigated if E2 also increased P-TEFb activity as a means of promoting transcriptional elongation of ERα-responsive genes. To do this, we performed kinase assays using endogenous immunoprecipitates of cyclin T1 from MCF-7 cells and examined the kinase activity of P-TEFb with a synthetic peptide substrate, CTD4 (YSPTSPS4). As shown in Figure 5A, E2 treatment increased P-TEFb activity in MCF-7 cells (compare lanes 2 and 5). We also observed that increased HEXIM1 expression in MCF-7 cells inhibited an E2-induced increase in P-TEFb activity (Figure 5A, compare lanes 2 and 5 to lanes 8 and 11). We confirmed that this inhibition was selective for P-TEFb by adding the CDK9 inhibitor, DRB, to an equal half of the kinase immunoprecipitates (Figure 5A, lanes 3, 6, 9 and 12) and that the input levels of cyclin T1 and HEXIM1 were evenly loaded (Supplemental Figure 4A). Quantitation of 32P incorporation into the CTD4 peptide verified that HEXIM1 significantly abrogates E2-induced P-TEFb activity in MCF-7 cells (Supplemental Figure 4B).
Figure 5. Increased HEXIM1 expression inhibits E2-induced P-TEFb activity and recruitment of serine 2 (hyperphosphorylated) RNA polymerase II to the coding region of ER-responsive genes.
A. Increased HEXIM1 expression decreased E2-induced CTD4 peptide phosphorylation. MCF-7 cells were transiently transfected with pCMV-Tag2B-HEXIM1 or empty vector and treated with ethanol vehicle or 100 nM E2 for 90 minutes. Cell lysates were subjected to immunoprecipitation with antibodies against cyclin T1 and rabbit IgG (as an IP control). The immunoprecipitates were divided into two halves with one half getting 2 μg of the CTD4 peptide added to the reaction (−/+ CTD4). Fifty μM DRB was also added to some immunoprecipitates as a kinase assay control. The kinase reactions were analyzed by SDS-PAGE using autoradiography. Equal volumes of kinase reactions were also analyzed by Western blot to check specificity of anti-cyclin T1 antibody in immunoprecipitation. Panels are representative of at least four independent experiments.
B. HEXIM1 inhibits E2-dependent recruitment of S2P RNAP II to pS2 ORF. MCF-7 cells were treated as described in Figure 4 and subjected to ChIP analysis with antibodies against serine 2 phosphorylated (S2P) RNAP II, serine 5 phosphorylated (S5P) RNAP II and the unphosphosphorylated form of RNAP II (8WG16). The regions of pS2 amplified by PCR are as indicated. Columns, mean of 3-5 independent replicates; bars, SE; **, P<0.005
C. HEXIM1 inhibits E2-dependent recruitment of S2P RNAP II to CCND1 ORF. MCF-7 cells were treated as described. The regions of CCND1 amplified by PCR are as indicated. Columns, mean of 3-4 independent replicates; bars, SE; *, P<0.05
We also observed that increased HEXIM1 expression did not inhibit basal P-TEFb activity in HEXIM1-transfected MCF-7 cells compared to non-transfected cells (Figure 5A, compare lanes 2 and 8). One reason for this could be attributed to the fact that at basal levels, the P-TEFb complex is thought to occur in two states, 50% free and 50% HEXIM1-7SK RNA bound (12) and in a study by He et al, gradually decreasing HEXIM1 expression by RNAi did not initially have much effect on this equilibrium because it was the “free” form of HEXIM1 that was being down-regulated, without any effect on the HEXIM1 protein associated with P-TEFb (11). In our experiments, it is possible that increasing HEXIM1 expression may not be significantly perturbing the P-TEFb equilibrium initially, but increases the availability of free HEXIM1 populations that subsequently diminishes any increases in E2 induced P-TEFb activity.
Since HEXIM1 inhibited the recruitment of cyclin T1 to the coding regions of ER-responsive genes and E2-induced P-TEFb activity, we determined the effect of increased HEXIM1 levels on the recruitment of S2P RNAP II as a mark of the modulation of transcription elongation (9). At the pS2 promoter, we found that E2 stimulated the enrichment of all forms of RNAP II, without any significant changes when HEXIM1 expression was increased (Figure 5B). This result differs with what has been observed in some studies, which show low S2P RNAP II occupancy in comparison to S5P RNAP II at the promoter of transcriptionally active genes (29, 37). However, it is possible for both S2P and S5P RNAP II to occupy similar regions on DNA (39), which likely marks the beginning of the transition to elongation, but it is unclear since we did not study proximal upstream or downstream regions to the pS2 promoter. In addition, there was no change in the recruitment of all forms of RNAP II recruitment to the CCND1 promoter in both control and HEXIM1-transfected cells, although it was not as sensitive as the pS2 promoter to E2 stimulation (Figure 5C).
However, at the pS2 and CCND1 coding regions, we found that increased HEXIM1 expression inhibited the E2-dependent recruitment of S2P RNAP II, without significant changes in the recruitment of S5P and unphosphorylated RNAP II forms (Figures 5B and 5C). These data indicate that in mammary epithelial cells, HEXIM1 does not affect transcription initiation, since the recruitment patterns of all forms of RNAP II to the promoter-regions of pS2 and CCND1 were unchanged regardless of HEXIM1 expression levels in the cell. However, increased HEXIM1 decreases transcription elongation since the recruitment of S2P RNAP II to the coding region was decreased.
Discussion
This study provides novel evidence for a physiological role of HEXIM1/P-TEFb interaction in attenuating E2/ERα driven transcription in the mammary gland and breast cancer cells. First, we demonstrated that increased HEXIM1 expression in the mammary gland of a transgenic mouse model decreased ductal branching, an E2-driven developmental process, due to changes in proliferation and apoptosis. We correlated these changes with a decrease in CCND1 and S2P RNAP II expression in vivo and in vitro. We also show that overexpression of HEXIM1 diminished E2-induced recruitment of ERα and cyclin T1 to the promoter and coding regions, respectively, of ER-responsive genes. Further, we show that E2 enhances the activity of the P-TEFb kinase, CDK9, which is inhibited by increased HEXIM1 expression. Surprisingly, increased HEXIM1 expression inhibited only E2-induced increases in P-TEFb activity and not basal P-TEFb kinase activity. In our studies, this inhibition of P-TEFb activity translates to a decrease in the recruitment of S2P RNAP II, and not other forms of RNAP II, to the coding regions of ER-responsive genes, pS2 and CCND1. These findings support a role for P-TEFb and transcription elongation in cell proliferation but more importantly, the data suggests a novel mechanism of action for HEXIM1 that can be recapitulated in vivo and a possible therapeutic role for HEXIM1 in hormone-dependent breast cancer.
Given that the regulation of CCND1 is complex (28), we do not assume that our findings represent a comprehensive explanation regarding E2-regulation of CCND1. Also, other sites within CCND1 contribute to the transcriptional output (21). We investigated the recruitment patterns of ERα, P-TEFb and RNAP II to two sites within the gene: an E2-responsive region of the promoter and the coding region. Perhaps, a more extensive analysis of different sites within the genes, pS2 and CCND1, and even other ER-responsive genes will reveal other insight into mechanism of HEXIM1 regulation of these genes. However, we believe that the information gathered from this study was sufficient to demonstrate the regulatory effects of HEXIM1 on ERα and P-TEFb recruitment to pS2 and CCND1, suggesting a role for HEXIM1 and P-TEFb in ERα transcriptional regulation of some, but not all ERα target genes.
Based on our previous (19) and current studies, we speculate that E2 enhances an ERα-P-TEFb interaction, and this increases the population of active P-TEFb at the gene locus of ER-responsive genes, which in turn phosphorylates the CTD of RNAP II. This phosphorylation event positions the gene in an active elongation state with increased S2P RNAP II occupancy at the coding regions, and enhanced recruitment of other forms of RNAP II marking the gene in an active transcription state (31, 39). However, increased HEXIM1 expression inhibits ERα and P-TEFb enrichment at the promoter and coding regions, respectively, of ER-responsive genes thus, decreasing the population of P-TEFb available to phosphorylate RNAP II to the serine 2 phosphorylated form associated with transcriptional elongation. In addition, we observed that increased HEXIM1 inhibits E2-induced P-TEFb activity and postulated that this was due to an increase in the “free” form of HEXIM1, which serves to diminish any subsequent increases in PTEFb activity. Taken together, this scenario could represent the mechanism by which HEXIM1 modulates ERα-mediated transcription in the context of some ER-responsive genes (See Figure 6 for proposed model).
Figure 6.
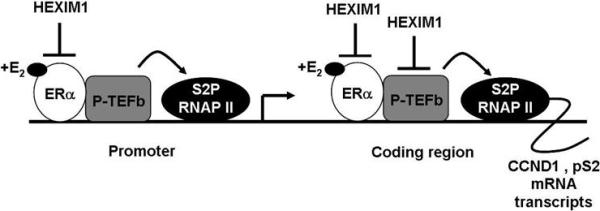
Proposed model for HEXIM1 action on ERα and P-TEFb at ER-responsive genes, pS2 and CCND1, in mammary cells.
The understanding of general mechanisms that control elongation stem from studies showing that HIV-1 harnesses P-TEFb as a cofactor to promote efficient mRNA transcript synthesis (12). These and other studies have raised questions about whether P-TEFb acts as a general transcription elongation factor or serves in a gene-selective or context-dependent manner. The P-TEFb complex components, cyclin T1 and CDK9, have not been shown to have sequence-specific DNA binding activity, but transcription factors interact with and recruit P-TEFb to their respective promoter targets (12, 40, 41). In addition, DNA microarray analyses of hearts from cyclin T1 transgenic mice indicate selective increases in subsets of genes, rather than a global increase in mRNA expression when compared to non-transgenic mice (42). Our studies suggest that in breast epithelial cells, P-TEFb can be modulated by E2/ERα and HEXIM1 in the context of some ERα target genes, although identical E2-induced recruitment patterns for P-TEFb to pS2 and CCND1 genes suggest a general transcription elongation mechanism is also involved. In addition, the interaction of P-TEFb and E2/ERα supports a positive aspect of ERα transcription elongation regulation, but E2/ERα also interacts with negative elongation factor (NELF) and this interaction inhibits ERα-mediated transcription (43).
HEXIM1 has been shown to have P-TEFb-independent action as seen with the glucocorticoid receptor (44). We have also reported on P-TEFb-independent action of HEXIM1 in cardiovascular development (45). Given this evidence, it is clear that HEXIM1 can inhibit transcription in a P-TEFb-dependent and -independent manner. Therefore, we cannot assume that the effect of HEXIM1 in the mammary gland is solely on ERα/P-TEFb, as other factors are involved in mammary gland development. However, we were able to demonstrate a specific inhibition pattern that HEXIM1 exerted on E2-induced events at ER-responsive genes, pS2 and CCND1, and increased HEXIM1 levels inhibited E2-induced P-TEFb activity. Thus, based on our data, the HEXIM1 inhibition patterns observed in this study is largely P-TEFb-dependent in both our cell culture and animal models. Several studies also support an emerging role for HEXIM1 as a regulator of cell growth and differentiation (12, 46). Conversely, deletion of CLP-1, the mouse HEXIM1 gene, leads to pathological cardiac hypertrophy and perinatal death (47).
In this study, a targeted increase in HEXIM1 expression in the mouse mammary gland driven by a mammary epithelial cell promoter (MMTV-LTR), led to a decrease in ductal branching (Figure 1A), an E2/ERα driven mammary gland developmental process (25). This observation was attributed to a decrease in proliferation and an increase in apoptosis (Figure 1B). The decrease in proliferation was linked to a concurrent decrease in CCND1 expression, further demonstrating that HEXIM1 regulates ER-responsive genes in vivo. Future studies will not only aim to address HEXIM1 regulation of other E2/ERα target genes, but also HEXIM1 regulation of other nuclear receptors relevant in mammary cell function and tumorigenesis.
Supplementary Material
Acknowledgements
We would like to Dr. Huayun Deng for his help with some of the animal work and advice on immunohistochemistry. We thank Dr. Lewis Chodosh (University of Pennsylvania) for the MTB mice and the pTet-Splice vector. This work was supported by National Institute of Health Grant CA92440 to M.M.M and a Department of Defense Predoctoral Fellowship W81XWH-06-1-0426 to N.O.
Abbreviations
- ERα
Estrogen receptor alpha
- E2
17-beta estradiol
- HEXIM1
Hexamethylene inducible gene-1
- P-TEFb
Positive transcription elongation factor b
- CTD
Carboxy-repeat terminal domain
- RNAP II
RNA polymerase II
- CCND1
cyclin D1
References
- 1.Bocchinfuso WP, Korach KS. Mammary gland development and tumorigenesis in estrogen receptor knockout mice. J Mammary Gland Biol Neoplasia. 1997;2:323–34. doi: 10.1023/a:1026339111278. [DOI] [PubMed] [Google Scholar]
- 2.Faulds MH, Olsen H, Helguero LA, Gustafsson JA, Haldosen LA. Estrogen receptor functional activity changes during differentiation of mammary epithelial cells. Mol Endocrinol. 2004;18:412–21. doi: 10.1210/me.2003-0290. [DOI] [PubMed] [Google Scholar]
- 3.Bardin A, Boulle N, Lazennec G, Vignon F, Pujol P. Loss of ERbeta expression as a common step in estrogen-dependent tumor progression. Endocr Relat Cancer. 2004;11:537–51. doi: 10.1677/erc.1.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hewitt SC, Harrell JC, Korach KS. Lessons in estrogen biology from knockout and transgenic animals. Annu Rev Physiol. 2005;67:285–308. doi: 10.1146/annurev.physiol.67.040403.115914. [DOI] [PubMed] [Google Scholar]
- 5.Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116:561–70. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kushner PJ, Agard DA, Greene GL, et al. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74:311–7. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- 7.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–82. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 8.Cheng AS, Jin VX, Fan M, et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 9.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–68. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 10.Svejstrup JQ. The RNA polymerase II transcription cycle: cycling through chromatin. Biochim Biophys Acta. 2004;1677:64–73. doi: 10.1016/j.bbaexp.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 11.He N, Pezda AC, Zhou Q. Modulation of a P-TEFb functional equilibrium for the global control of cell growth and differentiation. Mol Cell Biol. 2006;26:7068–76. doi: 10.1128/MCB.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev. 2006;70:646–59. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garriga J, Grana X. Cellular control of gene expression by T-type cyclin/CDK9 complexes. Gene. 2004;337:15–23. doi: 10.1016/j.gene.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Wittmann BM, Wang N, Montano MM. Identification of a novel inhibitor of breast cell growth that is down-regulated by estrogens and decreased in breast tumors. Cancer Res. 2003;63:5151–8. [PubMed] [Google Scholar]
- 15.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–82. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 16.Michels AA, Fraldi A, Li Q, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–19. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byers SA, Price JP, Cooper JJ, Li Q, Price DH. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J Biol Chem. 2005;280:16360–7. doi: 10.1074/jbc.M500424200. [DOI] [PubMed] [Google Scholar]
- 18.Yik JH, Chen R, Pezda AC, Zhou Q. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of transcription. J Biol Chem. 2005;280:16368–76. doi: 10.1074/jbc.M500912200. [DOI] [PubMed] [Google Scholar]
- 19.Wittmann BM, Fujinaga K, Deng H, Ogba N, Montano MM. The breast cell growth inhibitor, estrogen down regulated gene 1, modulates a novel functional interaction between estrogen receptor alpha and transcriptional elongation factor cyclin T1. Oncogene. 2005;24:5576–88. doi: 10.1038/sj.onc.1208728. [DOI] [PubMed] [Google Scholar]
- 20.Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–21. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 21.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–26. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunther EJ, Belka GK, Wertheim GB, et al. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB J. 2002;16:283–92. doi: 10.1096/fj.01-0551com. [DOI] [PubMed] [Google Scholar]
- 23.Kershnar E, Wu SY, Chiang CM. Immunoaffinity purification and functional characterization of human transcription factor IIH and RNA polymerase II from clonal cell lines that conditionally express epitope-tagged subunits of the multiprotein complexes. J Biol Chem. 1998;273:34444–53. doi: 10.1074/jbc.273.51.34444. [DOI] [PubMed] [Google Scholar]
- 24.Fraldi A, Varrone F, Napolitano G, et al. Inhibition of Tat activity by the HEXIM1 protein. Retrovirology. 2005;2:42. doi: 10.1186/1742-4690-2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heldring N, Pike A, Andersson S, et al. Estrogen Receptors: How do they signal and what are their targets. Physiol Rev. 2007;87:905–31. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- 26.Arnold A, Papanikolaou A. Cyclin D1 in breast cancer pathogenesis. J Clin Oncol. 2005;23:4215–24. doi: 10.1200/JCO.2005.05.064. [DOI] [PubMed] [Google Scholar]
- 27.Butt AJ, McNeil CM, Musgrove EA, Sutherland RL. Downstream targets of growth factor and oestrogen signalling and endocrine resistance: the potential roles of c-Myc, cyclin D1 and cyclin E. Endocr Relat Cancer. 2005;12(Suppl):S47–S59. doi: 10.1677/erc.1.00993. [DOI] [PubMed] [Google Scholar]
- 28.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–47. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 29.Komarnitsky P, Cho E, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–60. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho E, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001;15:3319–29. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3' end processing. Mol Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 32.Cicatiello L, Addeo R, Sasso A, et al. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol Cell Biol. 2004;24:7260–74. doi: 10.1128/MCB.24.16.7260-7274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–52. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 34.Métivier R, Penot G, Hübner MR, et al. Estrogen receptor alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–63. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 35.Burakov D, Crofts LA, Chang CB, Freedman LP. Reciprocal recruitment of DRIP/Mediator and p160 coactivator complexes in vivo by estrogen receptor. J Biol Chem. 2002;277:14359–62. doi: 10.1074/jbc.C200099200. [DOI] [PubMed] [Google Scholar]
- 36.Shao W, Krasnikas E, McDonnell DP, Brown M. Coactivator AIB1 links estrogen receptor transcriptional activity and stability. Proc Natl Acad Sci U S A. 2004;101:11599–604. doi: 10.1073/pnas.0402997101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris DP, Michelotti GA, Schwinn DA. Evidence that phosphorylation of the RNA polymerase II carboxy-terminal repeats is similar in yeast and humans. J Biol Chem. 2005;280:31368–77. doi: 10.1074/jbc.M501546200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Contreras X, Barboric M, Lenasi T, Peterlin BM. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathogens. 2007;3:e146. doi: 10.1371/journal.ppat.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Epinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–12. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iankova I, Petersen RK, Annicotte JS, et al. Peroxisome proliferator-activated receptor gamma recruits the positive transcription elongation factor b complex to activate transcription and promote adipogenesis. Mol Endocrinol. 2006;20:1494–505. doi: 10.1210/me.2005-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanazawa S, Soucek L, Evan G, Okamoto T, Peterlin BM. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene. 2003;22:5707–11. doi: 10.1038/sj.onc.1206800. [DOI] [PubMed] [Google Scholar]
- 42.Sano M, Abdellatif M, Oh H, et al. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat Med. 2002;8:1310–17. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 43.Aiyar SE, Sun J, Blair AL, et al. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 2004;18:2134–46. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimizu N, Ouchida R, Yoshikawa N, et al. HEXIM1 forms a transcriptionally abortive complex with glucocorticoid receptor without involving 7SK RNA and positive transcription elongation factor b. Proc Natl Acad Sci U S A. 2005;102:8555–60. doi: 10.1073/pnas.0409863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montano MM, Doughman YQ, Deng H, et al. Mutation of the HEXIM1 gene results in defects during heart and vascular development partly through down-regulation of vascular endothelial growth factor. Circ Res. 2008;102:415–22. doi: 10.1161/CIRCRESAHA.107.157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turano M, Napolitano G, Dulac C, Majello B, Bensaude O, Lania L. Increased HEXIM1 expression during erythroleukemia and neuroblastoma cell differentiation. J Cell Physiol. 2006;206:603–10. doi: 10.1002/jcp.20502. [DOI] [PubMed] [Google Scholar]
- 47.Huang F, Wagner M, Siddiqui MA. Ablation of the CLP-1 gene leads to down-regulation of the HAND1 gene and abnormality of the left ventricle of the heart and fetal death. Mech Dev. 2004;121:559–72. doi: 10.1016/j.mod.2004.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.