Abstract
In response to evolutionary selective pressure, prokaryotes have developed a rich array of secondary metabolites, some of which may be inhibitory to the innate immune system and the inflammatory response in vertebrates. We utilized the RAW264.7 macrophage cell line stimulated with LPS in a nitric oxide (NO) assay to screen for compounds with immunomodulatory activities from a library of marine natural products, and found that the malyngamide structure class, found commonly in the marine cyanobacterium Lyngbya majuscula, has potent activity. Several of the malyngamides were found to possess IC50 values of 5.4 – 18 μM. Malyngamide F acetate exhibited strong concentration-dependent anti-inflammatory activity in the NO assay with an IC50 of 7.1 μM and with no cytotoxicity at the concentrations tested. Subsequent real-time PCR of selected genes revealed a unique cytokine profile after LPS stimulation (TLR4) with decreased expression of iNOS, IL-1β, IL-6, and IL-10, but increased TNF-α expression. Additional experiments utilizing CpG and Poly I:C stimulation to selectively activate the MyD88-dependent and -independent pathways via TLR9 and TLR3 substantiated the finding that malyngamide F acetate selectively inhibits the MyD88-dependent pathway. To our knowledge, this is the first report of a natural product inhibiting the MyD88-dependent pathway.
Keywords: Malyngamide F acetate, NO (nitric oxide), MyD88-dependent, TNF-α (tumor necrosis factor alpha), IL-6 (interleukin 6), marine natural product
1. Introduction
An enormous demand exists for new and potent anti-inflammatory drugs because inflammation underlies a multitude of human diseases, including cancer, Alzheimer's disease, and arthritis (Mantovani et al., 2008; Villoslada et al., 2008). A potential source for inhibitors of inflammation includes the rich suite of secondary metabolites from diverse bacteria (Binks and Sriprakash, 2004; Renner et al., 1999). We reasoned, however, that many marine microbes might also produce anti-inflammatory factors since a large portion of marine life has not developed an adaptive immune response and rely mainly on their innate immune system as a defense against pathogens. Thus, marine bacteria might be selectively better at evading detection by the innate immune system than their terrestrial counterparts, and production of anti-inflammatory natural products might be an important part of this evolutionary strategy. Indeed, several examples of molecules with anti-inflammatory properties have been discovered in cyanobacteria (Appleton et al., 2002; Prinsep and Thomson, 1996; Stevenson et al., 2002).
The cyanobacterium Lyngbya majuscula is a prolific producer of secondary metabolites displaying significant structural diversity and biological activity (Edwards et al., 2004; Gerwick et al., 1994; Gerwick et al., 2001, Tidgewell et al., 2009). The malyngamides, one common class of metabolite from L. majuscula, are structurally intriguing because they combine polyketide portions with presumed amino acid subunits. Since 1978, over 30 different malyngamides have been isolated and possess a wide range of biological properties such as antifeedant activity, ichthyotoxicity, toxicity to other marine animals, cytotoxicity to cancer cells, anti-HIV, anti-leukemic, and anti-tumor activity (Appleton et al., 2002; Kan et al., 1998; Orjala et al., 1995; Tan et al., 2000; Todd and Gerwick, 1995; Wan and Erickson, 1999; Wu et al., 1997; Wylie and Paul, 1988). Surprisingly, a specific molecular target of the malyngamides has yet to be identified, but it is likely that there are several due to the diversity of biological effects these elated molecules display.
Lipopolysaccharide (LPS), an endotoxin, stimulates the production of pro-inflammatory mediators such as nitric oxide (NO), tumor necrosis factor-α (TNF-α), and interleukins (Hewett and Roth, 1993; Kubes and McCafferty, 2000; Watson et al., 1999). Excessive production of these pro-inflammatory mediators is implicated in several inflammatory-related diseases (Beck and Wallace, 1997; Carroll et al., 1998; Macmicking et al., 1997; Martel-Pelletier, 1998; Norrby-Teglund and Stevens, 1998). Therefore, inhibition of the over-production of these mediators is an attractive strategy by which to treat these disease conditions.
In this study we used an in vitro NO anti-inflammatory assay with murine macrophage cells (RAW264.7) to screen a diversity of marine cyanobacterial and algal natural products from a library housed at the Scripps Institution of Oceanography. Malyngamide F and F acetate were our most potent screening hits. Subsequently, we evaluated a broader range of malyngamides present in this library to gather initial structure-activity relationships. While several of these other malyngamides were found to be active, malyngamide F acetate was the most potent and hence was further characterized as to its mechanism of action in the RAW264.7 cell culture.
2. Materials and methods
2.1 Chemicals
Defined Fetal Bovine Serum (FBS) and penicillin-streptomycin were purchased from HyClone (Logan, UT). Dulbecco's modified Eagle medium (DMEM) was purchased from Gibco (Auckland, New Zealand). Lipopolysaccharide (LPS) (Escherichia coli, serotype 026:B6), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), PCR primers, CpG (5'-TCCATGACGTTCCTGACGTT-3'), Poly I:C, and sulfanilamide were purchased from Sigma-Aldrich (St. Louis, MO). N-(1-napthyl) ethylenediamine dihydrochloride (NED) was purchased from Ricca Chemical Company (Arlington, TX). NG-nitro-l-arginine-methyl ester hydrochloride (L-NAME) was purchased from Acros Organics (Morris Plains, NJ). Malyngamides C acetate, F, F acetate, H, I, J, K, L, and T were purified as previously described and accessed from the marine algal and cyanobacterial natural products library at Scripps (Ainslie et al., 1985; Gerwick et al., 1987; Nogle and Gerwick, 2003; Orjala et al., 1995; Todd and Gerwick, 1995; Wu et al., 1997).
2.2 Cell culture
Cells from the mouse macrophage cell line RAW264.7 (ATCC; Manassas, VA) were cultured in DMEM with 4 mM L-glutamine and 4.5 g/l glucose supplemented with 10% FBS, penicillin, and streptomycin. Unless otherwise stated RAW264.7 cells were seeded in 96-well plates (5 × 104 cells/well), and after 1 day were stimulated with 3 μg/ml LPS in the absence or presence of pure compound (1 to 10 μg/ml) for 24 hours in triplicate wells at 37 °C with 5% CO2.
2.3 Nitrite assay
The generation of NO was assessed in the supernatant of cell cultures by quantification of nitrite using the Griess reaction (Green et al., 1982). In brief, 50 μl of each supernatant were added to 96-well plates together with 50 μl 1% sulfanilamide in 5% phosphoric acid plus 50 μl 0.1% NED in water, and the optical density was measured at 570 nm. The IC50 value, the sample concentration resulting in 50% inhibition of NO production, was determined using non-linear regression analysis (% nitrite versus concentration).
2.4 MTT assay
Cell viability was determined by mitochondrial-dependent reduction of MTT to formazan quantified at 570 and 630 nm. Cells were incubated with 1 mg/ml MTT at 37 °C for 25 min, the medium was aspirated, and resuspended in 100 μl DMSO for solubilization of the formazan dye. The percent survival was determined by comparison with the control group.
2.5 RNA isolation and cDNA synthesis
RAW264.7 cells were seeded in 6-well plates (8 × 105 cells/well), and after 1 day each well was stimulated with either LPS (3 μg/mL), CpG (10 μg/mL), or Poly I:C (50 μg/mL) in the absence or presence of malyngamide F acetate (10 μg/ml) in triplicate at 37 °C with 5% CO2. Total cellular RNA was isolated from cells incubated for 6 h. Cells were harvested with 1 ml of TRIZol in a 1.5 ml microcentrifuge tube. Total RNA was extracted using the TRIZol protocol for isolating RNA from cell culture (Invitrogen, Carlsbad, CA) followed by cDNA synthesis using SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA). Total RNA (2500 ng/sample) were incubated with Superscript III at 55 °C for 1 h following manufacturers protocol for First Strand Synthesis to produce the cDNAs.
2.6 Quantitative real-time polymerase chain reaction (qRT-PCR)
PCR was performed using the selective primers for mouse genes 18S, TNF-α, IL-1β, IL-6, IL-10, and iNOS genes (Table 1). Real-time quantitative PCR assays were performed in 20 μl reaction mixtures containing 10 μl SYBR® Premix Ex Taq™, 0.4 μl of ROXII™ solution (Cat # RR041A, Takara Mirus Bio Inc, Madison, WI), with primers at 200 nM final concentration, 1 μl cDNA template at 1:20 dilution, and dH2O for a final reaction volume of 20 μl. Each sample was run in triplicate using the 96-well MX3000P QPCR thermocycler (Stratagene, La Jolla, CA). Thermocycling conditions were comprised of an initial polymerase activation step at 95 °C for 30 s, followed by 40 cycles at 95 °C for 15 s, at 62 °C for 15 s, and at 72 °C for 15 s. No template controls were run in tandem to ensure that PCR signals generated were devoid of contaminating genomic DNA. For each target gene, the qRT-PCR protocol was validated by checking melting curves for the absence of primer-dimers or other unwanted amplicons. Reaction efficiencies of 94% for 18S, 97% for TNF-α, 89% for IL-1β, 92% for IL-6, 94% for IL-10, and 82% for iNOS were determined from a dilution series of cDNA ranging from 1×102 to 1×106 molecules/μl for each gene, created using RNA isolated from samples treated with LPS only. Initial data were expressed as Ct (threshold cycle) values obtained from real-time PCR. The relative quantification of TNF-α, IL-1β, IL-6, IL-10, and iNOS target genes were performed by calibrating against the mean of the control sample and normalizing to the reference gene, 18S. The MxPro v4 (Stratagene, La Jolla, CA) software package was used to analyze raw data and Gnumeric was used to perform Student's t-tests of the means and to graph the resulting fold changes.
Table 1.
Primer sequences used to quantify gene expression by real-time PCR
| Gene | Sequences |
|---|---|
| TNF-α | 5'-CCACCACGCTCTTCTGTCTA-3' (f) |
| 5'-CACTTGGTGGTTTGCTACGA-3' (r) | |
| IL-1β | 5'-TGTGAAATGCCACCTTTTGA-3' (f) |
| 5'-TGTCCTCATCCTGGAAGGTC-3' (r) | |
| IL-6 | 5'-CCGGAGAGGAGACTTCACAG-3' (f) |
| 5'-TCCAGTTTGGTAGCATCCATC-3' (r) | |
| IL-10 | 5'-TGCTATGCTGCCTGCTCTTA-3' (f) |
| 5'-ATGTTGTCCAGCTGGTCCTT-3' (r) | |
| iNOS | 5'-AACGGAGAACGTTGGATTTG-3' (f) |
| 5'-TTCTGTGCTGTCCCAGTGAG-3' (r) | |
| 18S | 5'-CGGCTACCACATCCAAGGAA-3' (f) |
| 5'-GGGCCTCGAAAGAGAGACCTGT-3' (r) | |
Primers were designed, using the program Primer 3 (vO.4.0), taking into consideration minimal secondary structure (http://frodo.wi.mit.edu/). Sequences were analyzed using BLAST to verify specificity. All oligonucleotides were optimized for PCR assays.
2.7 ELISA assay
The secretion of mouse TNF-α and IL-6 were measured by ELISA using the Ready-Set-Go! kit obtained from eBioscience (San Diego, CA). Media from three separate wells of RAW264.7 cells after treatment with malyngamide F acetate and LPS for 6 h were serially diluted and assayed and the optical density was measured at 450 nm.
2.8 Statistics
All experiments were repeated at least three times. Data are presented as the mean ± standard deviation for the indicated number of independently performed experiments. Student's t-test was used for the determination of statistical significance with P < 0.05 being considered significant.
3. Results
3.1 The effect of malyngamides on NO production
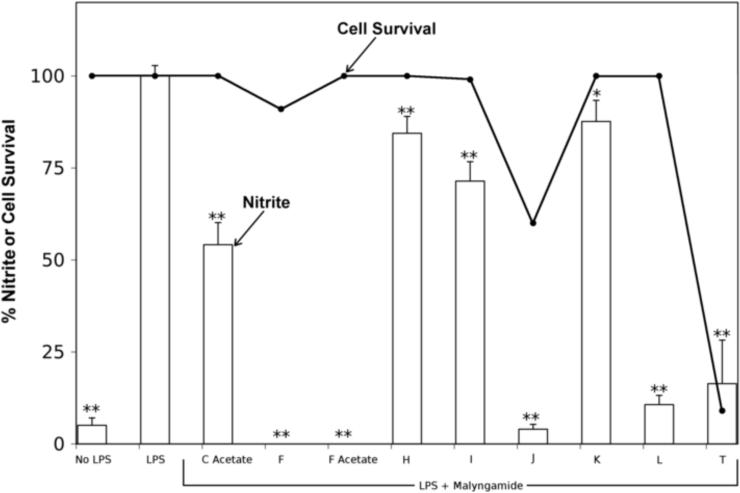
A screen of over 130 compounds from the Scripps Institution of Oceanography-UCSD library of marine natural products (Gerwick et al., 2008) for inhibition of NO production in LPS-stimulated RAW264.7 macrophage cells revealed malyngamide F and F acetate as the two most potent screening hits (0% nitrite production at 10 μg/ml of compound). Subsequently, a small library of different malyngamides (C acetate, F, F acetate, H, I, J, K, L, and T) was further screened in this assay (Fig. 1). Nitrite, one of two primary, stable, and nonvolatile breakdown products of NO, is commonly used as an indicator for NO production. In this assay system, nitrite was monitored in cultured LPS-stimulated RAW264.7 cells using the Griess reaction. LPS (3 μg/ml) markedly increased the amount of NO from basal level to 21.8 ± 0.6 μM (100% nitrite production) after 24 h of incubation (Fig. 2). Of the compounds tested, malyngamides F, F acetate, and L showed significant decreases in nitrite production. Malyngamides C acetate, H, I, and K showed a slight reduction of nitrite production, however, they were not significantly different from controls. The anti-inflammatory activity of malyngamides J and T could not be determined due to their strong cytotoxic effect (60 and 9 % cell survival, respectively).
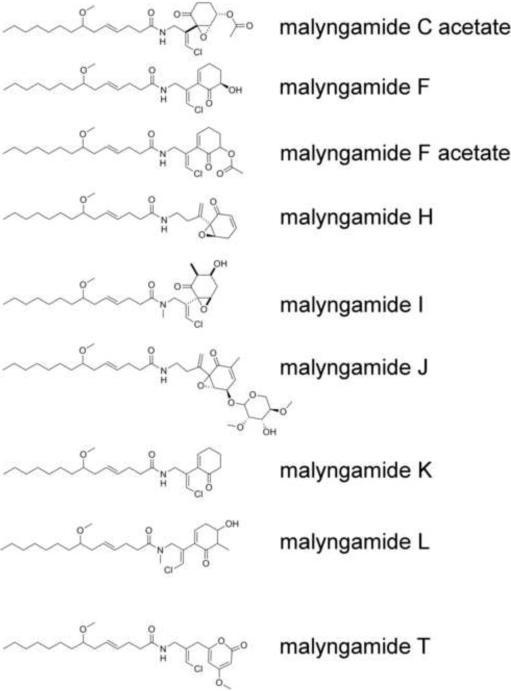
Fig. 1. Structures of the malyngamides tested in the NO and MTT assays.
The malyngamides are a structurally intriguing class of secondary metabolites which combine fatty acyl chains with polyketide synthase extended amino acid subunits, and which occur in nearly all tropical marine collections of the cyanobacterium Lyngbya majuscula.
Fig. 2. The anti-inflammatory effect of malyngamides.
Malyngamides C acetate, F, F acetate, H, I, J, K, L, and T (10 μg/ml) were incubated with RAW264.7 cells for 1 hour. Thereafter, lipopolysaccharide (LPS, 3 μg/ml) was added and the cells were incubated for 24 hours at 37 °C. The cell media was assayed for nitrite as a measure of LPS induced inflammatory response. The cells were assayed by MTT to determine cytotoxicity. Bars represent the mean ± standard deviation; N = 3. * P-value < 0.05 compared to LPS treatment alone. ** P-value < 0.01 compared to LPS treatment alone. Bars are % nitrite and ● are % cell survival.
3.2 Malyngamide F acetate, a potent inhibitor of NO production
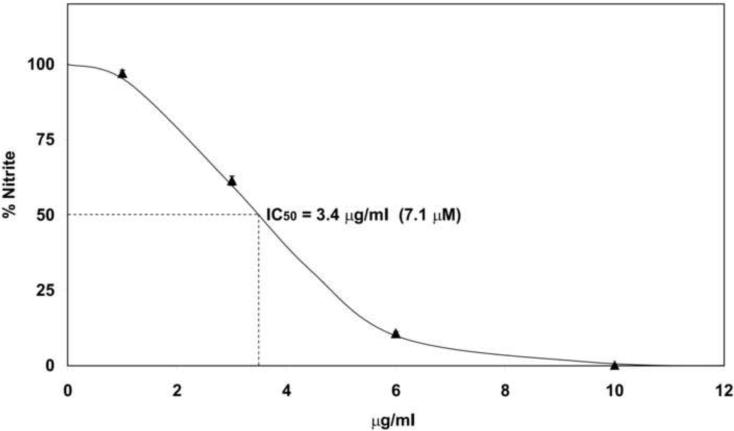
Various concentrations of malyngamide F acetate were analyzed for its effect on nitrite production in RAW264.7 cells stimulated with LPS to determine its dose-response characteristics and potency. NO production was significantly inhibited in a concentration-dependent manner with an IC50 of 3.4 μg/ml (7.1 μM) (Fig. 3). The slope of this curve suggests negative cooperativity and displayed a Hill coefficient of 0.43 an indication for a decrease in the affinity of the target enzyme for other malyngamide F acetate molecules. Malyngamides C acetate, F, J, and L also inhibited NO production in a concentration-dependent manner (supplemental data). These malyngamides had IC50 values of 18, 5.4, 7.7, and 15 μM respectively (Table 2). As an inhibitory control in this assay, L-NAME, a specific inhibitor of inducible nitric oxide synthase (iNOS), was tested and gave an IC50 of 193 μM.
Fig. 3. Concentration-dependent inhibition of NO by Malyngamide F acetate.
Malyngamide F acetate was incubated with RAW264.7 cells for 1 hour at various concentrations (10, 6, 3, and 1 μg/ml). Thereafter, lipopolysaccharide (LPS, 3 μg/ml) was added and the cells were incubated for 24 hours at 37 °C. The cell media was assayed for nitrite to determine the degree of inflammatory response. Bars represent the mean ± standard deviation; N = 3.
Table 2.
Potency of malyngamides in the Nitric Oxide Assay
| Compound | MW | IC50 (μM) |
|---|---|---|
| Malyngamide C Acetate | 497 | 18 |
| Malyngamide F | 439 | 5.4 |
| Malyngamide F Acetate | 481 | 7.1 |
| Malyngamide J | 607 | 7.7 |
| Malyngamide L | 425 | 15 |
| L-NAME | 270 | 193 |
Malyngamides C acetate, F, F acetate, J, and L were assayed as described in Figure 3. IC50 values were determined from the response curve generated from this data.
3.3 The cytokine expression pattern of malyngamide F acetate in RAW264.7 cells
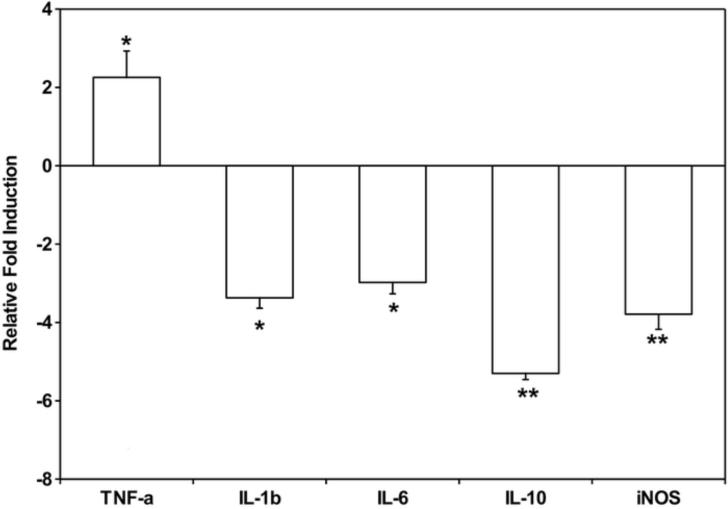
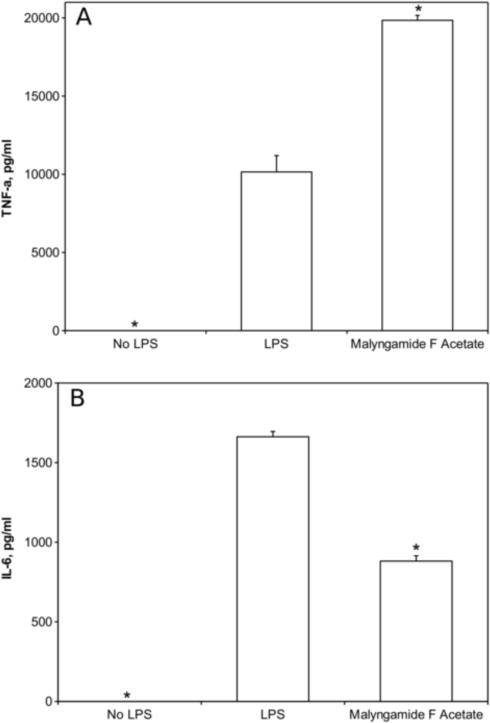
Further studies were performed to gain insight into the mechanism by which malyngamide F acetate inhibits NO production. Malyngamide F acetate was chosen over the other malyngamides tested because of its more potent NO inhibitory activity, lack of cytotoxicity, and availability. A select group of genes and proteins were chosen for analysis because they are common markers in the LPS-induced iNOS pathway. Specifically, the expression of TNF-α, IL-1β, IL-6, IL-10, and iNOS were investigated in RAW264.7 cells after treatment with malyngamide F acetate. Quantitative RT-PCR identified a decrease in expression of the iNOS transcript and showed that it was reduced almost 4-fold (Fig. 4). Interestingly, these studies revealed that IL-1β, IL-6, and IL-10 mRNA expression also decreased by about 3 – 5 fold, respectively. In contrast, TNF-α mRNA expression increased by 2-fold. ELISAs of TNF-α and IL-6 were performed on the cell media to demonstrate that the modulations observed at the transcriptional level corresponded to modulations in protein expression levels. Approximately 20 000 ± 320 pg/ml of TNF-α was detected in cell media containing LPS and malyngamide F acetate, an amount which was about twice as much as the LPS alone control (10 148 ± 1047 pg/ml), and thus was well correlated with the mRNA expression (Fig. 5A). IL-6 concentrations in cell media following treatment with LPS and malyngamide F acetate were measured at 881 ± 33 pg/ml. In LPS alone treated cells, IL-6 was detected at a concentration of 1662 ± 33 pg/ml, approximately twice the concentration as the samples that were treated with malyngamide F acetate, and thus also corroborated the mRNA expression data (Fig. 5B).
Fig. 4. Gene expression profiles of RAW264.7 cells after treatment with malyngamide F acetate as determined by quantitative RT-PCR.
Malyngamide F acetate (10 μg/ml) was incubated with RAW264.7 cells for 1 hour. Thereafter, LPS (3 μg/ml) was added and the cells were incubated for 6 hours at 37 °C. The media was aspirated, the cells were resuspended, and the RNA extracted using the TRIzol method. Next, cDNA was generated and a quantitative RTPCR was performed using primers for 18S, TNF-α, IL-1β, IL-6, IL-10, and iNOS. Samples were assayed in triplicate. Bars represent the mean ± standard deviation; N = 3. * P-value < 0.05 compared to LPS treatment alone. ** P-value < 0.01 compared to LPS treatment alone.
Fig. 5. Effects of malyngamide F acetate on TNF-α and IL-6 protein concentrations induced by LPS in RAW264.7 cells.
Cell media was isolated from the RAW264.7 murine macrophage cell line after treatment with malyngamide F acetate as described in Fig. 4 and analyzed by ELISA. Values are means ± standard deviation. * P < 0.01 compared to LPS treatment alone.
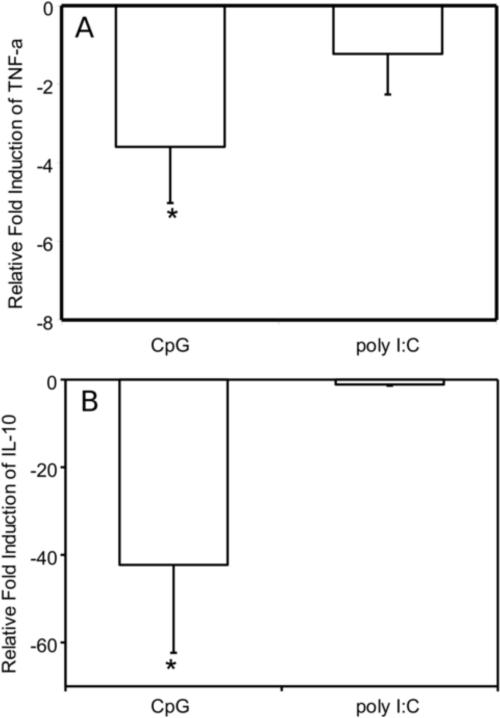
The unique cytokine profile of an increase in TNF-α transcription in contrast to decreases in IL-1β, IL-6, and IL-10 transcription suggests that malyngamide F acetate may be acting upon the MyD88-dependent inflammatory pathway. To explore this hypothesis, RAW264.7 macrophage cells were stimulated with either CpG (MyD88-dependent) or Poly I:C (MyD88-independent) with and without malyngamide F acetate. TNF-α transcripts experienced about a 4-fold reduction in expression upon treatment with malyngamide F acetate in cells stimulated with CpG while the samples that were treated with Poly I:C did not show a statistically significant change (Fig. 6A). Poly I:C treatment of the cells did exhibit an increase in TNF-α expression (Ct value of 22.8) compared to cells without any Poly I:C (Ct value of 26.9). Analysis of IL-10 transcripts showed that expression was reduced about 40-fold upon treatment with malyngamide F acetate in cells stimulated with CpG while samples that were treated with Poly I:C did not show a statistically significant fold change (Fig. 6B). In addition, the Ct values for the IL-10 transcript were very high (31) an indication of no real IL-10 activation by Poly:IC.
Fig. 6. Effects of malyngamide F acetate on TNF-α and IL-10 expression profiles in RAW264.7 cells after stimulation with CpG and Poly I:C as determined by quantitative RT-PCR.
Malyngamide F acetate (10 μg/ml) was incubated with RAW264.7 cells for 1 hour. Thereafter, CpG (10 μg/ml) or Poly I:C (50 μg/ml) was added and the cells were incubated for 6 hours at 37 °C. The media was aspirated, the cells were resuspended, and the RNA extracted using the TRIzol method. Next, cDNA was generated and a quantitative RT-PCR was performed using primers for 18S, TNF-α, and IL-10. Samples were assayed in triplicate. Bars represent the mean ± standard deviation; N = 3. * P-value < 0.01 compared to LPS treatment alone.
4. Discussion
A screening campaign of diverse marine natural products from marine algae and cyanobacteria for their potential anti-inflammatory activity, using the inhibition of NO formation assay in RAW264.7 cells, led to the initial discovery that malyngamide F and F acetate were relatively potent and had minimal cytotoxicity. Hence, a broader evaluation of the malyngamide structure class for anti-inflammatory activity was initiated, and an evaluation of cytokine modulation by the most potent in the series, malyngamide F acetate, was undertaken. The primary assay used in this investigation, LPS stimulation of the murine macrophage cell line RAW264.7, leads to NO formation resulting from iNOS activity. This end result is primarily initiated by signaling cascades in the toll-like receptor 4 (TLR4) pathways.
Following the initial screening hit, the next goal of this study was to determine the relative potency of the various malyngamides present in our library in the LPS-stimulated NO production assay. We found that malyngamides F, F acetate, J, L, and T displayed relatively potent inhibition of NO production (Fig. 2). Malyngamides J and T, however, were found to inhibit NO production largely as a consequence of their cytotoxicity. Structurally, the malyngamides tested are very similar in their overall molecular architecture in that they share the same methoxy-containing fatty acyl chain and have an oxidized cyclohexyl ring that appears to derive from an amino acid modified with several polyketide synthase (PKS) extensions (Fig. 1). It is quite clear that the differences in NO inhibition among the malyngamides are due to structural variations involving these latter PKS extended amino acid subunits. Furthermore, the activities of malyngamides F and F acetate can be specifically attributed to the hydroxyl or acetoxyl substituents at C-6 on the cyclohexenone ring because malyngamide K, which does not possess these substituents, lacked any ability to induce NO inhibition. These results suggest that the C-6 hydroxyl or acetoxyl group on a malyngamide skeleton is mechanistically involved in producing the observed anti-inflammatory activity in this structure class (Fig. 1). Furthermore, it is possible that malyngamide F acetate is a prodrug which is cleaved by cellular esterases to malyngamide F, and that its greater potency is a reflection of its increased cellular uptake.
Further investigation of the immunomodulatory properties of this structure class utilized malyngamide F acetate because of its greater potency as an inhibitor of NO production, lack of cytotoxicity, and adequate quantities available for further testing. Malyngamide F acetate was found to inhibit the production of NO in LPS-stimulated RAW264.7 cells in a concentration-dependent manner and was bound non-cooperatively (Fig. 3). Additionally, reduction in iNOS mRNA expression in the RAW264.7 cells after treatment with malyngamide F acetate suggests it acts upstream of the transcription of the iNOS enzyme (Fig. 4).
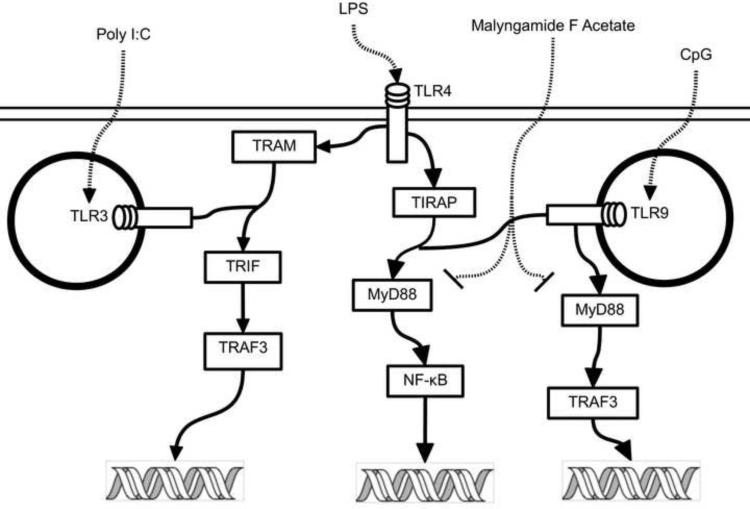
In addition to iNOS, the expression of a number of other potentially relevant cytokines was investigated. We hypothesized that expression of TNF-α, IL-1β, and IL-6 would decrease after treatment with these anti-inflammatory malyngamides in RAW264.7 cells (Chen et al., 2008). Cytokine expression with malyngamide F acetate, however, exhibited an interesting and unexpected pattern where IL-1β, IL-6 and IL-10 were all down-regulated while TNF-α was upregulated (Fig.s 4 and 5). Based on these observations, it is possible that malyngamide F acetate may be acting on a target in the MyD88-dependent pathway because it has been shown that LPS stimulation decreases IL-1β and increases TNF-α transcription in the MyD88 knockout mouse by means of the MyD88-independent pathway (TRIF-dependent) (Covert et al., 2005; Kawai et al., 2001; Kawai and Akira, 2007). In addition, high TNF-α transcript levels could be partially attributed to low transcriptional levels of IL-6 and IL-10 since both of these cytokines have been implicated in the inhibition TNF-α. (Benveniste et al., 1995, Rajasingh et al., 2006). To further decipher the nature of malyngamide F acetate's interaction with the MyD88-dependent and -independent pathways, transcription of TNF-α and IL-10 was measured in CpG and Poly I:C treated cells (Fig. 6). TNF-α and IL-10 were both down regulated at the transcriptional level in the CpG treated cells. If malyngamide F acetate is indeed inhibiting the MyD88-dependent pathway then activation of TLR9, which is exclusively activated by CpG and depends solely on the MyD88-dependent pathway (Boonstra et al, 2006, Kawai and Akira, 2007), would deliver a biological response with lower levels of TNF-α and IL-10 transcripts. Furthermore, when the macrophages were treated with Poly I:C, which activates TLR3 via a MyD88-independent pathway, no difference in TNF-α and IL-10 transcription between control and treated cells were seen. Indeed, there was no indication that Poly I:C treatment increased the transcription of IL-10 at all. This is in accordance with earlier observations by Ando et al. (2008). These results strongly suggest that the MyD88-dependent pathway was inhibited while the TRIF-dependent pathway was not (Fig. 7). Some examples of selective inhibition of MyD88-dependent and -independent pathway inhibition exist. For example tumor growth factor-β selectively inhibits the MyD88-dependent pathway (Naiki et al., 2005) and the natural product resveratrol has been shown to selectively inhibit the MyD88-independent pathway (TRIF) (Youn et al., 2005). To our knowledge though, this is the first report of a natural product that selectively inhibits the MyD88-dependent pathways of TLR4 and 9.
Figure 7. A simplified depiction of the intracellular pathways for TLR3, 4, and 9 that illustrates the selectivity of malyngamide F acetate inhibition.
Malyngamide F acetate acts as an inhibitor in the TRL4 and 9 MyD88-dependent pathways which are stimulated by LPS and CpG. The TLR3 MyD88-independent pathway, however, is not inhibited by malyngamide F acetate upon stimulation by Poly I:C.
Malyngamide F acetate should be further investigated when an additional supply of the compound is obtained, either from natural sources or chemical synthesis. These investigations should include activation and phosphorylation patterns of proteins involved in the TLR4 and the TNF-α MyD88-dependent pathways, such as MyD88, TRAF-3, TRAF-6 and IRF-3. In addition it will be useful to fluorescently label malyngamide F acetate, utilizing protocols developed by Hughes et al. (2009), in an effort to identify the protein target for this compound since we can envision that malyngamide F acetate could be a very useful tool in cell biology or as a potential immunomodulatory agent in the treatment of disease.
5. Conclusion
In summary, this study provides evidence that a select group of malyngamides inhibit NO production in LPS-stimulated murine macrophage cells (RAW264.7). Malyngamide F acetate from the marine cyanobacterium L. majuscula demonstrated the most potent concentration-dependent inhibition of NO. Further investigation of malyngamide F acetate showed that its engagement of TLR4 suppressed IL-1β, IL-6, IL -10, and iNOS transcription and increased TNF-α transcription. On the other hand CpG activation of TLR9 inhibited the transcription of all the cytokines tested including TNF-α. Furthermore, engagement of TLR3 (Poly I:C treatment) with and without malyngamide F acetate treatment did not change the transcription of TNF-α and IL-10 indicating that malyngamide F acetate selectively inhibits the MyD88-dependent pathway. The cytokine profile exhibited after malyngamide F acetate treatment is unique, and thus further use of this compound could be useful as a tool in cellular biology.
Acknowledgments
We would like to thank W. H. Gerwick for access to the Scripps Institution of Oceanography library of marine natural products. This work was supported in part by a NIH/NIGMS Institutional Research and Academic Career Development Award (IRACDA) to Francisco A. Villa, UCSD Chancellor's Research Scholarship to Kelly Lieske, and NIH grant CA100851.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ando M, Tu W, Nishijima, Iijima S. Siglec-9 enhances IL-10 production in macrophages via tyrosine-based motifs. Biochemical and Biophysical Research Communications. 2008;369:878–883. doi: 10.1016/j.bbrc.2008.02.111. [DOI] [PubMed] [Google Scholar]
- Ainslie RD, Barchi JJ, Kuniyoshi M, Moore RE, Mynderse JS. Structure of malyngamide-C. Journal of Organic Chemistry. 1985;50:2859–2862. [Google Scholar]
- Appleton DR, Sewell MA, Berridge MV, Copp BR. A new biologically active malyngamide from a New Zealand collection of the sea hare Bursatella leachii. Journal of Natural Products. 2002;65:630–631. doi: 10.1021/np010511e. [DOI] [PubMed] [Google Scholar]
- Beck PL, Wallace JL. Cytokines in inflammatory bowel disease. Mediators of Inflammation. 1997;6:95–103. doi: 10.1080/09629359791785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste EN, Tang PL, Law RM. Differential regulation of astrocyte TNF-α expression by the cytokines TGF-β, IL-6 and IL-10. International Journal of Developmental Neuroscience. 1995;13(3–4):341–349. doi: 10.1016/0736-5748(94)00061-7. [DOI] [PubMed] [Google Scholar]
- Binks M, Sriprakash KS. Characterization of a complement-binding protein, DRS, from strains of Streptococcus pyogenes containing the emm12 and emm55 genes. Infection and Immunity. 2004;72:3981–3986. doi: 10.1128/IAI.72.7.3981-3986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonstra A, Rajsbaum R, Holman M, Marques R, Asselin-Paturel C, Pereira JP, Bates EME, Akira S, Vieira P, Liu JY, Trinchieri G, O'Garra A. Macrophages and myeloid dendritic cells, but not plasmacytoid dendritic cells, produce IL-10 in response to MyD88- and TRIF-dependent TLR signals, and TLR-independent signals. Journal of Immunology. 2006;177:7551–7558. doi: 10.4049/jimmunol.177.11.7551. [DOI] [PubMed] [Google Scholar]
- Carroll G, Bell M, Wang H, Chapman H, Mills J. Antagonism of the IL-6 cytokine subfamily - a potential strategy for more effective therapy in rheumatoid arthritis. Inflammation Research. 1998;47:1–7. doi: 10.1007/s000110050235. [DOI] [PubMed] [Google Scholar]
- Chen D, Nie M, Fan MW, Bian Z. Anti-inflammatory activity of curcumin in macrophages stimulated by lipopolysaccharides from Porphyromonas gingivalis. Pharmacology. 2008;82:264–269. doi: 10.1159/000161127. [DOI] [PubMed] [Google Scholar]
- Covert MW, Leung TH, Gaston JE, Baltimore D. Achieving stability of lipopolysaccharide-induced NF-kappa B activation. Science. 2005;309:1854–1857. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscula. Chemistry & Biology. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- Gerwick WH, Coates RC, Engene N, Gerwick LG, Grindberg RV, Jones AC, Sorrels CM. Giant marine cyanobacteria offer exciting pharmaceutical potential. Microbe. 2008;3:277–284. [Google Scholar]
- Gerwick WH, Proteau PJ, Nagle DG, Hamel E, Blokhin A, Slate DL. Structure of Curacin-A, a novel antimitotic, antiproliferative, and brine shrimp toxic natural product from the marine cyanobacterium Lyngbya-majuscula. Journal of Organic Chemistry. 1994;59:1243–1245. [Google Scholar]
- Gerwick WH, Reyes S, Alvarado B. 2 Malyngamides from the Caribbean cyanobacterium Lyngbya-majuscula. Phytochemistry. 1987;26:1701–1704. [Google Scholar]
- Gerwick W, Tan L, Sitachitta N. Nitrogen-containing metabolites from marine cyanobacteria. Alkaloids Chem Biol. 2001;57:75–184. doi: 10.1016/s0099-9598(01)57003-0. [DOI] [PubMed] [Google Scholar]
- Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR. Analysis of nitrate, nitrite, and [N-15]-labeled nitrate in biological-fluids. Analytical Biochemistry. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- Hewett JA, Roth RA. Hepatic and extrahepatic pathobiology of bacterial lipopolysaccharides. Pharmacological Reviews. 1993;45:381–411. [PubMed] [Google Scholar]
- Hughes CC, Yang YL, Liu WT, Dorrestein PC, La Clair JJ, Fenical W. Marinopyrrole A target elucidation by acyl dye transfer. Journal of the American Chemical Society. 2009;131:12094–12096. doi: 10.1021/ja903149u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Y, Fujita T, Nagai H, Sakamoto B, Hokama Y. Malyngamides M and N from the Hawaiian red alga Gracilaria coronopifolia. Journal of Natural Products. 1998;61:152–155. doi: 10.1021/np970423n. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Signaling to NF-kappa B by Toll-like receptors. Trends in Molecular Medicine. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takeuchi O, Fujita T, Inoue J, Muhlradt PF, Sato S, Hoshino K, Akira S. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. Journal of Immunology. 2001;167:5887–5894. doi: 10.4049/jimmunol.167.10.5887. [DOI] [PubMed] [Google Scholar]
- Kubes P, McCafferty DM. Nitric oxide and intestinal inflammation. American Journal of Medicine. 2000;109:150–158. doi: 10.1016/s0002-9343(00)00480-0. [DOI] [PubMed] [Google Scholar]
- MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annual Review of Immunology. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- Martel-Pelletier J. Pathophysiology of osteoarthritis. Osteoarthritis and Cartilage. 1998;6:374–376. doi: 10.1053/joca.1998.0140. [DOI] [PubMed] [Google Scholar]
- Naiki Y, Michelsen KS, Wzhang W, Chen S, Doherty TM, Arditi M. Transforming growth factor-beta differentially inhibits MyD88-dependent, but not TRAM- and TRIF-dependent, lipopolysaccharide-induced TLR4 signaling. Journal of Biological Chemistry. 2005;280:5491–5495. doi: 10.1074/jbc.C400503200. [DOI] [PubMed] [Google Scholar]
- Nogle LM, Gerwick WH. Diverse secondary metabolites from a Puerto Rican collection of Lyngbya majuscula. Journal of Natural Products. 2003;66:217–220. doi: 10.1021/np020332c. [DOI] [PubMed] [Google Scholar]
- Norrby-Teglund A, Stevens DL. Novel therapies in streptococcal toxic shock syndrome: attenuation of virulence factor expression and modulation of the host response. Current Opinion in Infectious Diseases. 1998;11:285–291. doi: 10.1097/00001432-199806000-00004. [DOI] [PubMed] [Google Scholar]
- Orjala J, Nagle D, Gerwick WH. Malyngamide-H, an ichthyotoxic amide possessing a new carbon skeleton from the Caribbean cyanobacterium Lyngbya-majuscula. Journal of Natural Products-Lloydia. 1995;58:764–768. doi: 10.1021/np50119a019. [DOI] [PubMed] [Google Scholar]
- Prinsep MR, Thomson RA, West ML, Wylie BL. Tolypodiol, an anti-inflammatory diterpenoid from the cyanobacterium Tolypothrix nodosa. Journal of Natural Products. 1996;59:786–788. doi: 10.1021/np9602574. [DOI] [PubMed] [Google Scholar]
- Rajasingh J, Bord E, Luedemann C, Asai J, Hamada H, Thorne T, Qin G, Goukassian D, Zhu Z, Losordo DW, Kishore R. IL-10-induced TNF-alpha mRNA destabilization is mediated via IL-10 suppression of p38 MAP kinase activation and inhibition of HuR expression. FASEB Journal. 2006;20:2112–2114. doi: 10.1096/fj.06-6084fje. [DOI] [PubMed] [Google Scholar]
- Renner MK, Shen YC, Cheng XC, Jensen PR, Frankmoelle W, Kauffman CA, Fenical W, Lobkovsky E, Clardy J. Cyclomarins A-C, new antiinflammatory cyclic peptides produced by a marine bacterium (Streptomyces sp.) Journal of the American Chemical Society. 1999;121:11273–11276. [Google Scholar]
- Stevenson CS, Capper EA, Roshak AK, Marquez B, Grace K, Gerwick WH, Jacobs RS, Marshall LA. Scytonemin - a marine natural product inhibitor of kinases key in hyperproliferative inflammatory diseases. Inflammation Research. 2002;51:112–114. doi: 10.1007/BF02684014. [DOI] [PubMed] [Google Scholar]
- Tan LT, Okino T, Gerwick WH. Hermitamides A and B, toxic malyngamide-type natural products from the marine cyanobacterium Lyngbya majuscula. Journal of Natural Products. 2000;63:952–955. doi: 10.1021/np000037x. [DOI] [PubMed] [Google Scholar]
- Tidgewell K, Clark BR, Gerwick WH. The Natural Products Chemistry of Cyanobacteria. In: Moore B, Crews P, editors. Comprehensive Natural Products Chemistry. second ed. Elsevier; Oxford: 2009. In Press. [Google Scholar]
- Todd JS, Gerwick WH. Malyngamide-I from the tropical marine cyanobacterium Lyngbya-majuscula and the probable structure revision of Stylocheilamide. Tetrahedron Letters. 1995;36:7837–7840. [Google Scholar]
- Villoslada P, Moreno B, Melero I, Pablos JL, Martino G, Uccelli A, Montalban X, Avila J, Rivest S, Acarin L, Appel S, Khoury SJ, McGeer P, Ferrer I, Delgado M, Obeso J, Schwartz M. Immunotherapy for neurological diseases. Clinical Immunology. 2008;128:294–305. doi: 10.1016/j.clim.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Wan F, Erickson KL. Serinol-derived malyngamides from an Australian cyanobacterium. Journal of Natural Products. 1999;62:1696–1699. doi: 10.1021/np990291t. [DOI] [PubMed] [Google Scholar]
- Watson WH, Zhao YM, Chawla RK. S-adenosylmethionine attenuates the lipopolysaccharide-induced expression of the gene for tumour necrosis factor alpha. Biochemical Journal. 1999;342:21–25. [PMC free article] [PubMed] [Google Scholar]
- Wu M, Milligan KE, Gerwick WH. Three new malyngamides from the marine cyanobacterium Lyngbya majuscula. Tetrahedron. 1997;53:15983–15990. [Google Scholar]
- Wylie CR, Paul VJ. Feeding preferences of the surgeonfish Zebrasoma-flavescens in relation to chemical defenses of tropical algae. Marine Ecology-Progress Series. 1988;45:23–32. [Google Scholar]
- Youn HS, Lee JY, Fitzgerald KA, Young HA, Akira S, Hwang DH. Specific inhibition of MyD88-independent signaling pathways of TLR3 and TLR4 by resveratrol: Molecular targets are TBK1 and RIP1 in TRIF complex1. Journal of Immunology. 2005;175:3339–3346. doi: 10.4049/jimmunol.175.5.3339. [DOI] [PubMed] [Google Scholar]