Abstract
Background
Phthalates can alter steroidogenesis and peroxisome proliferator–activated receptor gamma (PPARγ)–mediated transcription in rodent tissues. The placenta offers a rich source of biomarkers to study these relationships in humans.
Objective
We evaluated whether gestational phthalate exposures in humans were associated with altered human placental steroidogenesis and trophoblast differentiation as measured by markers of mRNA transcription.
Methods
We measured seven target genes in placentas collected from 54 Dominican and African-American women at delivery in New York City using quantitative real-time polymerase chain reaction (qPCR), normalized to 18S rRNA. qPCR results for the target genes were log-transformed, converted to Z-scores, and grouped into two functional pathways: steroidogenesis (aromatase, cholesterol side chain cleavage enzyme, 17β-hydroxysteroid dehydrogenase type 1, and cytochrome P450 1B1) and trophoblast differentiation (PPARγ, aryl hydrocarbon receptor, and human chorionic gonadotropin). Repeated measures models were used to evaluate the association of phthalate metabolites measured in third-trimester urine samples with each group of target genes, accounting for correlation among the genes within a pathway.
Results
Higher urinary concentrations of five phthalate metabolites were associated with lower expression of the target genes reflecting trophoblast differentiation. Results were less consistent for genes in the steroidogenesis pathway and suggested a nonlinear dose–response pattern for some phthalate metabolites.
Conclusions
We observed a significant association between prenatal exposure to phthalates and placental gene expression within two pathways. Further studies are warranted to understand the significance of this association with respect to fetal development and placental function.
Keywords: epidemiology, gene expression, phthalates, placenta, pregnancy, prenatal, steroidogenesis, trophoblast differentiation
Phthalates are synthetic chemicals used in plastics, personal care products, and building materials. Phthalates are metabolized and excreted in urine 12–48 hr after exposure (Koch et al. 2005). Urinary phthalate metabolites measured have been used extensively as biomarkers of human exposure (Hauser and Calafat 2005).
The metabolites of at least four phthalates can induce a wide range of responses with potential relevance to human reproduction and development (Hauser and Calafat 2005; Swan 2008). When given to rats in utero, di(2-ethylhexyl) phthalate (DEHP), di-n-butyl phthalate (DnBP), diisobutyl phthalate (DiBP), and butylbenzyl phthalate (BBzP) altered steroidogenesis and produced a phenotype that involved reduced testosterone synthesis in fetal Leydig cells and concomitant effects on the male offspring resembling the testicular dysgenesis syndrome (Barlow et al. 2003; Ema et al. 2000; Mylchreest et al. 2000; Saillenfait et al. 2008; Sharpe 2001). Similarly, in female rats, DEHP, DnBP, and DiBP produced alterations in ovarian steroidogenesis, partly mediated by CYP19 (aromatase) inhibition and progesterone inhibition (Boberg et al. 2008; Davis et al. 1994a, 1994b; Lovekamp and Davis 2001).
The peroxisome proliferator–activated receptor protein gamma (PPARγ) is a master regulator of several reproductive and developmental pathways, including steroidogenesis, differentiation, fatty acid uptake and transport, and inflammation related to parturition (Froment et al. 2006; Schaiff et al. 2006). PPARγ is expressed in the human trophoblast and is essential to basic placental development and function (Fournier et al. 2007). Agonism of PPARγ by phthalate metabolites has been proposed as a mechanism of action (Lovekamp-Swan et al. 2003; Maloney and Waxman 1999). In vivo prenatal exposure to a metabolite of DEHP resulted in dose-dependent activation of PPARγ in rat placenta and changes in the expression of its downstream targets, including fatty acid transport protein and cytochrome oxidase-2 (Xu et al. 2008).
Prenatal exposure to DEHP has been associated with the timing of labor in four different cohorts, suggesting phthalate-related disruption in placental function (Adibi et al. 2009a; Latini et al. 2003; Whyatt et al. 2009; Wolff et al. 2008). We hypothesized that phthalate exposure could also be associated with alterations in human placental development and function, through PPARγ-mediated pathways (e.g., trophoblast differentiation).
We selected specific gene targets unified in two pathways based on their strong association with placental health and function and direct experimental evidence of their disruption by DEHP, DnBP, DiBP, or BBzP. Our criteria included targets downstream of those disrupted by phthalates and/or hypothesized to be disrupted by phthalates given their known interactions with structurally similar compounds (Schaiff et al. 2006). For steroidogenesis, target genes included CYP19, P450 cholesterol side chain cleavage enzyme (P450scc), 17β-hydroxysteroid dehydrogenase type 1 (17β-HSD), and cytochrome P450 1B1 (CYP1B1). For trophoblast differentiation, target genes included PPARγ, human chorionic gonadotropin (HCG), and the aryl hydrocarbon receptor (AhR).
Materials and Methods
Study participants
Fifty-four of 148 participants in the Columbia Center for Children’s Environmental Health (CCCEH) with placentas collected at delivery between May 2002 and June 2005 and with both maternal urine samples and medical record data were included this study. Placentas were sampled from 70% (148 of 211) of the 211 CCCEH births over this time; the remainder could not be obtained because of lack of notification of labor onset and other logistical obstacles. CCCEH participants were enrolled through the prenatal clinics at New York Presbyterian and Harlem Hospital Centers in New York City. To be eligible, the woman had to reside in the study area for at least 1 year, receive her first prenatal visit by the 20th week of pregnancy, and be free of diabetes, hypertension, and known HIV and drug or alcohol abuse (Perera et al. 2006; Whyatt et al. 2003). The institutional review boards of Columbia University, the Centers for Disease Control and Prevention (CDC), and the Harvard School of Public Health Human Subjects Committee approved the CCCEH study and substudies. Written informed consent was obtained from all study participants.
Placenta sampling
The 54 placentas were sampled between 4 min and 2 hr after delivery. Samples of chorionic villi were taken on the fetal side of the placenta. One sample was taken from the inner region proximal to the umbilical cord insertion point, and one from the outer region closer to the edge, yielding two samples per placenta. Methods and rationale for the sampling scheme are described in detail elsewhere (Adibi et al. 2009b). We attempted to control for within-placenta variability by collecting one sample from the inner and one from the outer region of each placenta. Care was taken in dissection to maximize the amount of villous tissue in the sample and to avoid membrane contamination as well as decidua contamination. Samples were preserved in RNAlater (Ambion, Austin, TX) to stabilize the RNA and stored at − 80°C.
RNA analysis
Total RNA was isolated from approximately 300 mg of tissue using the RNeasy Midi Kit (Qiagen, Valencia, CA). Genomic DNA contamination in the sample was minimized with a DNase digestion step (Rozen and Skaletsky 2000). Total RNA was measured by determining absorbance at 260 nm using an Ultrospec 2100 pro ultraviolet/visible spectrophotometer (GE Healthcare, Piscataway, NJ). RNA purity was assessed by the ratio of absorption at 260 nm to 280 nm and visually by agarose gel electrophoresis. Approximately 3 μg total RNA was used in a reverse transcription (RT) reaction to synthesize cDNA using the SuperScript First-Strand Synthesis System from Invitrogen (Carlsbad, CA). Finally, quantitative real-time polymerase chain reaction (qPCR) was used to quantitate mRNA levels in each sample for individual genes. Ribosomal RNA from 18S was selected as a housekeeping gene to serve as an internal control for quantity and quality of cDNA going into the RT reaction, based on the results of a previous analysis (Adibi et al. 2009b). All samples were analyzed using the ABI Prism 7500 Sequence Detection System (Applied Biosystems, Foster City, CA). Cycling conditions were the same for seven transcripts (18S, CYP19, PPARγ, AhR, CYP1B1, 17β-HSD, HCG): 95.0°C for 5 min for activation of the enzyme, 95.0°C for 30 sec for denaturation and 60.0°C for 1 min for annealing/extension for 40 cycles, followed by a dissociation step. Cycling conditions for P450scc were the same except the annealing/extension was carried out at 55.0°C for 1 min. Forward and reverse primers (Sigma, St. Louis, MO) were either designed by Primer3 (Rozen and Skaletsky 2000) or selected from the literature and referenced by a PubMed Identifier (PMID; Table 1). Each reaction used 2 μL or 90 ng cDNA assuming 90% efficiency of the cDNA synthesis reaction, forward and reverse primers at optimized concentrations, and SYBR Green PCR Master Mix kit (Applied Biosystems, Foster City, CA) for a total reaction volume of 25 μL.
Table 1.
Primer sets for qPCR analysis.
| Gene | Direction | Primer sequence | Base pairs | Reference |
|---|---|---|---|---|
| CYP19 | Forward | ATACCAGGTCCTGGCTACTG | 249 | PMID 11745463a |
| Reverse | TCTCATGCATACCGATGCACTG | |||
| PPARγ | Forward | GCTGTGCAGGAGATCACAGA | 225 | Designed in Primer3 |
| Reverse | GGGCTCCATAAAGTCACCAA | |||
| AhR | Forward | ACATCACCTACGCCAGTCGC | 101 | PMID 12520072b |
| Reverse | TCTATGCCGCTTGGAAGGAT | |||
| CYP1B1 | Forward | ACCGTTTTCCGCGAATTC | 196 | PMID 12040753c |
| Reverse | GTACGTTCTCCAAATCCAGCC | |||
| P450scc | Forward | CCTGCAGTGGCACTTGTATG | 418 | PMID 11997174d |
| Reverse | GGTCATCTCTAGCTCAGCGA | |||
| 17β-HSD type 1 | Forward | GCCGCGTGGACGTGCTGGTGTGTAAC | 201 | PMID 12242730e |
| Reverse | CCATCAATCCTCCCACGCTCCCGG | |||
| HCG | Forward | TCCCACTCCACTAAGGTCCAA | 106 | PMID 15081642f |
| Reverse | CCCCATTACTGTGACCCTGTT | |||
| 18S rRNA | Forward | CGGCTACCACATCCAAGGAA | 187 | PMID 14583453g |
| Reverse | GCTGGAATTACCGCGGCT |
Koh et al. 2002.
Pidoux et al. 2004.
Specificity and quantitation
Each sample was run in duplicate, and values not falling within 50% of their mean were rerun. Specificity of the PCR product was evaluated using the melting curve generated at the end of amplification and by running a 2% agarose gel to visualize the PCR product. Absolute quantitation of mRNA concentration in the original sample was achieved using a standard curve generated for each batch. Each standard curve included two nontemplate controls and eight serial dilutions covering the range of 103 to 107 molecules/μL. The standards for each gene were prepared as described previously (Bhat and Epelboym 2004). The R2 for the standard curve was between 0.98 and 1.00; the plate was rerun if it was < 0.95. The ratio of target gene mRNA molecules to 18S mRNA molecules was calculated for each.
Phthalate metabolite measurements
Maternal urine samples, collected in the early third trimester (n = 54), were analyzed for the four DEHP metabolites mono(2-ethylhexyl) phthalate (MEHP), mono(2-ethyl-5-oxohexyl) phthalate (MEOHP), mono(2-ethyl-5-hydroxyhexyl) phthalate (MEHHP), and mono-2-ethyl-5-carboxypentyl phthalate (MECPP); the DnBP metabolite mono-n-butyl phthalate (MnBP); the DiBP metabolite monoisobutyl phthalate (MiBP); and the BBzP metabolite monobenzyl phthalate (MBzP) at the CDC. Because of the high correlations among MEOHP, MEHHP, and MECPP (Spearman correlation r = 0.96–0.98), we limited our analysis to MEOHP. To represent total DEHP urinary concentration, we summed the four DEHP metabolites (MEHP, MEOHP, MEHHP, MECPP) in nanomoles per liter (∑DEHP).
The analytical approach involved enzymatic deconjugation of the phthalate metabolites from their glucuronidated form, solid-phase extraction, separation with high-performance liquid chromatography, and detection by isotope-dilution tandem mass spectrometry (Kato et al. 2005). To monitor for accuracy and precision, each analytical run included the unknown samples together with calibration standards, reagent blanks, and quality control materials of high and low concentration. The limits of detection (LODs) were (in nanograms per milliliter) MEHP, 0.9; MEOHP, 0.45; MEHHP, 0.32; MECPP, 0.25; MiBP, 0.26; MnBP, 0.40; and MBzP, 0.11. Concentrations below the LOD were set to one-half the LOD for statistical analysis. Specific gravity was measured at CDC using a PAL 10-S hand-held refractometer (Atago, Bellevue, WA). Urinary concentrations were adjusted for specific gravity using a modification of the formula by Hauser et al. (2004): Pc = P × [(1.016 − 1)/(SG − 1)], where Pc is the specific-gravity–corrected phthalate concentration, P is the observed phthalate concentration, and SG is the specific gravity of the urine samples.
Statistical analysis
Gene expression values and phthalate metabolite concentrations were log transformed to approximate a normal distribution. Z-scores were calculated individually for each gene to put them on the same scale. The Z-score, also called the standard score, uses the population mean and standard deviation to standardize or normalize sample values so that they share a common underlying distribution. Z-scores were modeled in two groups by functional pathways, steroidogenesis and trophoblast differentiation. Spearman correlation coefficients were used to estimate pairwise associations among gene transcripts. Variance component analysis was used to evaluate between- versus within-placenta variability.
Multivariate mixed effects models were used to estimate associations between gene expression and specific gravity–adjusted phthalate metabolite concentrations (Fitzmaurice et al. 2004; Verbeke and Molenberghs 2000). Gene expression Z-scores for multiple genes within a pathway were modeled as a correlated vector of eight responses within each placenta for the steroidogenesis pathway (i.e., two samples per placenta and four target genes measured on each sample) and six responses for trophoblast differentiation (i.e., two samples per placenta and three target genes measured on each sample) as a function of phthalate concentrations and other covariates. The mixed effect model approach allows for the analysis of the expression of each gene separately while taking into account differences between genes within a pathway, and simultaneously adjusting for the correlation between the two samples and three to four target genes measured on a single placenta. We assumed equal correlation between any two responses measured within a single placenta. We grouped the gene transcripts by pathway to most efficiently use information collected from each placenta and to increase our statistical power to detect associations with phthalate exposures. Urinary phthalate concentrations were considered both as log-transformed continuous measures (assuming linearity of dose response) and as grouped into quintiles to avoid the assumption of linearity. We used the fitted mixed effect model to calculate the predicted mean Z-scores and standard errors for each quintile of phthalate exposure, in order to illustrate dose–response relationships. The assumption that the dose–response pattern was the same across all target genes within a pathway was assessed by including additional interaction terms between type of gene and quintile of exposure; tests of interactions significant at p < 0.05 suggest a lack of agreement in dose–response patterns.
We evaluated model fit and potential confounding by sampling characteristics and other variables (season, demographic characteristics, maternal size and adiposity, smoking status, maternal health, pregnancy history, and fetal sex). Covariates other than specific-gravity–adjusted phthalate exposure levels and gene (categorical variable with a level for each gene in pathway) were retained in the model if significant at p < 0.05. In our final models, we controlled for qPCR plate, season of delivery, and level of education. In the steroidogenesis models, we also controlled for year of delivery. In the trophoblast differentiation model, we also controlled for mother’s ethnicity, net weight gain, and history of hypertension. Statistical significance was defined by a (two-sided) p-value ≤ 0.05. SAS version 9.1 (SAS Institute Inc., Cary, NC) software was used to conduct all analyses.
Results
The 54 women included in our study were a subset of CCCEH participants similar demographically to those in the parent study except with a larger proportion of Dominican women (83% vs. 74%) and a higher percentage of participants with a high school degree or GED (65% vs. 37%) (Table 2). The distributions of the urinary phthalate metabolites were similar to those reported in a larger sample from the same cohort (Table 3) (Adibi et al. 2008).
Table 2.
CCCEH sample characteristics (n = 54).
| Characteristic | Value |
|---|---|
| Gestational age (weeks)a | 39.5 ± 1.6 |
| Birth weight (g)b | 3,324 ± 466 |
| Maternal age (years) | 26.1 ± 4.5 |
| Gestational agea | |
| < 37 weeks | 3 (6) |
| 37–41 weeks | 45 (88) |
| > 41 weeks | 3 (6) |
| Race/ethnicity | |
| Dominican | 45 (83) |
| African American | 9 (17) |
| Marital status | |
| Never married | 35 (65) |
| Widowed, divorced, separated | 4 (7) |
| Married | 15 (28) |
| Education | |
| Less than high school | 6 (11) |
| High school or GED | 35 (65) |
| More than high school | 13 (24) |
| Parity | |
| 0 live births | 21 (39) |
| 1 live birth | 19 (35) |
| > 1 live births | 14 (26) |
| Cesarean sectionc | 14 (27) |
| Body mass indexd | |
| ≤ 24.9 | 29 (54) |
| > 24.9 and ≤ 29.9 | 12 (22) |
| > 29.9 | 10 (19) |
| Female sex, newborn (%) | 26 (48) |
| Season | |
| Summer | 13 (24) |
| Fall | 8 (15) |
| Winter | 17 (31) |
| Spring | 16 (30) |
| Year of delivery | |
| 2002 | 8 (15) |
| 2003 | 13 (24) |
| 2004 | 14 (26) |
| 2005 | 19 (35) |
Values are mean ± SD or no. (%).
Three missing for gestational age.
Three missing for birth weight.
Two missing for delivery method.
Three missing for body mass index.
Table 3.
Distribution of maternal urinary phthalate metabolites (ng/mL; n= 54).
| Measure | MEHP | MEOHP | ∑DEHP metabolitesa | MnBP | MiBP | MBzP |
|---|---|---|---|---|---|---|
| Geometric mean | 5.5 | 16.5 | 279.8 | 34.6 | 10.4 | 20.4 |
| Quintiles, specific-gravity adjusted | ||||||
| Quintile 1 | ≤ 2.2 | ≤ 10.2 | ≤ 112.2 | ≤ 19.6 | ≤ 6.2 | ≤ 5.7 |
| Quintile 2 | 2.3–4.8 | 10.3–15.0 | 112.3–221.1 | 19.7–37.9 | 6.3–11.1 | 5.8–15.7 |
| Quintile 3 | 4.9–9.6 | 15.1–26.0 | 221.2–371.5 | 38.0–52.8 | 11.2–14.1 | 15.8–32.1 |
| Quintile 4 | 9.7–18.8 | 26.1–47.5 | 371.6–735.2 | 52.9–73.0 | 14.2–24.4 | 32.2–88.9 |
| Quintile 5 | > 18.8 | > 47.5 | > 735.2 | > 73.0 | > 24.4 | > 88.9 |
Sum of MEHP, MEOHP, MEHHP, and MECPP, in nanomoles/liter.
The median yield of total RNA per placenta biopsy was 62 μg (mean ± SE = 72 ± 5 μg). Analytic gels showed two distinct bands at 18S and 28S with minimal signs of degradation. Of the 108 biopsies from 54 placentas, two samples had insufficient RNA/cDNA at the time of qPCR analysis for HCG, and one for 17β-HSD. One RNA/cDNA sample was missing at the time of analysis of 18S (housekeeping gene), which is the reason for the single missing value across all remaining transcripts (Table 4). The ranking of transcripts by median message level (mRNA/3 μg total RNA) was as follows: 18S rRNA (9.80 × 105), HCG (4.30 × 105), P450scc (3.94 × 104), CYP19 (3.30 × 104), 17β-HSD (7.47 × 103), PPARγ (3.56 × 103), AhR (5.03 × 102), and CYP1B1 (1.02 × 102).
Table 4.
Mean values of log-transformed gene transcripts and their corresponding mean Z-scores and Spearman correlations (95% confidence interval) between placental mRNA levels, adjusted for 18S mRNA, grouped by common pathway (n = 54 placentas).
| Gene transcript |
||||
|---|---|---|---|---|
| Measure | CYP19a | 17β-HSDb | P450scca | CYP1B1b |
| Steroidogenesis pathway | ||||
| Mean ± SD, log transformed | −3.4 ± 3.3 | −4.2 ± 1.6 | −2.4 ± 2.0 | −8.4 ± 1.6 |
| Mean Z-score ± SD | 0.01 ± 1.1 | −0.02 ± 1.2 | −0.01 ± 1.2 | −0.02 ± 1.2 |
| Spearman correlation (95% CI) | ||||
| CYP19 | 1.00 | 0.75 (0.65–0.82) | 0.85 (0.79–0.90) | 0.62 (0.49–0.72) |
| 17β-HSD | 1.00 | 0.87 (0.81–0.91) | 0.69 (0.57–0.78) | |
| P450scc | 1.00 | 0.63 (0.50–0.73) | ||
| PPARγa | AhRa | HCGb | ||
| Trophoblast differentiation pathway | ||||
| Mean ± SD, log transformed | −5.3 ± 2.1 | −7.1 ± 2.5 | −0.9 ± 2.7 | |
| Mean Z-score ± SD | −0.04 ± 1.1 | 0.02 ± 1.1 | 0.01 ± 1.1 | |
| Spearman correlation (95% CI) | ||||
| PPARγ | 1.00 | 0.41 (0.24–0.56) | 0.83 (0.75–0.88) | |
| AhR | 1.00 | 0.68 (0.56–0.77) | ||
One missing (n = 107).
Two missing (n = 106).
Between-placenta variability was higher than within-placenta variability for CYP19 (62% vs. 38%) and AhR (59% vs. 42%). For four other transcripts, the between-placenta variability was slightly lower than 50% (PPARγ, 47%; P450scc, 40%; 17β-HSD, 41%; HCG, 46%), and was 30% for CYP1B1. This was consistent with our previous analysis that demonstrated high within-placenta variability, which was mitigated by controlling for location within the chorionic plate (Adibi et al. 2009b). Transcripts were significantly correlated within the pathway groupings (Table 4).
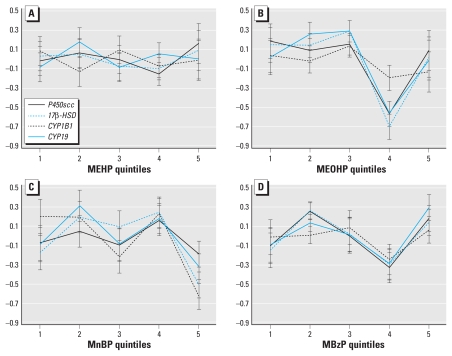
We fitted mixed effects models both with and without the assumption of linearity of phthalate exposure effects to evaluate associations with gene expression levels within each pathway (Tables 5 and 6). In the case of steroidogenesis, we found no significant associations of the phthalate metabolite levels with target genes in the models using log concentration levels (i.e., assuming linearity) (Table 5). We found a significant difference across quintiles of MnBP exposure (p = 0.001) (Table 6), with a trend toward a U-shaped dose–response pattern (Figure 1C). The Pearson correlation between the Z-score and the log-transformed gene expression value (ratio of target gene to 18S RNA) was 1.00. Even though Z-scores are not typically used in the presentation of qPCR data, they should be interpreted as a direct proxy for the mRNA levels measured in the sample.
Table 5.
Associations between pathway-specific placental gene expression and maternal urinary phthalate metabolites adjusted for specific gravity (log-transformed), using a linear model approach.
| β-Coefficient (SE), p-value |
||
|---|---|---|
| Phthalate metabolite | Steroidogenesis: CYP19, 17β-HSD, P450scc, CYP1B1a | Trophoblast differentiation: PPARγ, AhR, HCGb |
| MEHP | 0.01 (0.12), p = 0.90 | −0.15 (0.06), p = 0.02 |
| MEOHP | −0.04 (0.13), p = 0.74 | −0.28 (0.06), p < 0.0001 |
| ∑DEHP metabolitesc | −0.01 (0.13), p = 0.96 | −0.19 (0.08), p = 0.03 |
| MnBP | −0.19 (0.16), p = 0.25 | −0.16 (0.08), p = 0.05 |
| MiBP | −0.11 (0.14), p = 0.46 | −0.21 (0.09), p = 0.02 |
| MBzP | 0.03 (0.09), p = 0.78 | −0.14 (0.06), p = 0.02 |
n = 54, adjusted for gene, qPCR batch (CYP1B1), year of delivery, season of delivery, and level of education.
n = 54, adjusted for gene, qPCR batch (AhR, PPARγ), season of delivery, level of education, net weight gain, mother’s ethnicity, and history of hypertension.
The square root of specific gravity was included as an independent term in the model.
Table 6.
Associations between pathway-specific placental gene expression and maternal urinary phthalate metabolites, using a quintile model approach.
| β-Coefficient (SE), p-value |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Steroidogenesis: CYP19, 17β-HSD, P450scc, CYP1B1a |
Trophoblast differentiation: PPAR γ, AhR, HCGb |
|||||||||
| Phthalate metabolite | Quintile 2 | Quintile 3 | Quintile 4 | Quintile 5 | p-Valuec | Quintile 2 | Quintile 3 | Quintile 4 | Quintile 5 | p-Valuec |
| MEHP | 0.26 (0.35) | −0.28 (0.47) | −0.13 (0.43) | 0.30 (0.42) | 0.29 | 0.02 (0.29) | −0.15 (0.26) | −0.58 (0.30) | −0.43 (0.26) | 0.22 |
| MEOHP | −0.50 (0.38) | −0.34 (0.28) | −0.95 (0.43)* | −0.03 (0.39) | 0.06 | −0.59 (0.31) | −0.18 (0.24) | −0.92 (0.25)** | −0.83 (0.23)** | 0.002 |
| ∑DEHP metabolitesd | −0.36 (0.36) | 0.03 (0.42) | −0.40 (0.41) | −0.01 (0.47) | 0.54 | −0.13 (0.30) | −0.90 (0.33)* | −0.90 (0.28)** | −0.92 (0.27)** | 0.02 |
| MnBP | 0.72 (0.39) | 0.48 (0.46) | 0.83 (0.38)* | −0.61 (0.41) | 0.001 | 0.07 (0.29) | −0.52 (0.26)* | 0.33 (0.28) | 0.57 (0.21)** | 0.004 |
| MiBP | 0.14 (0.53) | 0.35 (0.61) | −0.18 (0.45) | −0.22 (0.44) | 0.71 | 0.43 (0.24) | 0.19 (0.36) | −0.92 (0.20)** | −0.44 (0.23) | 0.0002 |
| MBzP | 0.14 (0.49) | 0.09 (0.42) | −0.14 (0.43) | 0.12 (0.35) | 0.89 | 0.04 (0.30) | −0.52 (0.27) | −0.67 (0.24)** | −0.42 (0.26) | 0.01 |
n = 54, adjusted for gene, qPCR batch (CYP1B1), year of delivery, season of delivery, and level of education.
n = 54, adjusted for gene, qPCR batch (AhR, PPARγ), season of delivery, level of education, net weight gain, mother’s ethnicity, and history of hypertension.
The type 3 test of fixed effects, a four degrees of freedom test of whether there is any difference among the 5 groups. Quintile 1 is the referent group.
The square root of specific gravity was included as an independent term in the model.
p = 0.05,
p = 0.01; quintile 1 is the referent group in the regression models.
Figure 1.
Quintile plots of estimated mean Z-scores (± SE) of gene transcripts in the placental steroidogenesis pathway in relation to maternal urinary concentrations of MEHP (A), MEOHP (B), MnBP (C), and MBzP (D). C depicts a significant association between MnBP quintiles and steroidogenic gene expression (p = 0.001) and significantly different slopes among genes in relation to MnBP (p = 0.03).
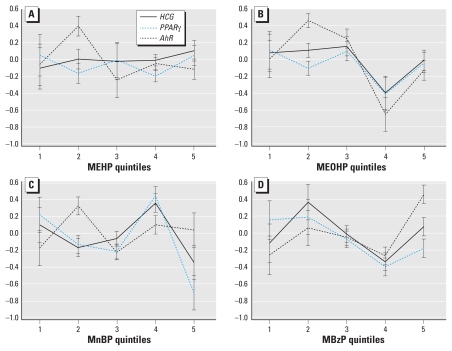
For the trophoblast differentiation pathway, higher levels of urinary metabolite concentrations were associated with significantly lower levels of gene expression for all five phthalate metabolites and ∑DEHP (Table 5). We also observed differences in gene expression across the quintiles of exposure for all metabolites except MEHP (Table 6, Figure 2). Although the trend was generally inverse, there was a suggestion of a U-shaped curve for all metabolites (Figure 2).
Figure 2.
Quintile plots of estimated mean Z-scores (± SE) of gene transcripts in the trophoblast differentiation pathway in relation to maternal urinary concentrations of MEHP (A), MEOHP (B), MnBP (C), and MBzP (D). B–D depict significant associations between MEOHP (p = 0.002), MnBP (p = 0.004), and MBzP (p = 0.01) quintiles and gene expression; C and D depict significantly different slopes among genes in relation to MnBP (p = 0.03) and MBzP (p = 0.01).
For most phthalate metabolites, additional tests of interaction between individual gene and metabolite levels were not significant and thus supported the assumption of a similar pattern of dose response for genes within the same pathway. That is, the shift in Z-scores for each quintile of exposure level did not depend on which of the genes we measured. However, there were isolated exceptions. For the steroidogenesis pathway, we found a significant difference in dose–response patterns of target genes across levels of MnBP exposure (p = 0.03). For the trophoblast pathway, we found a difference in dose–response patterns across target genes for MnBP (p = 0.03) and MBzP (p = 0.01; see Figure 2C,D).
Discussion
In a sample of 54 Dominican and African-American women in New York City, urinary phthalate metabolite concentrations were associated with placental biomarkers of gene expression in two pathways, steroidogenesis and trophoblast differentiation.
The consistent decreases in placental gene expression at the higher quintile concentrations of phthalate metabolites may mean that effects are concentrated at the higher doses or that women in the upper exposure quintiles are more susceptible to placental insults for other reasons that are correlated with phthalate exposures that we were unable to control for. This cohort, which is characterized by low income and high social disadvantage, has significantly higher urinary concentrations of DnBP and DiBP metabolites compared with other pregnant women in the U.S. general population and in another U.S. multicenter pregnancy cohort (Adibi et al. 2009a). Given that high exposures to chemicals may be accompanied by poor nutrition and co-exposures to other chemicals and other physical and psychosocial stress, these women and their fetuses may be in an especially high category of risk (Rauh et al. 2004).
Results differed slightly when we applied modeling strategies that assume linearity in the dose response versus those that do not. Inspection of the associations by level of exposure shows little evidence of linearity with most of the metabolites. Dose–response relationships within these pathways may be nonmonotonic, which would be expected given the nature of transcriptional regulation of nuclear receptors and endocrine signals and specifically with regard to the behavior of phthalates and other endocrine-disrupting compounds (Andrade et al. 2006; Li et al. 2007; Welshons et al. 2003).
There is no consensus on the molecular mechanism of phthalate actions, especially with regard to the steroidogenic effects (Feige et al. 2007; Hallmark et al. 2007; Wilson et al. 2004). Some data show effects at the level of mRNA transcription that are consistent with protein levels (Lovekamp and Davis 2001; McKinnell et al. 2005; Wilson et al. 2004), whereas others show effects at the level of protein expression but not transcription (Boberg et al. 2008; Lambrot et al. 2009). Attempts to recapitulate the steroidogenic effects of MEHP and MnBP in rodents using human testis explants have produced conflicting results (Hallmark et al. 2007; Lambrot et al. 2009), which could be due to differences in experimental systems and/or species. The association that we measured between phthalates and placental steroidogenesis was present only in the quintile models with MnBP. Interestingly, there is some evidence in humans that fetal testicular steroidogenesis and placental steroidogenesis may be linked (Akre et al. 2008).
The trophoblast differentiation pathway (PPARγ, HCG, AhR) was of interest to us as a well-studied PPARγ-mediated pathway in the human placenta (Tarrade et al. 2001). In fact, HCG is a marker of syncytium formation and is used as an indicator of placental function in clinical tests (Lepage et al. 2003; Yang et al. 2003). Beyond its role in xenobiotic metabolism, AhR is also believed to be a regulator of estrogen metabolism, vascularization, and hypoxic responses in the placenta (Detmar et al. 2008; Huuskonen et al. 2008). Trophoblast differentiation is most important early in pregnancy when the placenta is initially being constructed and assuming its spatial and physiologic orientation with respect to the fetus. Throughout pregnancy, a population of progenitor trophoblasts persists and undergoes renewal and differentiation (Tarrade et al. 2001). Toward late pregnancy, there is a shift toward higher proportions of syncytiotrophoblasts where PPARγ is localized (Borel et al. 2008; Rodie et al. 2005). Our ability to detect an association between phthalate metabolite urinary concentrations and trophoblast differentiation could mean that there was a disruption of this late-stage process. There may also have been effects on trophoblast differentiation and placental development early in pregnancy that were mirrored in these biomarkers measured at term.
It is difficult to draw conclusions with a limited set of gene targets per pathway. These targets and their relationship to phthalate metabolites should be pursued further by including additional gene targets and posttranscriptional markers. Nonetheless, correlations of maternal and fetal exposures with transcription may provide valuable information even if it is not possible to extrapolate to posttranscriptional phenotype. In most cases, our assumption that genes within a pathway were expressed in a parallel dose response was supported by the data.
In summary, we applied biomarkers of mRNA transcription in human placental tissue to test hypotheses on the association of prenatal phthalate exposure with placental development and function. Metabolites of DEHP, DnBP, DiBP, and BBzP were significantly associated with the joint expression of three gene targets known to be involved in trophoblast differentiation (PPARγ, AhR, HCG). These associations were robust to linear and nonlinear modeling strategies and to adjustment for urinary dilution. We did not detect robust associations of phthalate metabolites with the joint expression of four gene targets in the steroidogenic pathway (CYP19, 17β-HSD, P450scc, CYP1B1). This may suggest a null association of phthalates with placental steroidogenesis, or it may suggest lack of sensitivity in our methodology. It may also indicate that the model of phthalate-induced perturbation of steroidogenesis well described in rodent models cannot be directly translated to humans. In either case, we offer a novel approach to study the effects of endocrine-disrupting compounds on placental function.
Footnotes
This work was supported by National Institute of Environmental Health Sciences grants P50 ES09600, R01 ES013543, R01 ES08977, and R01 ES11158 and by U.S. Environmental Protection Agency grants R827027 and R82860901.
B.J.D. works for Millennium Pharmaceuticals, Cambridge, MA, which does not produce particular items in or related to this study. J.J.A. served as a 2008 Science Communication Fellow for and received an honorarium from Environmental Health Sciences, a foundation-funded journalism organization whose mission is to advance the public’s understanding of environmental health issues; the organization does not accept funds from interest groups. The remaining authors declare they have no competing financial interests.
We gratefully acknowledge the technical assistance of M. Silva, J. Reidy, E. Samandar, and J. Preau in measuring the urinary concentrations of phthalate metabolites and of A. Reyes, D. Holmes, and L. Qu for their help in data collection.
The findings expressed in this article are the opinions of the authors and do not necessarily reflect the official opinion of the Centers for Disease Control and Prevention.
References
- Adibi JJ, Hauser R, Williams PL, Whyatt RM, Calafat AM, Nelson H, et al. Maternal urinary metabolites of di-(2-ethylhexyl) phthalate in relation to the timing of labor in a US multicenter pregnancy cohort study. Am J Epidemiol. 2009a;169(8):1015–1024. doi: 10.1093/aje/kwp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adibi JJ, Hauser R, Williams PL, Whyatt RM, Thaker HM, Nelson H, et al. Placental biomarkers of phthalate effects on mRNA transcription: application in epidemiologic research. Environ Health. 2009b;8:20. doi: 10.1186/1476-069X-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adibi JJ, Whyatt RM, Williams PL, Calafat AM, Camann D, Herrick R, et al. Characterization of phthalate exposure among pregnant women assessed by repeat air and urine samples. Environ Health Perspect. 2008;116:467–473. doi: 10.1289/ehp.10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akre O, Boyd HA, Ahlgren M, Wilbrand K, Westergaard T, Hjalgrim H, et al. Maternal and gestational risk factors for hypospadias. Environ Health Perspect. 2008;116:1071–1076. doi: 10.1289/ehp.10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade AJ, Grande SW, Talsness CE, Grote K, Chahoud I. A dose-response study following in utero and lactational exposure to di-(2-ethylhexyl)-phthalate (DEHP): non-monotonic dose-response and low dose effects on rat brain aromatase activity. Toxicology. 2006;227(3):185–192. doi: 10.1016/j.tox.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Barlow NJ, Phillips SL, Wallace DG, Sar M, Gaido KW, Foster PM. Quantitative changes in gene expression in fetal rat testes following exposure to di(n-butyl) phthalate. Toxicol Sci. 2003;73(2):431–441. doi: 10.1093/toxsci/kfg087. [DOI] [PubMed] [Google Scholar]
- Bhat HK, Epelboym I. Quantitative analysis of total mitochondrial DNA: competitive polymerase chain reaction versus real-time polymerase chain reaction. J Biochem Mol Toxicol. 2004;18(4):180–186. doi: 10.1002/jbt.20024. [DOI] [PubMed] [Google Scholar]
- Boberg J, Metzdorff S, Wortziger R, Axelstad M, Brokken L, Vinggaard AM, et al. Impact of diisobutyl phthalate and other PPAR agonists on steroidogenesis and plasma insulin and leptin levels in fetal rats. Toxicology. 2008;250(2–3):75–81. doi: 10.1016/j.tox.2008.05.020. [DOI] [PubMed] [Google Scholar]
- Borel V, Gallot D, Marceau G, Sapin V, Blanchon L. Placental implications of peroxisome proliferator-activated receptors in gestation and parturition. PPAR Res. 2008;2008:758562. doi: 10.1155/2008/758562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BJ, Maronpot RR, Heindel JJ. Di-(2-ethylhexyl) phthalate suppresses estradiol and ovulation in cycling rats. Toxicol Appl Pharmacol. 1994a;128(2):216–223. doi: 10.1006/taap.1994.1200. [DOI] [PubMed] [Google Scholar]
- Davis BJ, Weaver R, Gaines LJ, Heindel JJ. Mono-(2-ethylhexyl) phthalate suppresses estradiol production independent of FSH-cAMP stimulation in rat granulosa cells. Toxicol Appl Pharmacol. 1994b;128(2):224–228. doi: 10.1006/taap.1994.1201. [DOI] [PubMed] [Google Scholar]
- Detmar J, Rennie MY, Whiteley KJ, Qu D, Taniuchi Y, Shang X, et al. Fetal growth restriction triggered by polycyclic aromatic hydrocarbons is associated with altered placental vasculature and AhR-dependent changes in cell death. Am J Physiol Endocrinol Metab. 2008;295(2):E519–E530. doi: 10.1152/ajpendo.90436.2008. [DOI] [PubMed] [Google Scholar]
- Ema M, Miyawaki E, Kawashima K. Effects of dibutyl phthalate on reproductive function in pregnant and pseudopregnant rats. Reprod Toxicol. 2000;14(1):13–19. doi: 10.1016/s0890-6238(99)00066-0. [DOI] [PubMed] [Google Scholar]
- Feige JN, Gelman L, Rossi D, Zoete V, Metivier R, Tudor C, et al. The endocrine disruptor monoethyl-hexyl-phthalate is a selective peroxisome proliferator-activated receptor gamma modulator that promotes adipogenesis. J Biol Chem. 2007;282(26):19152–19166. doi: 10.1074/jbc.M702724200. [DOI] [PubMed] [Google Scholar]
- Fitzmaurice G, Laird N, Ware J. Applied Longitudinal Analysis. San Francisco: John Wiley & Sons; 2004. [Google Scholar]
- Fournier T, Tsatsaris V, Handschuh K, Evain-Brion D. PPARs and the placenta. Placenta. 2007;28(2–3):65–76. doi: 10.1016/j.placenta.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Froment P, Gizard F, Defever D, Staels B, Dupont J, Monget P. Peroxisome proliferator-activated receptors in reproductive tissues: from gametogenesis to parturition. J Endocrinol. 2006;189(2):199–209. doi: 10.1677/joe.1.06667. [DOI] [PubMed] [Google Scholar]
- Hallmark N, Walker M, McKinnell C, Mahood IK, Scott H, Bayne R, et al. Effects of monobutyl and di(n-butyl) phthalate in vitro on steroidogenesis and Leydig cell aggregation in fetal testis explants from the rat: comparison with effects in vivo in the fetal rat and neonatal marmoset and in vitro in the human. Environ Health Perspect. 2007;115:390–396. doi: 10.1289/ehp.9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser R, Calafat AM. Phthalates and human health. Occup Environ Med. 2005;62(11):806–818. doi: 10.1136/oem.2004.017590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser R, Meeker JD, Park S, Silva MJ, Calafat AM. Temporal variability of urinary phthalate metabolite levels in men of reproductive age. Environ Health Perspect. 2004;112:1734–1740. doi: 10.1289/ehp.7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huuskonen P, Storvik M, Reinisalo M, Honkakoski P, Rysa J, Hakkola J, et al. Microarray analysis of the global alterations in the gene expression in the placentas from cigarette-smoking mothers. Clin Pharmacol Ther. 2008;83(4):542–550. doi: 10.1038/sj.clpt.6100376. [DOI] [PubMed] [Google Scholar]
- Kato K, Silva MJ, Needham LL, Calafat AM. Determination of 16 phthalate metabolites in urine using automated sample preparation and on-line preconcentration/high-performance liquid chromatography/tandem mass spectrometry. Anal Chem. 2005;77(9):2985–2991. doi: 10.1021/ac0481248. [DOI] [PubMed] [Google Scholar]
- Koch HM, Bolt HM, Preuss R, Angerer J. New metabolites of di(2-ethylhexyl)phthalate (DEHP) in human urine and serum after single oral doses of deuterium-labelled DEHP. Arch Toxicol. 2005;79(7):367–376. doi: 10.1007/s00204-004-0642-4. [DOI] [PubMed] [Google Scholar]
- Lambrot R, Muczynski V, Lecureuil C, Angenard G, Coffigny H, Pairault C, et al. Phthalates impair germ cell development in the human fetal testis in vitro without change in testosterone production. Environ Health Perspect. 2009;117:32–37. doi: 10.1289/ehp.11146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latini G, De Felice C, Presta G, Del Vecchio A, Paris I, Ruggieri F, et al. In utero exposure to di-(2-ethylhexyl)phthalate and duration of human pregnancy. Environ Health Perspect. 2003;111:1783–1785. doi: 10.1289/ehp.6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage N, Chitayat D, Kingdom J, Huang T. Association between second-trimester isolated high maternal serum human chorionic gonadotropin levels and obstetric complications in singleton and twin pregnancies. Am J Obstet Gynecol. 2003;188(5):1354–1359. doi: 10.1067/mob.2003.278. [DOI] [PubMed] [Google Scholar]
- Li L, Andersen ME, Heber S, Zhang Q. Non-monotonic dose-response relationship in steroid hormone receptor-mediated gene expression. J Mol Endocrinol. 2007;38(5):569–585. doi: 10.1677/JME-07-0003. [DOI] [PubMed] [Google Scholar]
- Lin P, Hu SW, Chang TH. Correlation between gene expression of aryl hydrocarbon receptor (AhR), hydrocarbon receptor nuclear translocator (Arnt), cytochromes P4501A1 (CYP1A1) and 1B1 (CYP1B1), and inducibility of CYP1A1 and CYP1B1 in human lymphocytes. Toxicol Sci. 2003;71(1):20–26. doi: 10.1093/toxsci/71.1.20. [DOI] [PubMed] [Google Scholar]
- Lovekamp TN, Davis BJ. Mono-(2-ethylhexyl) phthalate suppresses aromatase transcript levels and estradiol production in cultured rat granulosa cells. Toxicol Appl Pharmacol. 2001;172(3):217–224. doi: 10.1006/taap.2001.9156. [DOI] [PubMed] [Google Scholar]
- Lovekamp-Swan T, Jetten AM, Davis BJ. Dual activation of PPARalpha and PPARgamma by mono-(2-ethylhexyl) phthalate in rat ovarian granulosa cells. Mol Cell Endocrinol. 2003;201(1–2):133–141. doi: 10.1016/s0303-7207(02)00423-9. [DOI] [PubMed] [Google Scholar]
- Maloney EK, Waxman DJ. trans-Activation of PPARalpha and PPARgamma by structurally diverse environmental chemicals. Toxicol Appl Pharmacol. 1999;161(2):209–218. doi: 10.1006/taap.1999.8809. [DOI] [PubMed] [Google Scholar]
- McKinnell C, Sharpe RM, Mahood K, Hallmark N, Scott H, Ivell R, et al. Expression of insulin-like factor 3 protein in the rat testis during fetal and postnatal development and in relation to cryptorchidism induced by in utero exposure to di(n-butyl) phthalate. Endocrinology. 2005;146(10):4536–4544. doi: 10.1210/en.2005-0676. [DOI] [PubMed] [Google Scholar]
- Miyoshi Y, Ando A, Shiba E, Taguchi T, Tamaki Y, Noguchi S. Involvement of up-regulation of 17beta-hydroxysteroid dehydrogenase type 1 in maintenance of intratumoral high estradiol levels in postmenopausal breast cancers. Int J Cancer. 2001;94(5):685–689. doi: 10.1002/ijc.1525. [DOI] [PubMed] [Google Scholar]
- Mylchreest E, Wallace DG, Cattley RC, Foster PM. Dose-dependent alterations in androgen-regulated male reproductive development in rats exposed to di(n-butyl) phthalate during late gestation. Toxicol Sci. 2000;55(1):143–151. doi: 10.1093/toxsci/55.1.143. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Yoshitsugu H, Naito S, Hiraoka I. Evaluation of gene induction of drug-metabolizing enzymes and transporters in primary culture of human hepatocytes using high-sensitivity real-time reverse transcription PCR. Yakugaku Zasshi. 2002;122(5):339–361. doi: 10.1248/yakushi.122.339. [DOI] [PubMed] [Google Scholar]
- Perera F, Viswanathan S, Whyatt R, Tang D, Miller RL, Rauh V. Children’s environmental health research—highlights from the Columbia Center for Children’s Environmental Health. Ann NY Acad Sci. 2006;1076:15–28. doi: 10.1196/annals.1371.018. [DOI] [PubMed] [Google Scholar]
- Pidoux G, Gerbaud P, Laurendeau I, Guibourdenche J, Bertin G, Vidaud M, et al. Large variability of trophoblast gene expression within and between human normal term placentae. Placenta. 2002;25(5):469–473. doi: 10.1016/j.placenta.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Rauh VA, Whyatt RM, Garfinkel R, Andrews H, Hoepner L, Reyes A, et al. Developmental effects of exposure to environmental tobacco smoke and material hardship among inner-city children. Neurotoxicol Teratol. 2004;26(3):373–385. doi: 10.1016/j.ntt.2004.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodie VA, Young A, Jordan F, Sattar N, Greer IA, Freeman DJ. Human placental peroxisome proliferator-activated receptor delta and gamma expression in healthy pregnancy and in preeclampsia and intrauterine growth restriction. J Soc Gynecol Investig. 2005;12(5):320–329. doi: 10.1016/j.jsgi.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Saillenfait AM, Sabate JP, Gallissot F. Diisobutyl phthalate impairs the androgen-dependent reproductive development of the male rat. Reprod Toxicol. 2008;26(2):107–115. doi: 10.1016/j.reprotox.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Schaiff WT, Barak Y, Sadovsky Y. The pleiotropic function of PPARgamma in the placenta. Mol Cell Endocrinol. 2006;249(1–2):10–15. doi: 10.1016/j.mce.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Sharpe RM. Hormones and testis development and the possible adverse effects of environmental chemicals. Toxicol Lett. 2001;120(1–3):221. doi: 10.1016/s0378-4274(01)00298-3. [DOI] [PubMed] [Google Scholar]
- Swan SH. Environmental phthalate exposure in relation to reproductive outcomes and other health endpoints in humans. Environ Res. 2008;108(2):177–184. doi: 10.1016/j.envres.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrade A, Schoonjans K, Guibourdenche J, Bidart JM, Vidaud M, Auwerx J, et al. PPAR gamma/RXR alpha heterodimers are involved in human CG beta synthesis and human trophoblast differentiation. Endocrinology. 2001;142(10):4504–4514. doi: 10.1210/endo.142.10.8448. [DOI] [PubMed] [Google Scholar]
- Verbeke G, Molenberghs G. Linear Mixed Models for Longitudinal Data. New York: Springer; 2000. [Google Scholar]
- Vissac-Sabatier C, Coxam V, Déchelotte P, Picherit C, Horcajada MN, Davicco MJ, et al. Phytoestrogen-rich diets modulate expression of Brca1 and Brca2 tumor suppressor genes in mammary glands of female Wistar rats. Cancer Res. 2003;63(20):6607–6612. [PubMed] [Google Scholar]
- Welshons WV, Thayer KA, Judy BM, Taylor JA, Curran EM, vom Saal FS. Large effects from small exposures. I. Mechanisms for endocrine-disrupting chemicals with estrogenic activity. Environ Health Perspect. 2003;111:994–1006. doi: 10.1289/ehp.5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyatt RM, Adibi JJ, Calafat AM, Camann DE, Rauh V, Bhat HK, et al. Prenatal di(2-ethylhexyl) phthalate exposure and length of gestation among an inner-city cohort. Pediatrics. 2009;124(6):e1213–e1220. doi: 10.1542/peds.2009-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyatt RM, Barr DB, Camann DE, Kinney PL, Barr JR, Andrews HF, et al. Contemporary-use pesticides in personal air samples during pregnancy and blood samples at delivery among urban minority mothers and newborns. Environ Health Perspect. 2003;111:749–756. doi: 10.1289/ehp.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson VS, Lambright C, Furr J, Ostby J, Wood C, Held G, et al. Phthalate ester-induced gubernacular lesions are associated with reduced insl3 gene expression in the fetal rat testis. Toxicol Lett. 2004;146(3):207–215. doi: 10.1016/j.toxlet.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Wolff MS, Engel SM, Berkowitz GS, Ye X, Silva MJ, Zhu C, et al. Prenatal phenol and phthalate exposures and birth outcomes. Environ Health Perspect. 2008;116:1092–1097. doi: 10.1289/ehp.11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Agrawal S, Cook TJ, Knipp GT. Maternal di-(2-ethylhexyl)-phthalate exposure influences essential fatty acid homeostasis in rat placenta. Placenta. 2008;29(11):962–969. doi: 10.1016/j.placenta.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Lei ZM, Rao C. The central role of human chorionic gonadotropin in the formation of human placental syncytium. Endocrinology. 2003;144(3):1108–1120. doi: 10.1210/en.2002-220922. [DOI] [PubMed] [Google Scholar]
- Yu L, Romero DG, Gomez-Sanchez CE, Gomez-Sanchez EP. Steroidogenic enzyme gene expression in the human brain. Mol Cell Endocrinol. 2002;190(1–2):9–17. doi: 10.1016/s0303-7207(02)00041-2. [DOI] [PubMed] [Google Scholar]